Atad1 Is a Potential Candidate Gene for Prepulse Inhibition
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Acoustic Startle and Prepulse Inhibition
2.3. Single-Trait Mapping for Behavioral QTLs
2.4. Criteria for Identification of Candidate Genes
2.5. Gene Expression Datasets
2.6. Functional Analysis
2.7. Correlation and PheWAS Analyses
2.8. Protein–Protein Interaction Network Analysis
2.9. Validation of Candidate Genes Using Human RNA Sequencing Data
3. Results
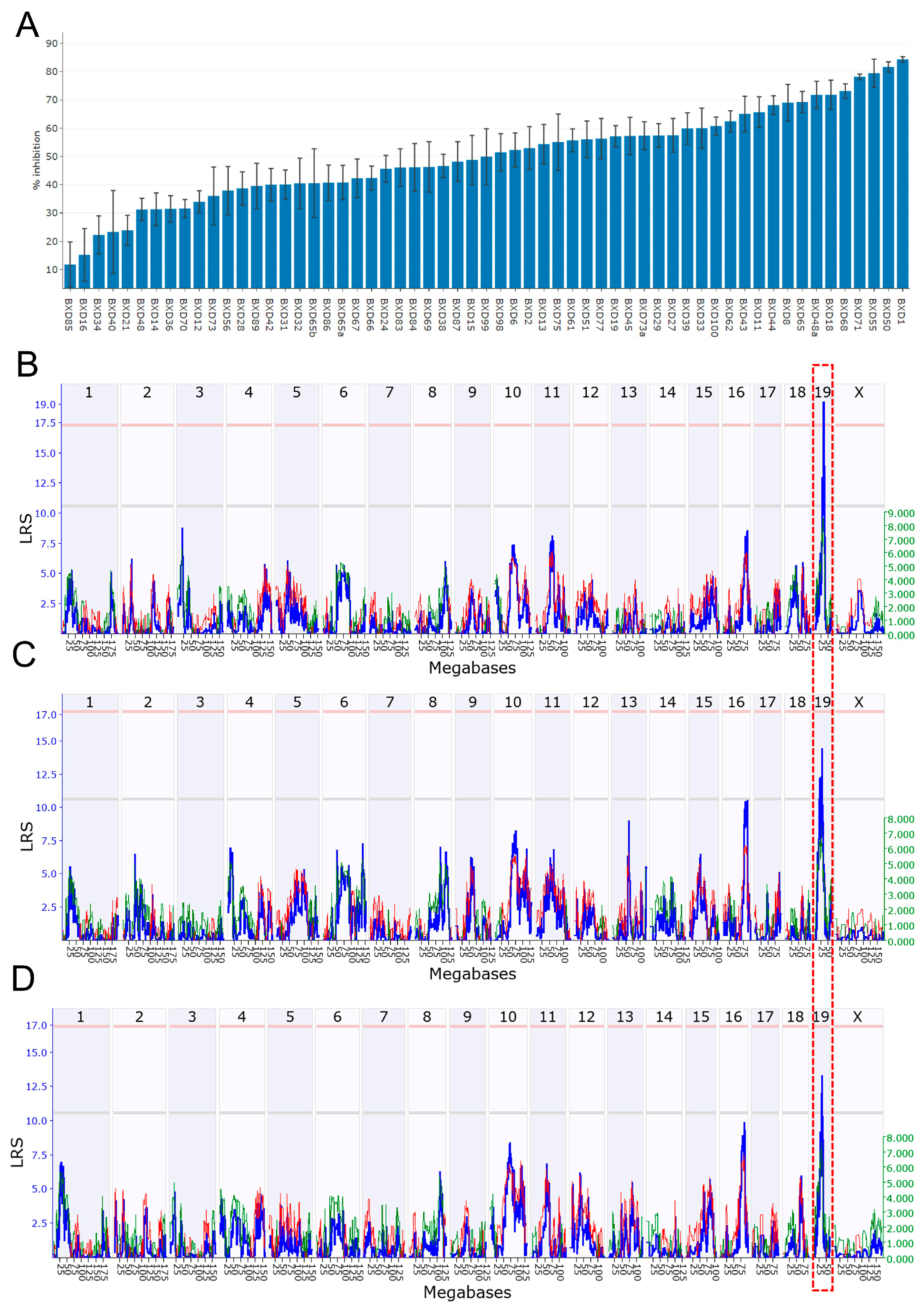
3.1. Prepulse Inhibition
3.2. Identification of Candidate Genes
3.3. Genetic, Partial, and Literature Correlations for Identified Candidate and Gene Sets
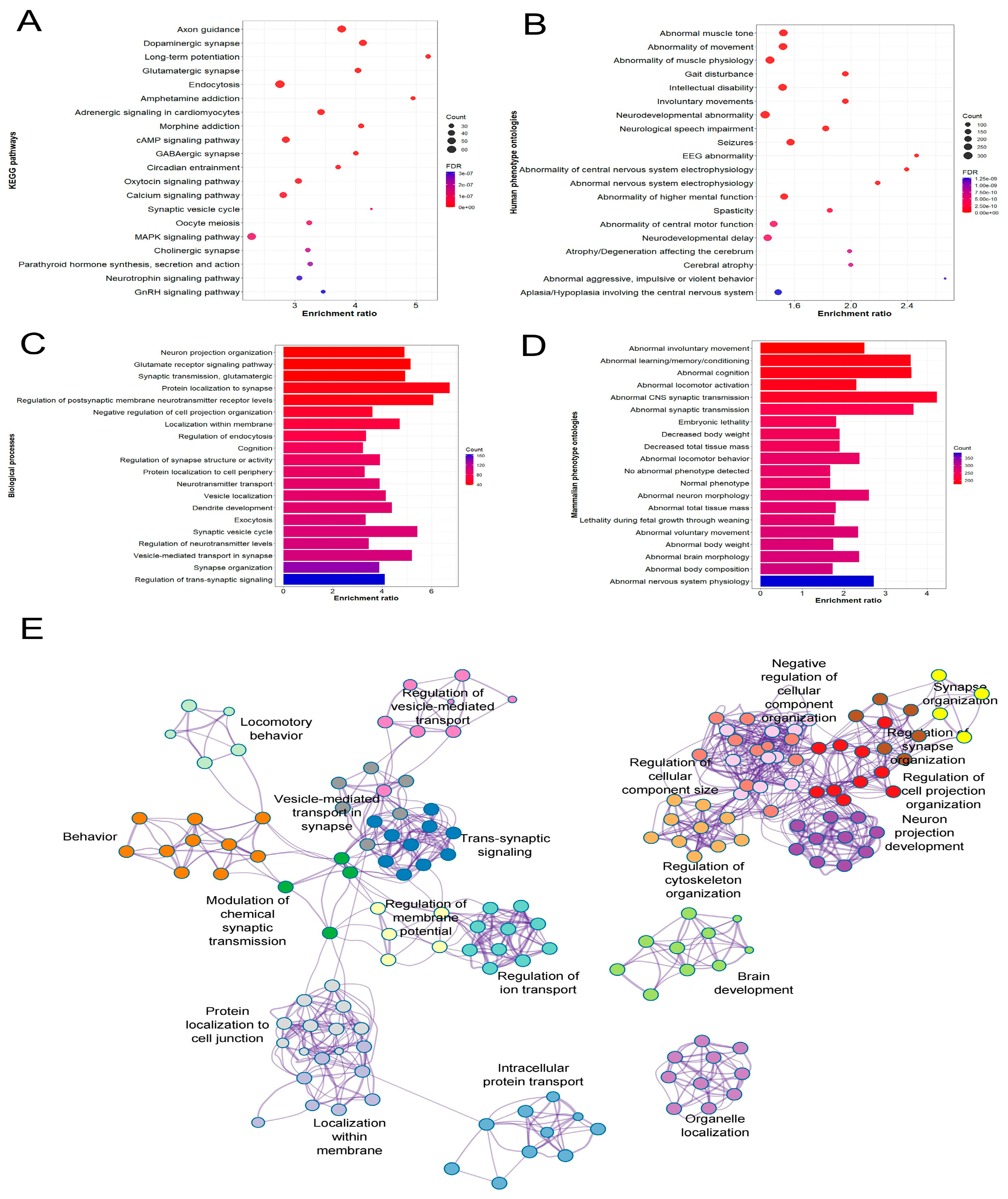
3.4. Gene Enrichment Analysis
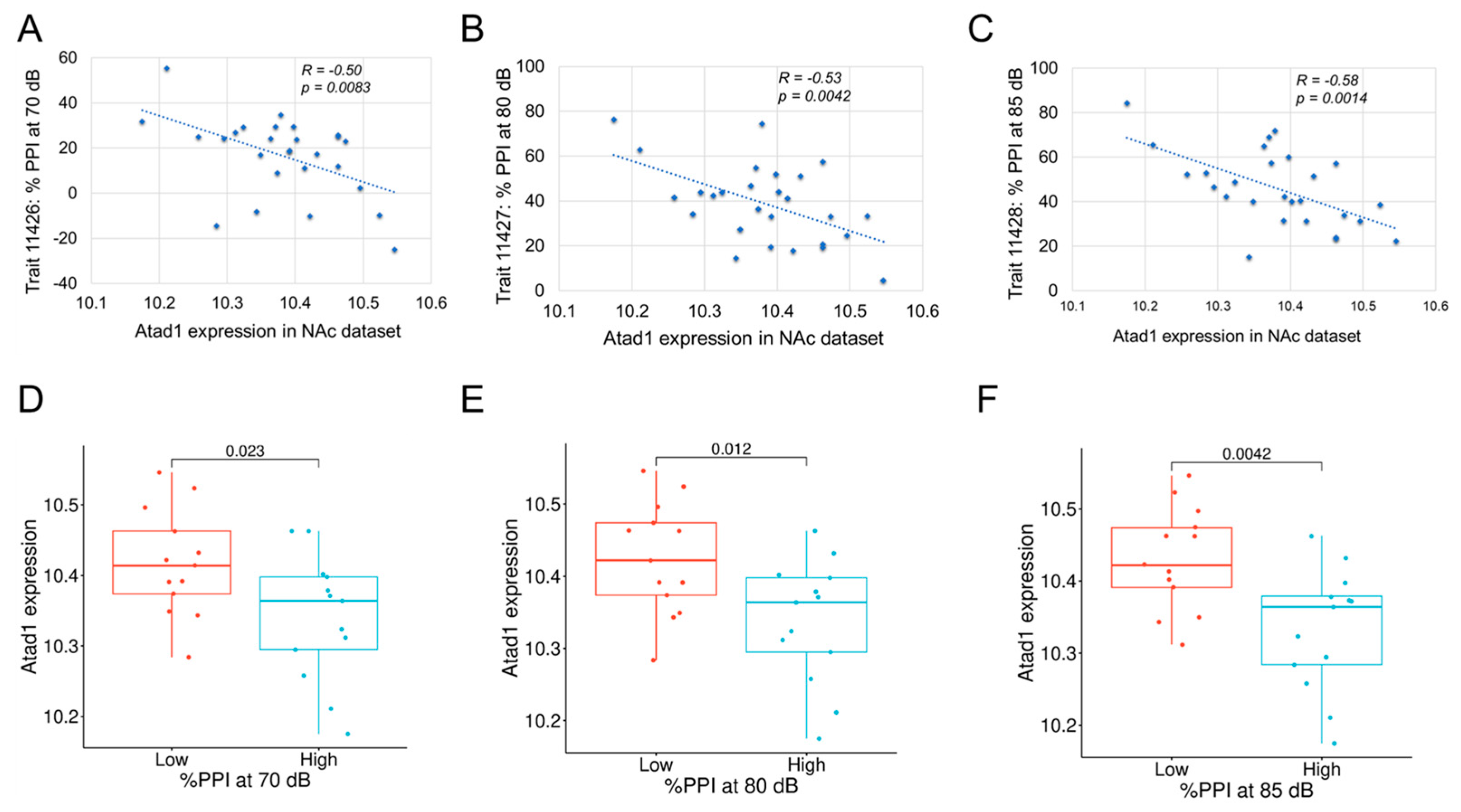
3.5. Phenotype Correlation
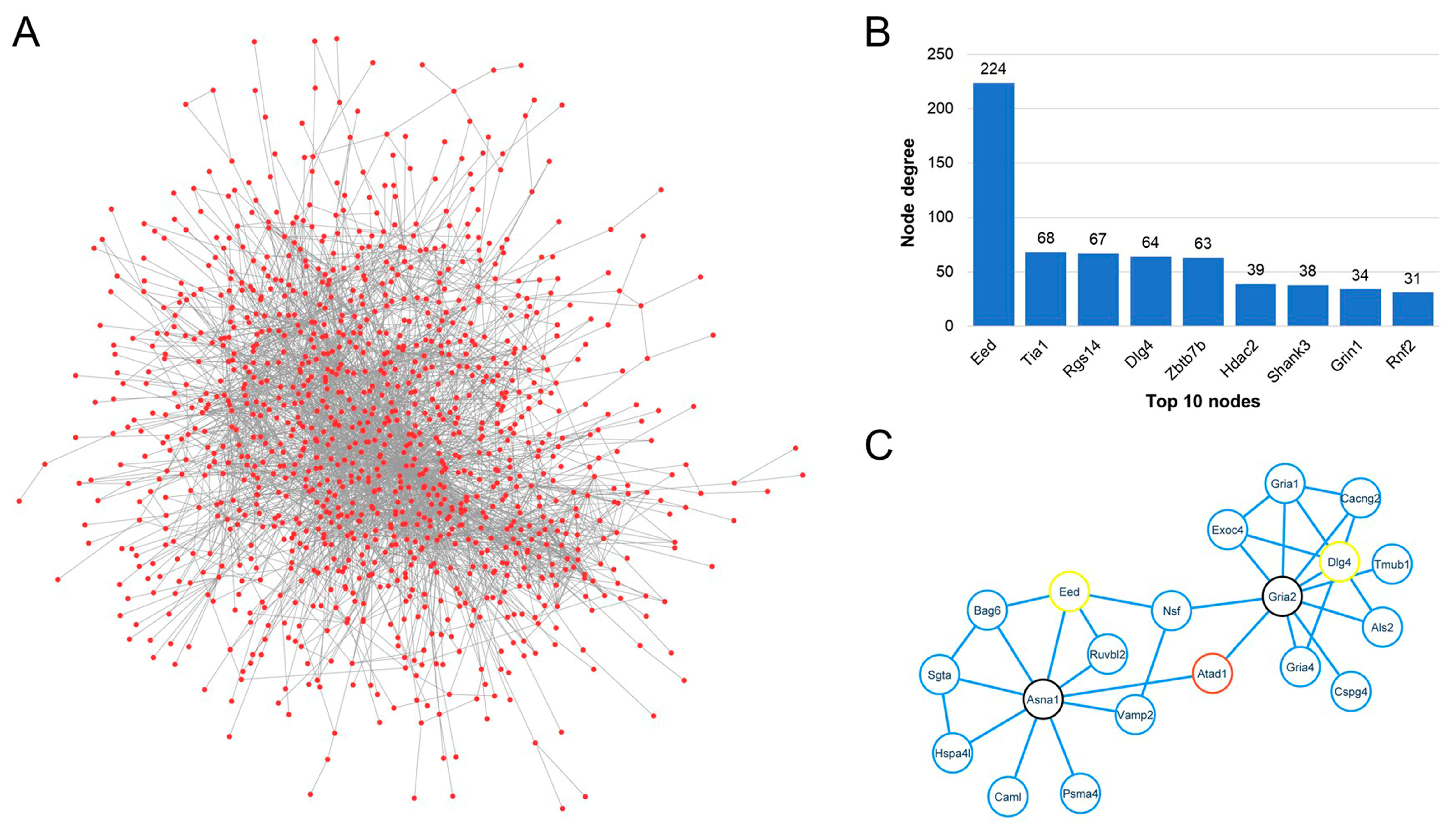
3.6. Protein–Protein Interaction Network
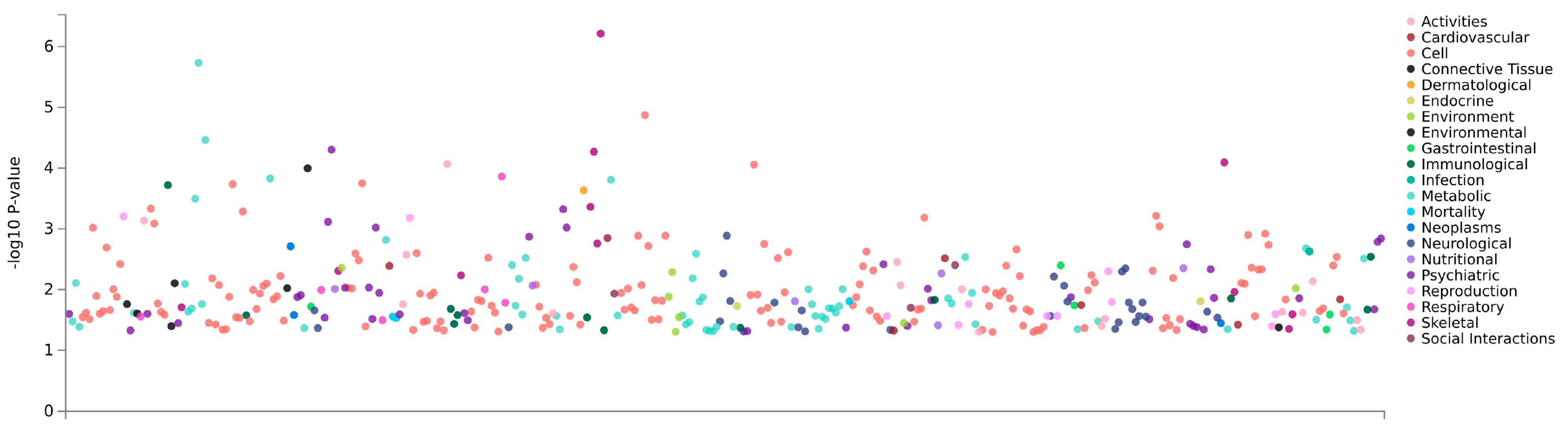
3.7. PheWAS Analysis
3.8. Candidate Variants in Atad1
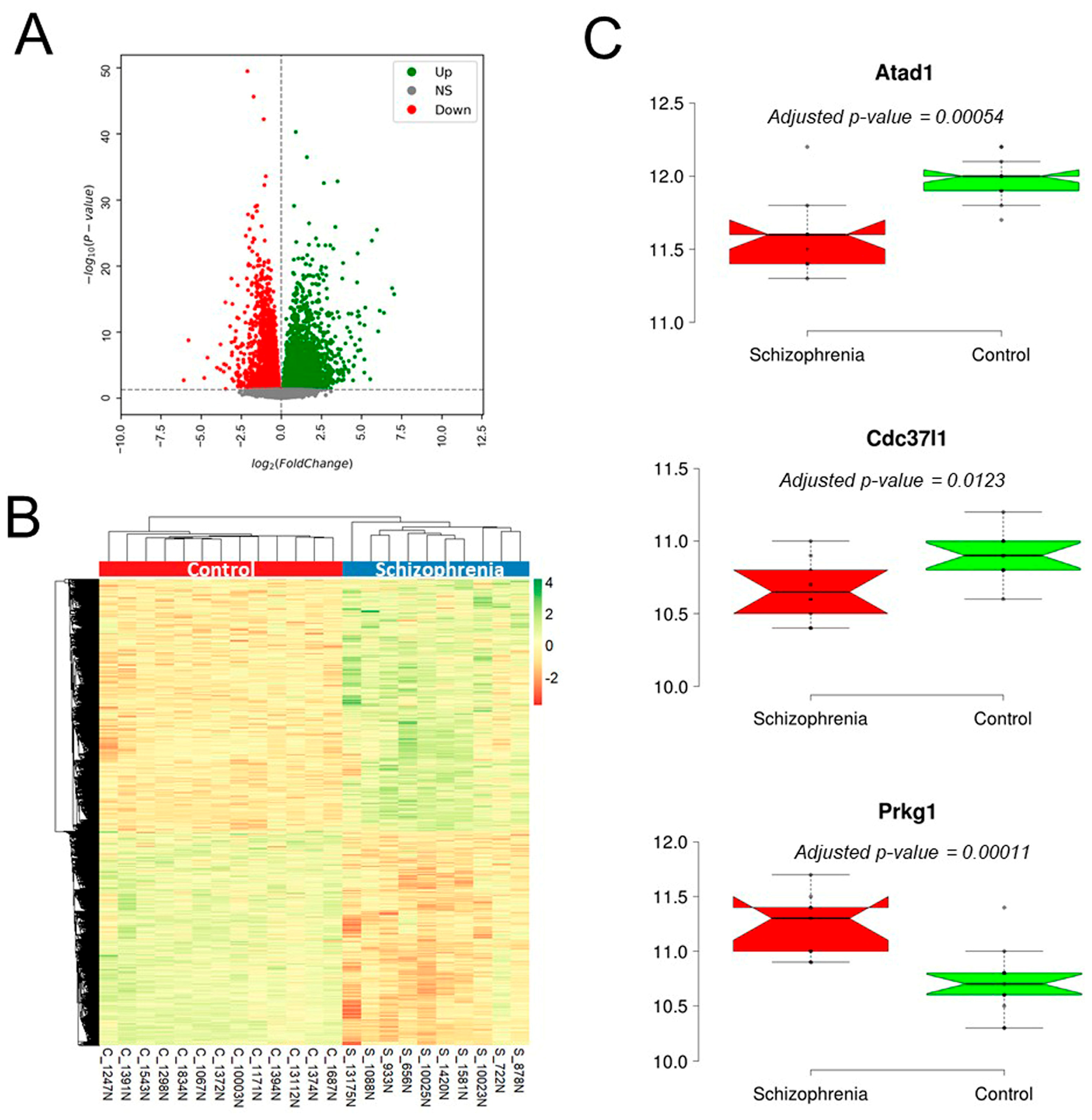
3.9. Validation of the Candidate Gene in Schizophrenia Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PPI | Prepulse Inhibition |
| QTL | Quantitative Trait Locus |
| BXD RI | B6 × D2 Recombinant Inbred Strain |
| B6 | C57/BL6 |
| D2 | DBA2/J |
| PheWAS | Phenome-Wide Association Study |
| GN | GeneNetwork |
| LRS | Likelihood Ratio Statistic |
| Atad1 | ATPase family, AAA domain containing 1, also known as thorase |
| PFC | Prefrontal Cortex |
| HIPP | Hippocampus |
| STR | Striatum |
| NAc | Nucleus Accumbens |
| MDB | Midbrain |
| VTA | Ventral Tegmental Area |
| AMYG | Amygdala |
| HYPO | Hypothalamus |
| GRIA2 | Glutamate Ionotropic Receptor AMPA type 2 subunit |
| ASNA1 | Arsenical Pump-Driving ATPase 1 |
| GWAS | Genome-Wide Association Study |
| Disc1 | Disrupted in Schizophrenia 1 |
| Nrg1 | Neuregulin 1 |
| Chr | Chromosome |
| SNP | Single-Nucleotide Polymorphism |
| GEMMA | Genome-Wide Efficient Mixed-Model Analysis |
| LOD | Logarithm Odds |
| MGI | Mouse Genome Informatics |
| RGD | Rat Genome Database |
| IMPC | International Mouse Phenotyping Consortium |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| mRNA | Messenger RNA |
| RMA | Robust Multi-Array Average |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| GEO | Gene Expression Omnibus |
| Prkg1 | Protein Kinase cGMP-dependent type 1 |
| Rnls | Renalase, FAD Dependent Amine Oxidase |
| Asah2 | N-acylsphingosine amidohydrolase 2 |
| Papss2 | 3′-phosphoadenosine 5′ phosphosulfate synthase 2 |
| Ifit1 | Interferon-induced protein with tetratricopeptide repeats 1 |
| Cdc37l1 | Cell Division cycle 37 homolog-like 1 |
| Ak3 | Adenylate Kinase 3 |
| Ranbp6 | RAN binding protein 6 |
| Ifit1bl2 | Interferon-induced protein with tetratricopeptide repeats 1B like 2 |
| MPO | Mammalian Phenotype Ontology |
| GO-BP | Gene Ontology Biological Processes |
| HPO | Human Phenotype Ontology |
| FDR | False Discovery Rate |
| ND | Node Degree |
| EED | Embryonic Ectoderm Development |
| TIA1 | Cytotoxic Granule-Associated RNA binding protein 1 |
| RGS14 | Regulator of G protein Signaling 14 |
| DLG4 | Discs Large Homolog 4 |
| ZBTB7B | Zinc finger and BTB domain containing 7B |
| GRIA2 | Glutamate Ionotropic Receptor AMPA type subunit 2 |
| bp | Base pair |
| UTR | Untranslated Region |
| PCA | Principal Component Analysis |
| Fabp7 | Fatty acid binding protein 7 |
| Ptpn5 | Tyrosine-Protein-Phosphatase Non-receptor Type 5 |
| APBB1lP | Amyloid Beta Precursor Protein Binding Family B Member 1 Interacting Protein |
| AMPAR | AMPA receptor |
| LTD | Long-Term Depression |
| cKO | Conditional Knockout |
| GRIP1 | AMPAR/Glutamate Interacting-Protein 1 |
References
- Regier, D.; Narrow, W.E.; Rae, D.S.; Manderscheid, R.W.; Locke, B.Z.; Goodwin, F.K. The de facto mental and addictive disorders service system. Epidemiologic catchment area prospective 1-year prevalence rates of disorders and services. Arch. Gen. Psychiatry 1993, 50, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Vawter, M.P.; Mamdani, F.; Macciardi, F. An integrative functional genomics approach for discovering biomarkers in schizophrenia. Brief. Funct. Genom. 2011, 10, 387–399. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. What Is Schizophrenia. 2018. Available online: https://www.psychiatry.org/patients-families/schizophrenia/what-is-schizophrenia (accessed on 13 April 2018).
- Crowley, J.J.; Sakamoto, K. Psychiatric Genomics: Outlook for 2015 and challenges for 2020. Curr. Opin. Behav. Sci. 2015, 2, 102–107. [Google Scholar] [CrossRef]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Trubetskoy, V.; Pardiñas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.-Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef]
- Cardno, A.G.; Marshall, E.J.; Coid, B.; Macdonald, A.M.; Ribsheter, T.R.; Davies, N.J.; Venturi, P.; Jones, L.A.; Lewis, S.W.; Sham, P.C.; et al. Heritability estimates for psychotic disorders: The Maudsley Twin Psychosis Series. Arch. Gen. Psychiatry 1999, 56, 162–168. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Kendler, K.S.; Neale, M.C. Schizophrenia as a complex trait. Arch. Gen. Psychiatry 2003, 60, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Hilker, R.; Helenius, D.; Fagerlund, B.; Skytthe, A.; Christensen, K.; Werge, T.M.; Nordentoft, M.; Glenthøj, B. Heritability of schizophrenia and schizophrenia spectrum based on the nationwide Danish twin register. Biol. Psychiatry 2017, 83, 492–498. [Google Scholar] [CrossRef]
- Roussos, P.; Giakoumaki, S.G.; Zouraraki, C.; Fullard, J.F.; Karagiorga, V.-E.; Tsapakis, E.-M.; Petraki, Z.; Siever, L.J.; Lencz, T.; Malhotra, A.; et al. The Relationship of Common risk variants and polygenic risk for schizophrenia to sensorimotor gating. Biol. Psychiatry 2015, 79, 988–996. [Google Scholar] [CrossRef]
- Anokhin, A.P.; Health, A.C.; Myers, E.; Ralano, A.; Wood, S. Genetic influences on prepulse inhibition of startle reflex in humans. Neurosci. Lett. 2003, 353, 45–48. [Google Scholar] [CrossRef]
- Hasenkamp, W.; Epstein, M.P.; Green, A.; Wilcox, L.; Boshoven, W.; Lewison, B.; Duncan, E. Heritability of acoustic startle magnitude, prepluse inhibition, and startle latency in schizophrenia and control families. Psychiatry Res. 2010, 175, 236–243. [Google Scholar] [CrossRef]
- Takahashi, H.; Hashimoto, R.; Iwase, M.; Ishii, R.; Kamio, Y.; Takeda, M. Prepulse inhibition of startle response: Recent advances in human psychiatric disease. Clin. Psychopharmacol. Neurosci. 2011, 9, 102–111. [Google Scholar] [CrossRef]
- Light, G.; Greenwood, T.A.; Swerdlow, N.R.; Calkins, M.E.; Freedman, R.; Green, M.F.; Gur, R.E.; Gur, R.C.; Lazzeroni, L.C.; Nuechterlein, K.H.; et al. Comparison of the heritability of schizophrenia and endophenotypes in the COGS-1 family study. Schizophr. Bull. 2014, 4, 1404–1411. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, T.A.; Light, G.A.; Swerdlow, N.R.; Calkins, M.E.; Green, M.F.; Gurr, R.E.; Gur, R.C.; Lazzeroni, L.C.; Nuechterlein, K.H.; Olincy, A.; et al. Gating deficit heritability and correlation with increased clinical severity in schizophrenia with positive family history. Am. J. Psychiatry 2016, 173, 385–391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- San-Martin, R.; Zimiani, M.I.; Vendramini de Ávila, M.A.; Shumama, R.; Del-Ben, C.M.; Menezes, P.R.; Fraga, F.J.; Salum, C. Early Schizophrenia and bipolar disorder patients display reduced neural prepulse inhibition. Brain Sci. 2022, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Desbonnet, L.; Waddington, J.L.; O’Tuatiagh, C.M.P. Mutant models for genes associated with schizophrenia. Biochem. Soc. Trans. 2009, 37, 308–312. [Google Scholar] [CrossRef]
- Swerdlow, N.R.; Braff, D.L.; Geyer, M.A. Sensorimotor gating of the startle reflex: What we said 25 Years Ago, what has happened since then, and what comes next. Psychopharmacology 2016, 30, 1072–1081. [Google Scholar] [CrossRef]
- Jafari, Z.; Kolb, B.E.; Mohajerani, M.H. Prepusle inhibition of the acoustic startle reflex and P50 gating in aging and alzheimer’s disease. Ageing Res. Rev. 2020, 59, 101028. [Google Scholar] [CrossRef]
- Kohl, S.; Heekeren, K.; Klosterköotter, J.; Kuhn, J. Prepulse inhibition in psychiatric disorders-apart from schizophrenia. J. Psychiatr. Res. 2013, 47, 445–452. [Google Scholar] [CrossRef]
- Crawley, J.N.; Paylor, R. A proposed test battery and constellations of specific behavioral paradigms to Investigate the behavioral phenotypes of transgenic and knockout mice. Horm. Behav. 1997, 31, 197–211. [Google Scholar] [CrossRef]
- Swerdlow, N.R.; Geyer, M.A. Using an animal model of deficient sensorimotor gating to study the pathophysiology and new treatments of schizophrenia. Schizophr. Bull. 1998, 24, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Petryshen, T.L.; Kirby, A.; Hammer, R.P., Jr.; Purcell, S.; O’Leary, S.B.; Singer, J.B.; Hill, A.E.; Nadeau, J.H.; Daly, M.J.; Sklar, P. Two quantitative trait loci for prepulse inhibition of startle identified on mouse chromosome 16 using chromosome substitution strains. Genetics 2005, 171, 1896–1904. [Google Scholar] [CrossRef]
- Leussis, M.P.; Frayne, M.L.; Saitor, M.; Berry, E.M.; Aldinger, K.A.; Rockwell, G.N.; Hammer , R.P., Jr.; askin-Hill, A.E.; Singer, J.B.; Nadeau, J.H.; et al. Genomic survey of prepulse inhibition in mouse chromosome substitution strains. Genes Brain Behav. 2009, 8, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Joober, R.; Zarate, J.-M.; Rouleau, G.-A.; Skamene, E.; Boska, P. Provisional mapping of quantitative trait loci modulating the acoustic startle response and prepulse inhibition of acoustic startle. Neuropsychopharmacology 2002, 27, 765–781. [Google Scholar] [CrossRef] [PubMed]
- Hitzemann, R.; Malmanger, B.; Belknap, J.; Darakjian, P.; McWeeny, S. Short-term selective breeding for high and low prepulse inhibition of the acoustic startle response: Pharmacological characterization and QTL mapping in the selected lines. Pharmacol. Biochem. Behav. 2008, 90, 525–533. [Google Scholar] [CrossRef]
- Watanabe, A.; Toyota, T.; Owada, Y.; Hayashi, T.; Iyayama, Y.; Matsumata, M.; Ishitsuka, Y.; Nakaya, A.; Maekawa, M.; Ohnishi, T.; et al. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 2007, 5, e297. [Google Scholar] [CrossRef]
- Sittig, L.J.; Carbonetto, P.; Engel, K.A.; Krauss, K.S.; Palmer, A.A. Integration of genome-wide association and extant brain expression QTL identifies candidate genes influencing prepuluse inhibition in F1 mice. Genes Brain Behav. 2016, 15, 260–270. [Google Scholar] [CrossRef]
- Samocha, K.E.; Lim, J.E.; Cheng, R.; Sokoff, G.; Palmer, A.A. Fine Mapping of QTL for Prepulse inhibition in LG/J and SM/J mice using F2 and advance intercross lines. Genes Brain Behav. 2010, 9, 759–767. [Google Scholar] [CrossRef]
- Loos, M.; Staal, J.; Pattij, T.; Neuro-BSK Mouse Phenome Consortium; Smit, A.B.; Spijker, S. Independent genetic loci for sensorimotor gating and attentional performance in BXD recombinant inbred strains. Genes Brain Behav. 2012, 11, 147–156. [Google Scholar] [CrossRef]
- Belknap, J.K.; Mitchell, S.R.; O’Toole, L.A.; Helms, M.L.; Crabbe, J.C. Type I and Type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav. Genet. 1997, 26, 149–160. [Google Scholar] [CrossRef]
- Peirce, J.; Lu, L.; Gu, J.; Silver, L.M.; Williams, R.W. A new set of BXD recombinant inbred lines from intercross populations in mice. BMC Genet. 2004, 5, 7. [Google Scholar] [CrossRef]
- Philip, V.M.; Duvvuru, S.; Gomero, B.; Ansah, T.A.; Blaha, C.D.; Cook, M.N.; Hamre, K.M.; Lariviere, W.R.; Matthews, D.B.; Mittleman, G.; et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010, 9, 129–159. [Google Scholar] [CrossRef]
- Cook, M.N.; Dunning, J.P.; Wiley, R.G.; Chesler, E.J.; Johnson, D.K.; Miller, D.R.; Goldowitz, D. Neurobehavioral mutants identified in an ENU mutagenesis project. Mamm. Genome 2007, 18, 559–572. [Google Scholar] [CrossRef]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef]
- Sul, J.H.; Kostem, E.; Furlotte, N.; He, D.; Eskin, E. Accounting for population structure in gene-by-environment interactions in genome-wide association studies using mixed models. PLoS Genet. 2015, 12, e1005849. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Alberts, R.; Schughart, K. QTLminer: Identifying genes regulating quantitative traits. BMC Bionform. 2010, 11, 516. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, Y.; Baker, J.A.; Gu, Q.; Hamre, K.M.; Yue, J.; Jones, B.C.; Cook, M.N.; Lu, L. The effect of alcohol on the differential expression of cluster of differentiation 14 genes, associated pathways, and genetic network. PLoS ONE 2017, 12, e0178689. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.N.; Baker, J.A.; Heldt, S.; Williams, R.W.; Hamre, K.M.; Lu, L. Identification of candidate genes that underlie the QTL on chromomsome 1 mediates genetic differences in stress-ethanol interactions. Physiol. Genom. 2015, 47, 308–317. [Google Scholar] [CrossRef][Green Version]
- Smith, C.L.; Blake, J.A.; Kadin, J.A.; Richardson, J.E.; Bult, C.J. Mouse Genome Database Group. Mouse Genome Database (MGD) -2018: Knowledgebase for the laboratory mouse. Nucleic Acids Res. 2018, 46, D836–D842. [Google Scholar] [CrossRef]
- Smith, J.R.; Hayman, G.T.; Wang, S.-J.; Faulederkind, S.J.F.; Hoffman, M.J.; Kaldunski, M.L.; Tutaj, M.; Thota, J.; Nalabolu, H.S.; Ellanki, S.L.R.; et al. The year of the rat: The rat genome database at 20: A multi-species knowledgebase and analysis platform. Nucleic Acids Res. 2020, 48, D731–D742. [Google Scholar] [CrossRef]
- Groza, T.; Gomez, F.L.; Mashhadi, H.H.; Munõz-Fuentes, V.; Gunes, O.; Wilson, R.; Cacheiro, P.; Frost, A.; Keskivali-Bond, P.; Vardal, B.; et al. The International Mouse Phenotyping Consortium: Comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2023, 51, D1045–D1308. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Kaneshia, M.; Goto, S.; Kawashima, S.; Nakaya, A. The KEGG database at GenomeNet. Nucleic Acids Res. 2002, 30, 42–46. [Google Scholar] [CrossRef]
- Homayouni, R.; Heinrich, K.; Wei, K.; Berry, M.W. Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics 2005, 21, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Gao, J.; Bergman, S.; Sims, A.C.; Ashbrook, D.G.; Baric, R.S.; Cui, Y.; Jonsson, C.B.; Li, K.; Williams, R.W.; et al. Genetic Dissection of the Regulatory Mechanisms of Ace2 in the Infected Mouse Lung. Front. Immunol. 2020, 11, 3332. [Google Scholar] [CrossRef] [PubMed]
- Chundri, A.; Watson, P.M.; Ashbrook, D.G. New Insights on Gene by Environment Effects of Drug Abuse in Animal Models Using GeneNetwork. Genes 2022, 13, 614. [Google Scholar] [CrossRef]
- Watanabe, K.; Stringer, S.; Frei, O.; Mirkov, M.U.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global view of pleitropy and genetic architecture in complex traits. Nat Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D60–D612. [Google Scholar] [CrossRef]
- Bajpai, A.K.; Davuluri, S.; Tiwary, K.; Narayanan, S.; Oguru, S.; Basavaraju, K.; Dayalan, D. Systematic comparison of the protein-protein interaction databases from a user’s perspective. J. Biomed. Inform. 2020, 103, 103380. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 11, 2450–2498. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; White, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Ketchesin, K.D.; Zong, W.; Hildebrand, M.A.; Scott, M.R.; Seney, M.L.; Cahill, K.M.; Shankar, V.G.; Glausier, J.R.; Lewis, D.A.; Tseng, G.C.; et al. Diurnal Alterations in Gene Expression Across striatal subregions in psychosis. Biol. Psychiatry 2022, 93, 137–148. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, N.R.; Light, G.A.; Thomas, M.L.; Sprock, J.; Calkins, M.E.; Green, M.F.; enwood, T.A.; Gur, R.E.; Gur, R.C.; Lazzeroni, L.C.; et al. Deficient prepulse inhibition in schizophrenia in a multi-site cohort: Internal replication and extension. Schizophr. Res. 2007, 17, S0920–S9964. [Google Scholar] [CrossRef]
- Uezato, A.; Kimura-Sato, J.; Yamamoto, N.; Iijima, Y.; Kunugi, H.; Nishikawa, T. Further evidence for a male-selective genetic association of synapse-associated protein 97 (SAP97) gene with schizophrenia. Behav. Brain Funct. 2012, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.R.; Longo-Guess, C.; Gagnon, L.H.; Yu, H.; Zheng, Q.Y. A locus on distal chromosome 11 (ahl8) and it interactions with Cd23 ahl underlie the early onset, age-related hearing loss in DBA/2J mice. Genomics 2008, 92, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.Y.; Kui, L.; Xu, F.; Zheng, T.; Li, B.; McCarty, M.; Sun, Z.; Zhang, A.; Liu, L.; Starland-Davenport, A.; et al. An Age-Related Hearing Protection Locus on Chromosome 16 of BXD Strain Mice. Neural Plast. 2020, 2020, 8889264. [Google Scholar] [CrossRef]
- Nagtegaal, A.P.; Spijker, S.; Crins, T.T.H.; Neuro-Bsik Mouse Phenomics Consortium; Bort, J.G.G. A novel QTL underlying early-onset frequency hearing loss in BXD recombinant inbred strains. Genes Brain Behav. 2014, 11, 911–920. [Google Scholar] [CrossRef]
- Johnson, K.R.; Longo-Guess, C.; Gagnon, L.H. A QTL on Chr 5 modified hearing loss associated with the fascin-2 variant of DBA/2J mice. Mamm. Genome 2015, 26, 338–347. [Google Scholar] [CrossRef]
- Pelov, I.; Teltsh, O.; Greenbaum, L.; Rigbi, A.; Kanyas-Sarner, K.; Lerer, B.; Lombroso, P.; Kohn, Y. Involvement of PTPN5, the gene encoding the STriatal-Enriched protein tyrosine Phosphatase (STEP), in schizophrenia and cognition. Psychiatr. Genet. 2012, 22, 168–176. [Google Scholar] [CrossRef]
- Luo, X.; Huang, L.; Jia, P.; Li, M.; Su, B.; Zhao, Z.; Gan, L. Protein-protein interaction pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (chromatin) assembly genes in schizophrenia susceptibility loci. Schizophr. Bull. 2014, 40, 39–49. [Google Scholar] [CrossRef][Green Version]
- Ludewig, K.; Geyer, M.A.; Etzensberger, M.; Vollenweider, F.X. Stability of the acoustic startle reflex: Prepulse inhibition, and habituation in schizophrenia. Schizophr. Res. 2002, 55, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Rovnỷ, R.; Besterciová, D.; Riečanskỷ, I. Genetic determinants of gating functions: Do we get closer to understanding schizophrenia etiopathogenesis? Front. Psychiatry 2020, 11, 550225. [Google Scholar] [CrossRef]
- Kirby, B.P.; Waddington, J.L.; O’Tuathiagh, C.M.P. Advancing a functional genomics for schizophrenia: Psychopathophysiological and cognitive phenotypes in mutants with gene disruption. Brain Res. Bull. 2010, 83, 162–176. [Google Scholar] [CrossRef]
- O’Tuathiagh, C.M.P.; Waddington, J.L. Mutant mouse models: Phenotypic relationships to domains of psychopathology and pathobiology in schizophrenia. Schizophr. Bull. 2010, 36, 243–245. [Google Scholar] [CrossRef][Green Version]
- Greenwood, T.A.; Light, G.A.; Swerdlow, N.R.; Radant, A.D.; Braff, D.L. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PLoS ONE 2012, 7, e29630. [Google Scholar] [CrossRef]
- Kauppi, K.; Rosenthal, S.B.; Lo, M.-T.; Sanyani, N.; Jiang, M.; Abagyan, R.; McEvoy, L.K.; Andreassen, O.A.; Chen, C.-H. Revisiting antipsychotic drug actions through gene networks associated with schizophrenia. Am. J. Psychiatry 2018, 175, 674–682. [Google Scholar] [CrossRef]
- Mistry, M.; Gillis, J.; Pavlidis, P. Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls. BMC Neurosci. 2013, 14, 105. [Google Scholar] [CrossRef] [PubMed]
- Balan, S.; Ohnishi, T.; Watanabe, A.; Ohba, H.; Iwayama, Y.; Toyoshima, M.; Hara, T.; Hisano, Y.; Miyasaka, Y.; Toyota, T.; et al. Role of an atypical cadherin gene, Cdh23 in prepulse inhibition, and implication of CDH23 in schizophrenia. Schizophr. Bull. 2021, 47, 1190–1200. [Google Scholar] [CrossRef]
- Ashbrook, D.G.; Cahill, S.; Hager, R. A cross-species systems genetic analysis links APBB1lP as a candidate gene for schizophrenia and prepulse inhibition. Front. Behav. Neurosci. 2019, 13, 266. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, Y.; Li, S.Y.; Liang, X.; Ren, Z.; Yang, Z.; Zhang, B.; Hu, Y.; Yang, X. Systematic discovery of signaling pathways linking immune activation to schizophrenia. iScience 2021, 24, 103209. [Google Scholar] [CrossRef] [PubMed]
- Hayashi-Takagi, A.; Sawa, A. Disturbed synaptic connectivity in schizophrenia: Convergence of genetic risk factors during neurodevelopment. Brain Res. Bull. 2010, 83, 140–146. [Google Scholar] [CrossRef]
- Kessels, W.; Malinow, R. Synaptic AMPA receptor plasticity and behavior. Neuron 2009, 61, 340–350. [Google Scholar] [CrossRef]
- Yan, H.-H.; He, J.-J.; Fu, C.; Chen, J.-H.; Tang, A.-H. ATAD1 regulates neuronal development and synapse formation through tuning mitochondrial function. Int. J. Mol. Sci. 2025, 26, 44. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Chi, Z.; Keuss, M.J.; Pai, Y.-M.E.; Kang, H.C.; Shin, J.-H.; Bugayenko, A.; Wang, H.; Xiong, Y.; et al. The AAA+ ATPase, thorase regulates AMPA receptor-dependent synaptic plasticity and behavior. Cell 2011, 145, 284–299. [Google Scholar] [CrossRef]
- Keifer, J.; Zheng, Z. AMPA receptor trafficking and learning. Eur. J. Neurosci. 2010, 32, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, M.; Umanah, G.K.E.; Ribeiro, S.P.; Dawson, V.L.; Dawson, T.; Bonci, A. Synaptic plasticity onto dopamine neurons shapes fear learning. Neuron 2017, 93, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Umanah, G.K.E.; Pignatelli, M.; Yin, X.; Chen, R.; Crawford, J.; Neifert, S.; Scarffe, L.; Behensky, A.A.; Guiberson, N.; Chang, M.; et al. Thorase variants are associated with defects in glutamatergic neurotransmission that can be rescued by perampanel. Sci. Transl. Med. 2017, 9, eeah4985. [Google Scholar] [CrossRef]
- Steri, M.; Idda, M.L.; Whalen, M.B.; Orrù, V. Genetic variants in mRNA untranslated regions. Wiley Interdiscip. Rev. RNA 2018, 9, e1474. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, H.; Yang, J.; Cai, M.; Yang, Q.; Wang, H.; Xu, Y.; Chen, H.; Hu, Y.; He, W.; et al. ATPase Thorase deficiency causes α-Synucleinopathy and Parkinson’s Disease-like behavior. Cells 2023, 11, 2990. [Google Scholar] [CrossRef]
- Schaefer, N.; Harvey, R.J.; Vilmann, C. Startle Disease: New Molecular Insights into an Old Neurological Disorder. Neuroscientist 2023, 29, 767–781. [Google Scholar] [CrossRef]
- Saini, A.G.; Pandey, S. Hyperexkplexia and other startle syndromes. J. Neurol. Sci. 2020, 416, 117051. [Google Scholar] [CrossRef]
- Craddock, K.E.; McKee, J.L.; Fitzgerald, M.; Ahrens-Nicklas, R.; Agarwal, S. Neonatal Hypertonia and Progressive Respiratory Failure due to Novel Heterozygous Mutation in ATAD1. Pediatr. Neurol. 2023, 142, 56–57. [Google Scholar] [CrossRef]
- Bunod, R.; Doummar, D.; Whalen, S.; Keren, B.; Chantot-Bastaraud, S.; Maincent, K.; Villy, M.-C.; Mayer, M.; Rodriguez, D.; Burglen, L.; et al. Congentital immobility and stiffness related to biallelic ATAD1 variants. Neurol. Genet. 2020, 6, e520. [Google Scholar] [CrossRef] [PubMed]
- Wolf, N.I.; Zschocke, J.; Jakobs, C.; Rating, D.; Hoffmnn, G.F. ATAD1 encephalopathy and stiff baby syndrome: A recognizable clinical presentation. Brain 2018, 141, e49. [Google Scholar] [CrossRef] [PubMed]
| Official Symbol | Mean Expression ≥ 8 (Score = 1) | Non-Syn SNPs/Indels (Score = 2) | Cis-Regulation (Score = 2) | Sig. Phenotype Correlation (Score = 2) | Functional Significance (Score = 3) | Total (Score = 10) |
|---|---|---|---|---|---|---|
| Atad1 | Yes | Yes | Yes | Yes | Yes | 10 |
| Rnls | -- | Yes | Yes | Yes | Yes | 9 |
| Prkg1 | Yes | Yes | -- | Yes | Yes | 8 |
| Asah2 | -- | Yes | -- | Yes | Yes | 7 |
| Ifit1 | -- | Yes | -- | Yes | Yes | 7 |
| Cdc3711 | Yes | -- | -- | Yes | Yes | 6 |
| Ak3 | Yes | -- | -- | Yes | Yes | 6 |
| Papps2 | Yes | -- | Yes | Yes | -- | 5 |
| Ifit1bl2 | -- | -- | -- | Yes | Yes | 5 |
| Ranbp6 | Yes | -- | -- | Yes | -- | 3 |
| Atlas ID | PMID | Year | Trait | p-Value | N |
|---|---|---|---|---|---|
| 12 | 24280982 | 2014 | Schizophrenia/bipolar disorder | 0.040 | 39,202 |
| 15 | 28439101 | 2017 | Post-traumatic stress disorder | 0.013 | 9223 |
| 30 | 21173776 | 2012 | Agreeableness (NEO-FFI) | 0.047 | 17,375 |
| 1141 | 27494321 | 2016 | Chronotype | 0.001 | 128,266 |
| 1142 | 27494321 | 2016 | Sleep duration | 0.045 | 128,266 |
| 1173 | 26955885 | 2016 | Chronotype (continuous) | 4.98 × 10−5 | 100,420 |
| 1174 | 26955885 | 2016 | Extreme chronotype | 0.032 | 100,420 |
| 2018 | 24369049 | 2014 | Lithium response in Bipolar I patients—Alda Scale of 7 to 8 | 0.042 | 294 |
| 2025 | 22952603 | 2012 | 10 mg response to amphetamine | 0.025 | 381 |
| 2043 | 27329760 | 2016 | Bipolar disorder | 0.036 | 34,950 |
| 3230 | 31427789 | 2019 | Morning/evening person (chronotype) | 0.040 | 345,148 |
| 3235 | 31427789 | 2019 | Current tobacco smoking | 0.009 | 386,150 |
| 3262 | 31427789 | 2019 | Average weekly red wine intake | 0.025 | 274,058 |
| 3287 | 31427789 | 2019 | Sensitivity/hurt feelings | 0.042 | 375,272 |
| 3292 | 31427789 | 2019 | Worry too long after embarrassment | 0.002 | 370,660 |
| 3295 | 31427789 | 2019 | Guilty feelings | 0.000 | 376,361 |
| 3296 | 31427789 | 2019 | Risk-taking | 0.031 | 372,651 |
| 3297 | 31427789 | 2019 | Frequency of depressed mood in last 2 weeks | 0.001 | 370,017 |
| 3394 | 31427789 | 2019 | Ever unenthusiastic/disinterested for a whole week | 0.024 | 123,848 |
| 3567 | 31427789 | 2019 | Why stopped smoking: illness or ill health | 0.021 | 94,509 |
| 3655 | 31427789 | 2019 | Smoking status: previous vs. current | 0.014 | 177,025 |
| 3729 | 31427789 | 2019 | Depression—age at first episode of depression | 0.009 | 65,776 |
| 3744 | 31427789 | 2019 | Cannabis use—ever taken cannabis | 0.012 | 126,632 |
| 3770 | 31427789 | 2019 | Depression—trouble falling or staying asleep or sleeping too much | 0.001 | 126,545 |
| 3772 | 31427789 | 2019 | Depression—recent feelings of tiredness or low energy | 0.030 | 126,540 |
| 3775 | 31427789 | 2019 | Traumatic events—able to pay rent/mortgage as an adult | 0.014 | 124,944 |
| 3795 | 29942085 | 2018 | Neuroticism | 0.015 | 390,278 |
| 3796 | 29942085 | 2018 | Depressive symptoms | 0.011 | 381,455 |
| 3982 | 29483656 | 2018 | Schizophrenia | 0.002 | 105,318 |
| 3993 | 29500382 | 2018 | Irritability (IRR) | 0.048 | 260,369 |
| 3999 | 29500382 | 2018 | Worry too long after embarrassment (WORR-EMB) | 0.001 | 261,094 |
| 4002 | 29500382 | 2018 | Guilty feelings (GUILT) | 0.001 | 265,139 |
| 4014 | 29700475 | 2018 | Major depressive disorder | 0.004 | 173,005 |
| 4040 | 29906448 | 2018 | Schizophrenia/bipolar disorder | 0.037 | 107,620 |
| 4087 | 29255261 | 2018 | Neuroticism | 0.010 | 329,821 |
| 4294 | 30696823 | 2019 | Chronotype | 0.029 | 449,732 |
| 4316 | 30643251 | 2019 | Smoking cessation | 0.005 | 312,821 |
| 4368 | 30150663 | 2018 | Cannabis use | 0.013 | 162,082 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajpai, A.K.; Freels, T.G.; Lu, L.; Cook, M.N. Atad1 Is a Potential Candidate Gene for Prepulse Inhibition. Genes 2025, 16, 1139. https://doi.org/10.3390/genes16101139
Bajpai AK, Freels TG, Lu L, Cook MN. Atad1 Is a Potential Candidate Gene for Prepulse Inhibition. Genes. 2025; 16(10):1139. https://doi.org/10.3390/genes16101139
Chicago/Turabian StyleBajpai, Akhilesh K., Timothy G. Freels, Lu Lu, and Melloni N. Cook. 2025. "Atad1 Is a Potential Candidate Gene for Prepulse Inhibition" Genes 16, no. 10: 1139. https://doi.org/10.3390/genes16101139
APA StyleBajpai, A. K., Freels, T. G., Lu, L., & Cook, M. N. (2025). Atad1 Is a Potential Candidate Gene for Prepulse Inhibition. Genes, 16(10), 1139. https://doi.org/10.3390/genes16101139






