Genetic Diversity of Fresh Maize Germplasm Revealed by Morphological Traits and SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Field Experiment Design, Phenotypic Trait Investigation, and Seed Character Evaluation Methods
2.2.2. Statistical Methods for Phenotypic Traits
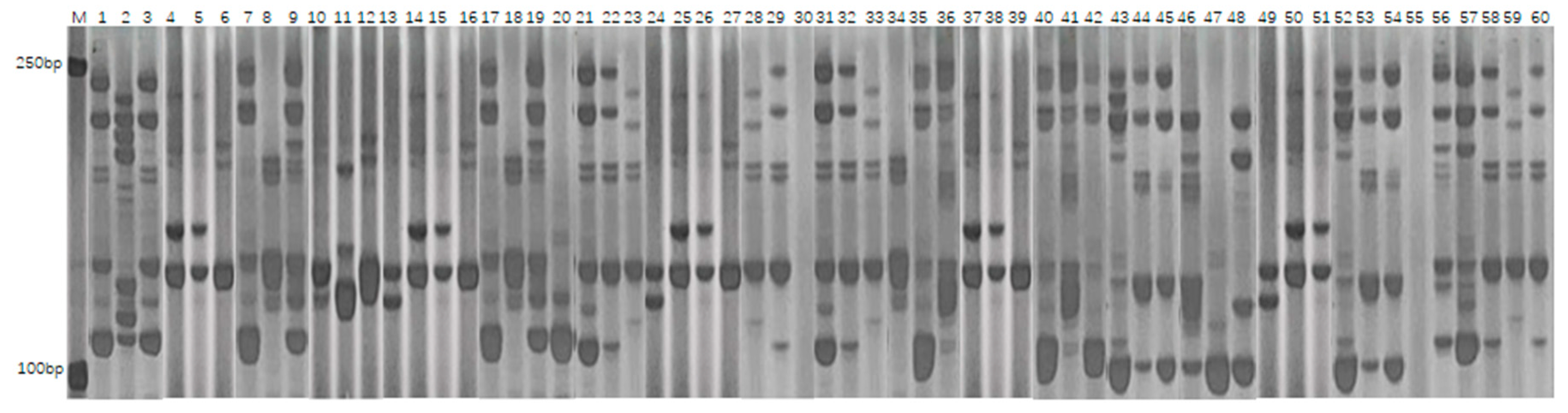
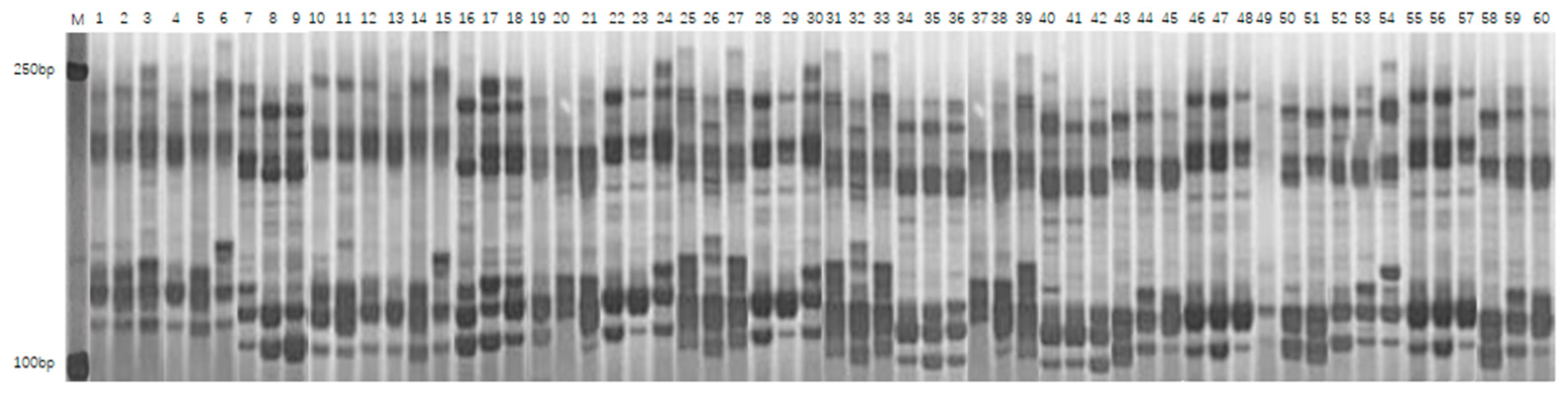
2.2.3. SSR Analysis and Data Processing Methods
- (1)
- In the analysis of genetic diversity, bands obtained through SSR silver staining were scored using a co-dominant scoring system. Homozygous loci were recorded with their corresponding allele fragment sizes (e.g., “215/215”), while heterozygous loci were recorded with both allele fragment sizes (e.g., “215/221”). Missing data were denoted as “9”. This scoring facilitated the construction of a comprehensive database. The genetic similarity coefficient (GS) and genetic distance (GD) between fresh maize inbred lines were calculated using Nei’s formula [18]. The specific formulas are expressed as follows: GSij = 2Nij/(Ni + Nj) and GDij = 1 − GSij. Here, Nij represents the number of bands shared between inbred lines *i* and *j*, while Ni and Nj denote the total number of bands present in inbred lines *i* and *j*, respectively. These equations are critical for understanding the genetic similarities and differences among various inbred lines. After completing the calculations, a cluster analysis was performed using the UPGMA method to construct a dendrogram. The polymorphism information content (PIC) of each polymorphic locus was calculated according to the formula proposed by Smith et al. [19]: PIC = 1 − ∑fi2, where fi represents the allele frequency at locus i.
- (2)
- The IBM SPSS Statistics software (version 27.0) was employed to compare the phenotypic traits of fresh maize inbred lines for significant differences.
3. Results
3.1. Phenotypic Cluster Analysis
3.2. Genetic Distance Analysis
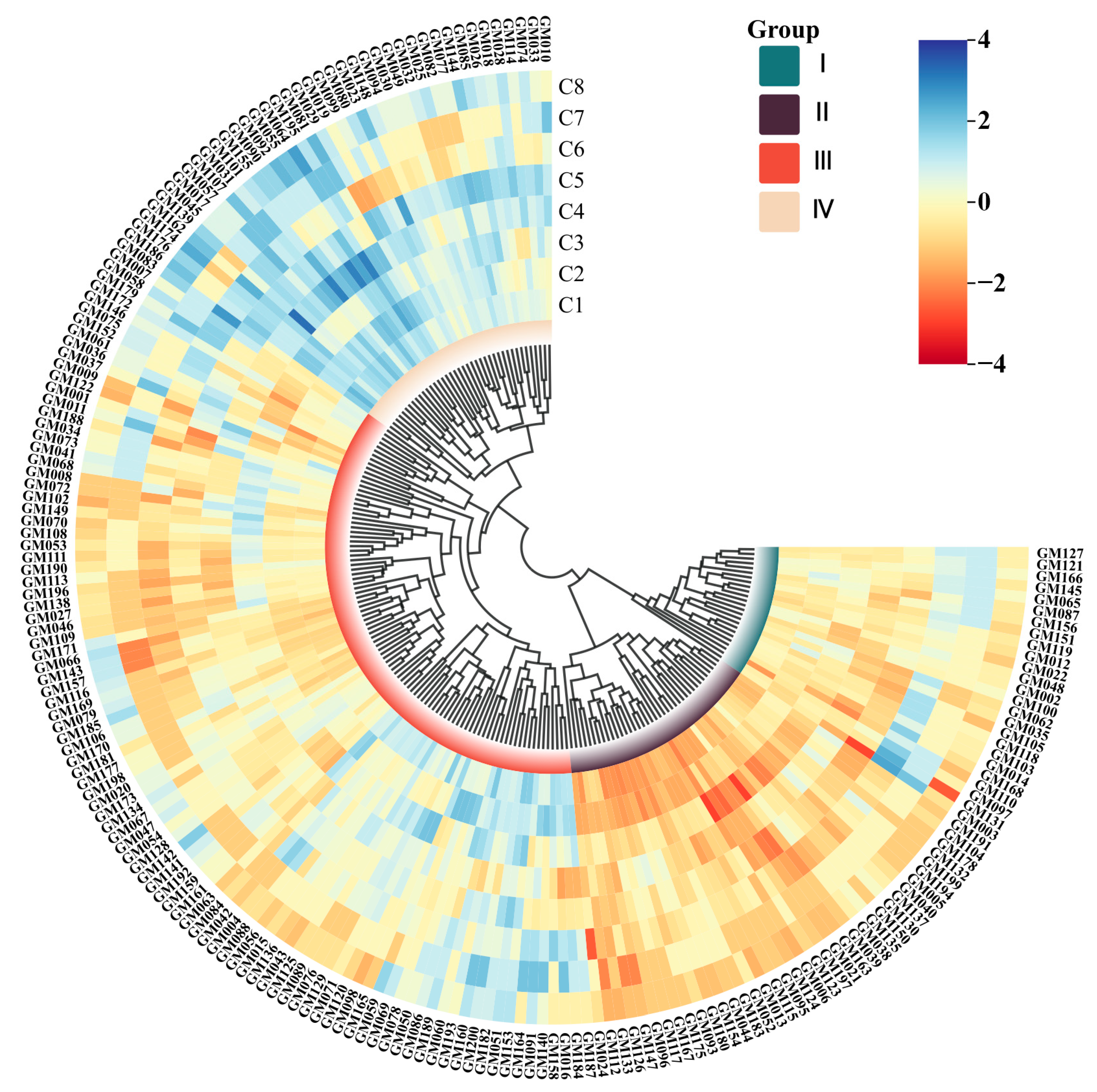
3.3. SSR Genetic Diversity and Cluster Analysis
3.3.1. Polymorphism Analysis of SSR Markers
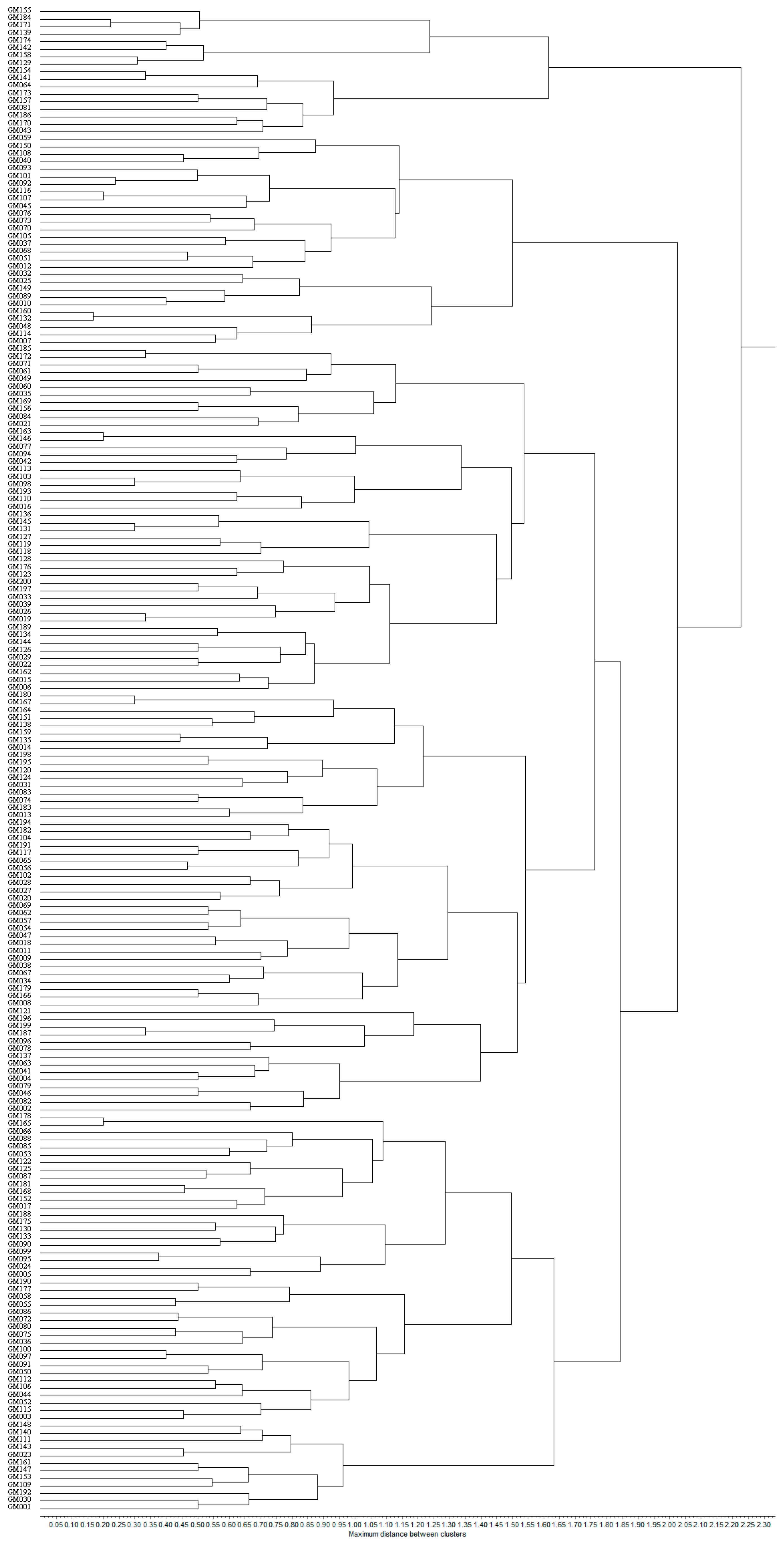
3.3.2. SSR Marker-Based Cluster Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SSR | Simple Sequence Repeat |
| PIC | Polymorphism Information Content |
| GS | Genetic Similarity Coefficient |
| GD | Genetic Distance |
Appendix A
| Number | Plant Height (cm) | Ear Height (cm) | Tassel Length (cm) | Tassel Branch Number (Number) | Ear Length (cm) | Ear Diameter (cm) | Kernel Row Number (Rows) | Kernel Number Per Row (Kernels) |
|---|---|---|---|---|---|---|---|---|
| GM001 | 184 | 69 | 28 | 6 | 8.2 | 3.9 | 16 | 21 |
| GM002 | 176 | 61 | 21 | 7 | 14 | 4 | 14 | 20 |
| GM003 | 165 | 54 | 26 | 5 | 6 | 5.3 | 18 | 18 |
| GM004 | 195 | 72 | 31 | 12 | 15.3 | 4.9 | 12 | 18 |
| GM005 | 155 | 52 | 19 | 11 | 12.2 | 3.9 | 14 | 16 |
| GM006 | 111 | 35 | 11 | 4 | 12.9 | 3.3 | 12 | 14 |
| GM007 | 190 | 58 | 27 | 9 | 16 | 4.8 | 16 | 23 |
| GM008 | 145 | 55 | 35 | 12 | 9.6 | 4.3 | 12 | 16 |
| GM009 | 168 | 63 | 21 | 18 | 15.2 | 3.1 | 14 | 15 |
| GM010 | 175 | 68 | 31 | 17 | 16.1 | 4.2 | 18 | 22 |
| GM011 | 180 | 71 | 31 | 16 | 18.1 | 3.6 | 18 | 28 |
| GM012 | 146 | 58 | 28 | 9 | 13.2 | 4.1 | 14 | 18 |
| GM013 | 134 | 40 | 24 | 4 | 11.6 | 3.7 | 12 | 14 |
| GM014 | 168 | 57 | 26 | 10 | 11.1 | 4.3 | 14 | 20 |
| GM015 | 197 | 65 | 31 | 12 | 14.1 | 4.2 | 14 | 18 |
| GM016 | 210 | 85 | 29 | 4 | 11.9 | 4.6 | 18 | 20 |
| GM017 | 221 | 95 | 30 | 17 | 16.8 | 4.8 | 16 | 25 |
| GM018 | 187 | 65 | 30 | 15 | 17 | 4.6 | 14 | 25 |
| GM019 | 186 | 89 | 29 | 16 | 18.2 | 3.1 | 14 | 22 |
| GM020 | 145 | 50 | 27 | 12 | 13.4 | 4.2 | 12 | 24 |
| GM021 | 123 | 52 | 17 | 5 | 7.8 | 3.2 | 12 | 15 |
| GM022 | 156 | 54 | 28 | 8 | 12.5 | 4 | 14 | 16 |
| GM023 | 176 | 77 | 30 | 16 | 20 | 3.5 | 16 | 32 |
| GM024 | 126 | 40 | 25 | 6 | 12.8 | 2.6 | 14 | 18 |
| GM025 | 190 | 83 | 35 | 12 | 17.5 | 3.5 | 12 | 28 |
| GM026 | 195 | 70 | 32 | 14 | 17.1 | 4.3 | 14 | 26 |
| GM027 | 167 | 61 | 31 | 8 | 12.5 | 3.1 | 12 | 19 |
| GM028 | 184 | 70 | 25 | 14 | 18.2 | 4.5 | 16 | 28 |
| GM029 | 182 | 91 | 32 | 14 | 18.7 | 3.1 | 16 | 27 |
| GM030 | 196 | 79 | 33 | 10 | 16.2 | 3.8 | 14 | 24 |
| GM031 | 221 | 88 | 31 | 20 | 16.5 | 4.6 | 18 | 26 |
| GM032 | 190 | 81 | 32 | 13 | 16.4 | 3.5 | 12 | 26 |
| GM033 | 185 | 70 | 35 | 15 | 17.7 | 3.8 | 16 | 23 |
| GM034 | 175 | 75 | 33 | 9 | 13.3 | 3.9 | 16 | 22 |
| GM035 | 164 | 60 | 30 | 7 | 10.3 | 4.2 | 12 | 19 |
| GM036 | 155 | 51 | 33 | 9 | 17.2 | 3.9 | 16 | 24 |
| GM037 | 143 | 47 | 30 | 11 | 15.7 | 3.7 | 18 | 22 |
| GM038 | 134 | 55 | 18 | 11 | 14 | 3.2 | 14 | 20 |
| GM039 | 170 | 56 | 11 | 9 | 10 | 3 | 12 | 16 |
| GM040 | 172 | 60 | 20 | 13 | 10 | 3.4 | 14 | 18 |
| GM041 | 169 | 66 | 35 | 9 | 12.2 | 3.6 | 16 | 25 |
| GM042 | 210 | 65 | 27 | 15 | 14.5 | 4.6 | 12 | 16 |
| GM043 | 205 | 72 | 32 | 15 | 13.5 | 4 | 14 | 15 |
| GM044 | 118 | 35 | 21 | 7 | 11.5 | 3.9 | 14 | 16 |
| GM045 | 215 | 85 | 32 | 16 | 17.3 | 4.8 | 16 | 25 |
| GM046 | 164 | 65 | 31 | 6 | 10.6 | 3.4 | 12 | 18 |
| GM047 | 174 | 75 | 29 | 9 | 12.2 | 4.1 | 14 | 26 |
| GM048 | 176 | 71 | 21 | 10 | 13 | 4.2 | 14 | 20 |
| GM049 | 198 | 85 | 35 | 13 | 15.5 | 4.1 | 14 | 24 |
| GM050 | 183 | 101 | 28 | 11 | 13.4 | 4 | 16 | 23 |
| GM051 | 224 | 86 | 37 | 13 | 18.1 | 4.4 | 16 | 28 |
| GM052 | 124 | 46 | 25 | 8 | 11.8 | 3.7 | 12 | 20 |
| GM053 | 168 | 64 | 32 | 7 | 11.5 | 3.2 | 14 | 19 |
| GM054 | 178 | 75 | 31 | 9 | 12.1 | 4 | 14 | 22 |
| GM055 | 198 | 94 | 37 | 21 | 18.5 | 4.1 | 16 | 32 |
| GM056 | 201 | 71 | 28 | 10 | 14.5 | 3.6 | 14 | 16 |
| GM057 | 210 | 87 | 29 | 16 | 15.1 | 5.1 | 18 | 25 |
| GM058 | 185 | 67 | 24 | 9 | 12.9 | 4.9 | 16 | 28 |
| GM059 | 216 | 89 | 25 | 14 | 13.9 | 4.2 | 12 | 30 |
| GM060 | 226 | 83 | 37 | 8 | 16 | 4.4 | 14 | 23 |
| GM061 | 138 | 36 | 21 | 9 | 16.2 | 4.1 | 14 | 24 |
| GM062 | 158 | 75 | 31 | 6 | 9.8 | 4.6 | 12 | 18 |
| GM063 | 195 | 95 | 36 | 8 | 11 | 3.8 | 14 | 16 |
| GM064 | 210 | 109 | 40 | 23 | 16.3 | 4.5 | 18 | 35 |
| GM065 | 172 | 80 | 29 | 10 | 11.8 | 4.4 | 16 | 22 |
| GM066 | 143 | 52 | 26 | 9 | 11.3 | 3.1 | 10 | 28 |
| GM067 | 161 | 63 | 29 | 8 | 12.2 | 4.1 | 14 | 24 |
| GM068 | 171 | 69 | 33 | 9 | 11 | 3.9 | 16 | 24 |
| GM069 | 186 | 102 | 26 | 10 | 12.8 | 4.1 | 14 | 27 |
| GM070 | 156 | 56 | 35 | 12 | 13.5 | 3.6 | 14 | 16 |
| GM071 | 194 | 66 | 34 | 14 | 15.6 | 4.3 | 14 | 20 |
| GM072 | 153 | 54 | 33 | 15 | 12.7 | 3.1 | 12 | 16 |
| GM073 | 173 | 81 | 32 | 9 | 13 | 4 | 16 | 22 |
| GM074 | 189 | 72 | 31 | 14 | 16.8 | 3.9 | 16 | 27 |
| GM075 | 170 | 65 | 22 | 7 | 14.2 | 4.7 | 14 | 26 |
| GM076 | 191 | 67 | 33 | 12 | 13.5 | 4.2 | 12 | 18 |
| GM077 | 195 | 95 | 35 | 13 | 18.9 | 4.1 | 12 | 24 |
| GM078 | 187 | 102 | 31 | 13 | 12.4 | 3.9 | 14 | 23 |
| GM079 | 144 | 51 | 29 | 11 | 14.1 | 3.5 | 12 | 26 |
| GM080 | 194 | 87 | 33 | 17 | 15.3 | 3.5 | 14 | 28 |
| GM081 | 204 | 100 | 33 | 19 | 13 | 4.1 | 18 | 33 |
| GM082 | 206 | 85 | 37 | 17 | 17.8 | 3.8 | 12 | 24 |
| GM083 | 187 | 62 | 26 | 11 | 17.3 | 5.4 | 16 | 30 |
| GM084 | 193 | 98 | 34 | 7 | 13 | 3.5 | 12 | 18 |
| GM085 | 185 | 73 | 38 | 16 | 18.2 | 3.9 | 14 | 28 |
| GM086 | 181 | 98 | 31 | 11 | 13.2 | 4.3 | 16 | 25 |
| GM087 | 154 | 73 | 30 | 8 | 10 | 4.1 | 16 | 19 |
| GM088 | 197 | 70 | 30 | 13 | 14.5 | 4.7 | 14 | 18 |
| GM089 | 214 | 83 | 35 | 13 | 13.6 | 3.9 | 12 | 18 |
| GM090 | 228 | 103 | 35 | 20 | 15.3 | 4.1 | 16 | 30 |
| GM091 | 189 | 96 | 29 | 14 | 12.3 | 4.7 | 18 | 27 |
| GM092 | 211 | 99 | 37 | 23 | 16.7 | 4 | 16 | 33 |
| GM093 | 119 | 42 | 27 | 11 | 13.4 | 3.6 | 14 | 18 |
| GM094 | 176 | 76 | 31 | 12 | 13.7 | 3.6 | 14 | 24 |
| GM095 | 128 | 40 | 25 | 7 | 10.4 | 3.8 | 12 | 16 |
| GM096 | 114 | 42 | 23 | 11 | 12.1 | 3.5 | 12 | 16 |
| GM097 | 119 | 58 | 26 | 9 | 13.7 | 4.9 | 16 | 20 |
| GM098 | 223 | 69 | 40 | 13 | 15.5 | 3.9 | 14 | 14 |
| GM099 | 200 | 81 | 35 | 16 | 16.2 | 3.3 | 16 | 24 |
| GM100 | 149 | 56 | 22 | 7 | 13.6 | 3.9 | 14 | 22 |
| GM101 | 189 | 81 | 30 | 23 | 19.2 | 4.5 | 16 | 28 |
| GM102 | 178 | 61 | 37 | 12 | 11.3 | 3.7 | 12 | 14 |
| GM103 | 170 | 63 | 25 | 9 | 11 | 4.7 | 14 | 20 |
| GM104 | 132 | 65 | 22 | 11 | 9.5 | 3.6 | 14 | 18 |
| GM105 | 155 | 79 | 26 | 6 | 9.5 | 4.7 | 14 | 15 |
| GM106 | 147 | 56 | 25 | 11 | 14.2 | 3.6 | 12 | 24 |
| GM107 | 229 | 99 | 29 | 19 | 17 | 4.5 | 18 | 32 |
| GM108 | 165 | 63 | 31 | 9 | 12.6 | 3.6 | 14 | 17 |
| GM109 | 168 | 61 | 34 | 7 | 11.1 | 3.3 | 12 | 18 |
| GM110 | 157 | 60 | 20 | 3 | 10.9 | 4.4 | 16 | 15 |
| GM111 | 160 | 60 | 33 | 6 | 12.3 | 3.2 | 14 | 17 |
| GM112 | 118 | 41 | 24 | 9 | 10.1 | 3.3 | 10 | 14 |
| GM113 | 181 | 73 | 27 | 6 | 12.9 | 3.2 | 12 | 22 |
| GM114 | 190 | 83 | 25 | 17 | 16.9 | 3.9 | 14 | 24 |
| GM115 | 135 | 40 | 25 | 5 | 11.3 | 3.6 | 12 | 16 |
| GM116 | 146 | 50 | 24 | 10 | 11.2 | 3.7 | 12 | 26 |
| GM117 | 118 | 39 | 24 | 10 | 13.4 | 3.9 | 12 | 16 |
| GM118 | 166 | 54 | 26 | 9 | 10 | 4.7 | 14 | 21 |
| GM119 | 152 | 61 | 26 | 9 | 12.3 | 4.3 | 14 | 19 |
| GM120 | 180 | 60 | 38 | 13 | 14.6 | 4.4 | 14 | 17 |
| GM121 | 159 | 64 | 25 | 9 | 13.3 | 4.3 | 16 | 20 |
| GM122 | 206 | 63 | 23 | 15 | 14.3 | 3.5 | 14 | 14 |
| GM123 | 124 | 56 | 15 | 5 | 9.1 | 3.8 | 14 | 17 |
| GM124 | 127 | 44 | 26 | 7 | 12.1 | 3.9 | 12 | 15 |
| GM125 | 196 | 71 | 29 | 15 | 13.5 | 4.2 | 14 | 18 |
| GM126 | 149 | 43 | 27 | 6 | 10.2 | 3.8 | 10 | 15 |
| GM127 | 156 | 60 | 26 | 9 | 13.5 | 4.5 | 16 | 20 |
| GM128 | 179 | 78 | 32 | 9 | 12 | 4.1 | 14 | 24 |
| GM129 | 182 | 65 | 36 | 12 | 15.3 | 4.2 | 12 | 18 |
| GM130 | 145 | 48 | 16 | 7 | 11.3 | 3.5 | 14 | 22 |
| GM131 | 162 | 53 | 27 | 9 | 12 | 5.2 | 16 | 8 |
| GM132 | 138 | 60 | 20 | 13 | 11 | 3.5 | 12 | 16 |
| GM133 | 143 | 44 | 27 | 7 | 10.6 | 3.2 | 12 | 14 |
| GM134 | 178 | 73 | 31 | 9 | 13.3 | 4.2 | 12 | 26 |
| GM135 | 138 | 45 | 22 | 9 | 13.4 | 3.3 | 14 | 22 |
| GM136 | 199 | 67 | 29 | 12 | 14 | 4 | 14 | 16 |
| GM137 | 143 | 55 | 19 | 9 | 11 | 3.2 | 14 | 20 |
| GM138 | 166 | 65 | 29 | 5 | 12.6 | 3.4 | 12 | 19 |
| GM139 | 211 | 90 | 31 | 24 | 16.5 | 4.2 | 12 | 30 |
| GM140 | 188 | 100 | 27 | 10 | 12.7 | 4.5 | 18 | 23 |
| GM141 | 155 | 69 | 22 | 11 | 12.6 | 3.8 | 12 | 26 |
| GM142 | 177 | 70 | 24 | 10 | 11 | 4.1 | 12 | 31 |
| GM143 | 148 | 55 | 26 | 9 | 13.6 | 3.4 | 10 | 27 |
| GM144 | 195 | 79 | 36 | 13 | 18.5 | 3.9 | 14 | 31 |
| GM145 | 175 | 69 | 29 | 10 | 12.2 | 4.4 | 16 | 19 |
| GM146 | 173 | 62 | 18 | 9 | 11.5 | 4.7 | 14 | 24 |
| GM147 | 145 | 45 | 22 | 5 | 11.2 | 3.4 | 10 | 15 |
| GM148 | 190 | 79 | 31 | 14 | 15.7 | 3.9 | 14 | 26 |
| GM149 | 167 | 63 | 34 | 9 | 12 | 3.7 | 12 | 19 |
| GM150 | 126 | 46 | 16 | 9 | 12.2 | 3.5 | 14 | 16 |
| GM151 | 169 | 59 | 28 | 7 | 11.8 | 4.4 | 16 | 17 |
| GM152 | 171 | 68 | 22 | 12 | 14.6 | 4.6 | 14 | 26 |
| GM153 | 223 | 85 | 39 | 11 | 16.6 | 4.1 | 16 | 28 |
| GM154 | 121 | 40 | 26 | 8 | 11.3 | 3.9 | 14 | 15 |
| GM155 | 211 | 100 | 30 | 19 | 17.1 | 3.9 | 16 | 30 |
| GM156 | 160 | 58 | 29 | 6 | 13.5 | 4.2 | 16 | 19 |
| GM157 | 135 | 50 | 25 | 10 | 13.2 | 3.3 | 10 | 27 |
| GM158 | 213 | 95 | 32 | 8 | 12.9 | 4.9 | 14 | 20 |
| GM159 | 170 | 60 | 22 | 10 | 11.9 | 4.3 | 12 | 22 |
| GM160 | 198 | 83 | 30 | 11 | 16 | 4.3 | 18 | 28 |
| GM161 | 175 | 64 | 20 | 11 | 12.4 | 4.1 | 12 | 22 |
| GM162 | 193 | 92 | 39 | 17 | 17.6 | 5.2 | 14 | 32 |
| GM163 | 125 | 51 | 17 | 7 | 7.5 | 3.1 | 12 | 15 |
| GM164 | 205 | 79 | 25 | 12 | 12.9 | 4.7 | 18 | 20 |
| GM165 | 188 | 67 | 40 | 11 | 13.6 | 3.9 | 14 | 16 |
| GM166 | 177 | 70 | 26 | 11 | 13.8 | 4.3 | 16 | 22 |
| GM167 | 116 | 37 | 22 | 12 | 13.3 | 3.7 | 12 | 16 |
| GM168 | 166 | 56 | 24 | 5 | 11.3 | 4.9 | 14 | 18 |
| GM169 | 155 | 56 | 26 | 10 | 12.5 | 3.3 | 12 | 25 |
| GM170 | 148 | 59 | 23 | 8 | 15.1 | 3.6 | 12 | 24 |
| GM171 | 158 | 54 | 29 | 11 | 10.5 | 3.4 | 14 | 26 |
| GM172 | 174 | 69 | 22 | 13 | 14.3 | 4.2 | 16 | 26 |
| GM173 | 146 | 57 | 28 | 13 | 13.9 | 4.1 | 14 | 24 |
| GM174 | 220 | 87 | 36 | 15 | 16.9 | 4.7 | 12 | 34 |
| GM175 | 113 | 39 | 23 | 13 | 11.2 | 3.4 | 14 | 16 |
| GM176 | 234 | 90 | 39 | 15 | 17.2 | 4.6 | 14 | 30 |
| GM177 | 142 | 56 | 31 | 12 | 11.4 | 3.9 | 12 | 22 |
| GM178 | 133 | 61 | 20 | 10 | 9.6 | 3.8 | 14 | 16 |
| GM179 | 188 | 69 | 23 | 9 | 12.9 | 4.9 | 18 | 24 |
| GM180 | 117 | 33 | 26 | 12 | 13.5 | 3.7 | 14 | 15 |
| GM181 | 165 | 59 | 27 | 11 | 15.2 | 4.2 | 12 | 19 |
| GM182 | 200 | 84 | 34 | 13 | 17.1 | 4.2 | 16 | 26 |
| GM183 | 129 | 43 | 22 | 7 | 13.2 | 4 | 14 | 16 |
| GM184 | 206 | 92 | 31 | 5 | 12.2 | 4.7 | 16 | 20 |
| GM185 | 145 | 52 | 29 | 11 | 14 | 3.5 | 12 | 29 |
| GM186 | 222 | 88 | 39 | 15 | 18.2 | 4.5 | 14 | 32 |
| GM187 | 205 | 93 | 35 | 5 | 12.3 | 4.3 | 16 | 20 |
| GM188 | 168 | 65 | 35 | 9 | 11.1 | 3.1 | 16 | 19 |
| GM189 | 203 | 79 | 38 | 12 | 11.7 | 4.6 | 16 | 28 |
| GM190 | 161 | 60 | 32 | 5 | 12.4 | 3.4 | 14 | 19 |
| GM191 | 137 | 65 | 23 | 13 | 9.4 | 3.5 | 14 | 16 |
| GM192 | 176 | 67 | 23 | 12 | 14.3 | 4.3 | 12 | 24 |
| GM193 | 198 | 91 | 35 | 7 | 15.3 | 3.9 | 16 | 22 |
| GM194 | 151 | 60 | 23 | 9 | 11 | 3.8 | 14 | 16 |
| GM195 | 213 | 104 | 37 | 39 | 13.6 | 4.7 | 18 | 28 |
| GM196 | 169 | 67 | 28 | 6 | 10.2 | 3.1 | 12 | 18 |
| GM197 | 125 | 39 | 14 | 5 | 9.6 | 3.5 | 12 | 16 |
| GM198 | 152 | 58 | 30 | 9 | 12.9 | 4.1 | 12 | 22 |
| GM199 | 147 | 62 | 21 | 9 | 11 | 4.3 | 14 | 16 |
| GM200 | 193 | 85 | 31 | 8 | 14.5 | 4.2 | 18 | 26 |
References
- Li, Y.L.; Suo, H.C. Research progress on endosperm mutant genes and molecular breeding in waxy maize. Chin. Agric. Sci. Bull. 2019, 35, 21–27. [Google Scholar]
- Zhao, J.R.; Lu, B.S.; Shi, Y.X.; Xu, L. Current status of waxy maize breeding and industry development in China. J. Maize Sci. 2016, 24, 67–71. [Google Scholar]
- Song, Q.H.; Kong, L.L.; Liu, J.F.; Zhang, Y.; Yang, Y.H. Creation and identification techniques of sweet-waxy double recessive genotype maize materials. China Veg. 2018, 5, 28–32. [Google Scholar]
- Shi, Z.S.; Zhong, X.M. Principles and technical skills for breeding new waxy maize varieties. J. Maize Sci. 2016, 24, 1–5. [Google Scholar]
- Guo, J.T.; Zhang, L.; Li, Y.X.; Zhang, H.; Ping, X.M. Development status and countermeasures of fresh maize industry in Yunnan Province. Agric. Technol. 2024, 44, 177–180. [Google Scholar]
- Song, X.B. Quantitative Definition of Phenotypic Traits and Genetic Analysis of Chrysanthemum Cultivars; Beijing Forestry University: Beijing, China, 2018. [Google Scholar]
- Du, Z.Z.; Li, X.X.; Song, J.P.; Wu, Y.H.; Zhao, Q.; Xu, T.; Zhang, X.H.; Hellier, B.; Hu, J.G.; Wang, H.P. Phenotypic diversity analysis and preliminary evaluation of adaptability in 228 introduced garlic resources. J. Plant Genet. Resour. 2019, 20, 1186–1196. [Google Scholar]
- Hu, B.L.; Wan, Y.; Li, X.; Lei, J.G.; Luo, X.D.; Yan, W.G.; Xie, J.K. Genetic diversity analysis and comprehensive evaluation of phenotypic traits in rice core collections. Acta Agron. Sin. 2012, 38, 829–839. [Google Scholar] [CrossRef]
- Rêgo, E.R.; Rêgo, M.M.; Cruz, C.D.; Finger, F.L.; Casali, V.W.D. Phenotypic diversity, correlation and importance of variables for fruit quality and yield traits in Brazilian peppers (Capsicum baccatum). Genet. Resour. Crop Evol. 2011, 58, 909–918. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, L.; Li, C.H.; Zhang, D.Y.; Fu, J.; Cheng, Z.Q. Construction of SSR fingerprint database and genetic diversity analysis of rice resources in southern China. Mol. Plant Breed. 2020, 18, 6502–6517. [Google Scholar]
- Wu, R.G.; Pei, Y.T.; Zhang, C.; Fan, Y.Q.; Jin, Y.R.; Liu, P.; Jia, D.X.; Dai, Z.M. Diversity analysis and evaluation of wheat agronomic traits under salt stress. J. Triticeae Crops 2019, 39, 1029–1037. [Google Scholar]
- Liu, H.Z.; Song, W.; Wang, B.Q.; Wang, J.H.; Zhang, Q.G.; Zhang, D.M.; Li, X.H.; Wei, J.F.; Li, R.G. Genetic diversity analysis of 120 European and American maize inbred lines. J. Plant Genet. Resour. 2018, 19, 676–684. [Google Scholar]
- Chen, H.L.; Hu, L.L.; Yang, Y.; Wang, S.; Wang, L.X.; Cheng, X.Z. Evaluation of agronomic traits and bruchid resistance in 481 mungbean germplasms. J. Plant Genet. Resour. 2020, 21, 549–559. [Google Scholar]
- Wang, L.X.; Cheng, X.Z.; Wang, S.H.; Zhu, X.; Liu, Z.X. Phenotypic variation and ecological adaptability evaluation of Chinese mungbean core collection under different environments. Acta Agron. Sin. 2014, 40, 739–744. [Google Scholar]
- Bojinov, B.M.; Danailov, Z.P. Applicability of ISSRs for Genotype Identification in a Tomato Breeding Collection. Acta Hortic. 2009, 830, 63–70. [Google Scholar] [CrossRef]
- Shu, G.; Cao, G.; Li, N.; Li, J.; Zhao, Y.; Liu, C.; Wang, R.; Chen, C.; Zhang, Y.; Tian, H.; et al. Genetic variation and population structure in China summer maize germplasm. Sci. Rep. 2021, 11, 8012. [Google Scholar] [CrossRef]
- Tuo, M.Q.; Gao, X.; Gao, X.F.; Li, X.; Wang, J.; Zhang, Y.; Li, J.H. Current status and perspectives of maize germplasm research in China. Xinjiang Agric. Reclam. Sci. Technol. 2021, 44, 68–69. [Google Scholar]
- Liu, J.W.; Zhang, R.P.; Shi, D.K.; Zheng, Q.D.; Li, F.J.; Wei, F.; Wei, X.Y.; Ma, J.F.; Wang, J.M.; Ma, Y.; et al. Genetic diversity analysis and heterotic grouping of 43 maize inbred lines. Seed 2024, 43, 100–104. [Google Scholar]
- Smith, J.S.C.; Chin, E.C.L.; Shu, H.; Smith, O.S.; Wall, S.J.; Senior, M.L.; Mitchell, S.E.; Kresovich, S.; Ziegle, J. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L): Comparisons with data from RFLPs and pedigree. Theor. Appl. Genet. 1997, 95, 163–173. [Google Scholar] [CrossRef]
- Sakthipriya, M.; Vishnu, S.S.; Sujith, S.; Jelly, C.; Anu, S.; Nair, A.S. Analysis of genetic diversity of Centella asiatica using SSR markers. Int. J. Appl. Sci. Biotechnol. 2018, 6, 103–109. [Google Scholar] [CrossRef]
- Zhao, X.H.; Wang, C.B.; Dong, Y.H.; Li, Y.F. Research progress of SSR molecular markers in maize breeding. Mod. Agric. Sci. Technol. 2016, 7, 15–16. [Google Scholar]
- Wang, Y.M.; Yin, D.P.; Zhang, C.X.; Li, M.S.; Zhao, J.R.; Lu, Y.Q.; Wang, Y.D.; Wang, C.Y. Genetic diversity analysis of 35 waxy maize germplasms using SSR markers. J. Maize Sci. 2014, 22, 27–31. [Google Scholar]
- Li, R.; Bai, J.R.; Wang, X.H.; Zhang, C.Z.; Zhang, X.M.; Yan, L.; Yang, R.J. Genetic diversity analysis of 144 sweet maize populations. Crops 2018, 2, 17–24. [Google Scholar]
- Huang, J.; Feng, F.Q.; Wang, Q.F.; Zhang, J.; Zhou, P.; Li, X.Q. SSR-based genetic diversity analysis of 54 sweet maize inbred lines. J. South China Agric. Univ. 2012, 33, 1–4. [Google Scholar]
- Lu, Y.; Ai, W.D.; Han, Q.; Wang, Y.F.; Li, H.Y.; Qu, Y.J.; Shi, B.; Shen, X.F. Genetic diversity and population structure analysis of waxy maize inbred lines using SSR markers. Acta Agron. Sin. 2019, 45, 214–224. [Google Scholar] [CrossRef]
- Zhao, Y. Genetic Diversity Analysis and Heterosis Study of Waxy Maize Inbred Lines Using SSR Markers; Jilin Agricultural University: Changchun, China, 2022. [Google Scholar]
- Tan, C.R.; Lei, Y.H.; Chen, D.G.; Wang, Y.B.; Li, J.H.; Wang, C.L. Genetic diversity analysis of agronomic traits and elite combination screening in 61 maize hybrids. China Seed Ind. 2025, 3, 105–110. [Google Scholar]
- Marwa, G.; Ruslan, K.; Hoda, B.; Abdelhady, A.A.M.; Eltobgy, D.S.; El-Malky, M.M.; Nagaty, M.A.; Saker, M.M.; Al-Doss, A.A.; Ibrahim, S.D. High-throughput retrotransposon-based genetic diversity of maize germplasm assessment and analysis. Mol. Biol. Rep. 2020, 47, 1589–1603. [Google Scholar]
- Qiu, L.W.; Yang, Y.; Fan, Y.M.; Tian, H.L.; Yi, H.M.; Wang, L.; Ren, J.; Ge, J.R.; Wang, F.G.; Lu, D.L. Analysis of quality, agronomic traits and SSR-based genetic diversity of waxy maize in southeast China. Jiangsu Agric. Sci. 2022, 50, 130–135. [Google Scholar]
- Wang, F.G.; Tian, H.L.; Zhao, J.R.; Wang, L.; Yi, H.M.; Song, W.; Gao, Y.Q.; Yang, G.H. Genetic diversity analysis of 328 maize varieties using SSR markers. Sci. Agric. Sin. 2014, 47, 856–864. [Google Scholar]
- Kumari, P.; Chauhan, P.; Kumar, P.; Ravika. Genetic diversity and resistance assessment among maize genotypes against banded leaf and sheath blight (caused by Rhizoctonia solani f. sp. sasakii) utilizing SSR markers. J. Phytopathol. 2023, 171, 642–655. [Google Scholar] [CrossRef]
- Mukesh, C.; Alla, S.; Das, M.M.; Pardeep, K.; Ritu, N.; Vishal, S.; Bhupender, K.; Sujay, R. Morpho-physiological traits and SSR markers-based analysis of relationships and genetic diversity among fodder maize landraces in India. Mol. Biol. Rep. 2023, 50, 6829–6841. [Google Scholar]
- Shete, S.G.; Nikam, D.S.; Surbhaiyya, D.S.; Shinde, D.A.; Patil, S.S.; Khandagale, S.G. Analysis of Genetic Diversity in Maize (Zea mays L.) Variety Using SSR Markers. Int. J. Plant Soil Sci. 2023, 35, 205–212. [Google Scholar] [CrossRef]
- Nikola, G.; Nenad, D.; Milan, S.; Milan, S.; Jovan, P.; Milos, C.; Marko, M.; Nemanja, N. Genetic distance of maize inbred lines based on SSR markers for prediction of heterosis and combining ability. Genetika 2018, 50, 359–368. [Google Scholar] [CrossRef]
- Jiang, Y.F.; Yin, F.X.; Li, W.Z.; Xing, F.; Li, Z.W.; Guo, R.J.; Wang, J.; Fan, J.; Zhang, Y.D.; Fan, X.M.; et al. Predicting heterosis via genetic distance and the number of SNPs in selected segments of chromosomes in maize. Front. Plant Sci. 2023, 14, 1111961. [Google Scholar] [CrossRef]
- Cui, Q.X.; Tang, B.J.; Chen, H.Q.; Liu, Y.X. Genetic relationship between high-oil maize and normal maize inbred lines revealed by SSR markers. J. Maize Sci. 2011, 19, 14–18. [Google Scholar]
- Gao, S.; Liu, H.W.; He, H.; Lü, Q.X.; Zhou, D.L.; Xing, Y.X.; Zhao, X.Y.; Zhang, Z.J. Genetic diversity and population structure analysis using SNP chips and new variety breeding in maize. J. Maize Sci. 2021, 29, 39–45. [Google Scholar]
| Traits | Minimum Value | Maximum Value | Mean Value |
|---|---|---|---|
| Plant height (cm) | 104 | 236 | 172 |
| Ear height (cm) | 28 | 114 | 67 |
| Tassel length (cm) | 8 | 46 | 28 |
| Tassel branch number | 3 | 22 | 11 |
| Ear length (cm) | 8.1 | 22.4 | 13.3 |
| Ear diameter (cm) | 2.6 | 6.4 | 4 |
| Kernel row number | 8 | 20 | 14 |
| Kernel number per row | 6 | 36 | 21 |
| Forms | Quantities | Serial Number |
|---|---|---|
| Cluster I | 25 | GM003, GM127, GM121, GM166, GM145, GM065, GM087, GM156, GM151, GM119 |
| GM012, GM022, GM048, GM002, GM100 | ||
| GM062, GM035, GM105, GM118, GM103, GM014, GM168, GM110, GM097, GM131 | ||
| Cluster II | 38 | GM005, GM191, GM104, GM178, GM132, GM199, GM194, GM040, GM137, GM130, GM150, GM135, GM038, GM039, GM163 |
| GM021, GM197, GM123, GM006, GM124, GM095, GM115, GM013, GM052, GM183, GM044, GM154, GM180, GM093, GM175, GM167, GM117, GM096, GM147, GM126, GM133, GM112, GM024 | ||
| Cluster III | 97 | GM004, GM187, GM184, GM016, GM158, GM140, GM091, GM153, GM051, GM182 |
| GM200, GM160, GM193, GM060, GM086 | ||
| GM050, GM078, GM069, GM059, GM165, GM098, GM071, GM129, GM076, GM089 | ||
| GM125, GM043, GM136, GM056, GM088, | ||
| GM007, GM042, GM084, GM063, GM161, GM192, GM141, GM142, GM128, GM054, GM047, GM067, GM173, GM020, GM198 | ||
| GM177, GM181, GM170, GM106, GM079, | ||
| GM169, GM116, GM157, GM143, GM066, GM171, GM046, GM027, GM138, GM196 | ||
| GM113, GM190, GM111, GM108, GM070 | ||
| GM149, GM102, GM072, GM008, GM068 | ||
| GM073, GM034, GM188, GM011, GM001, GM122, GM009, GM036, GM061, GM152, GM075, GM146, GM172, GM179, GM164 | ||
| GM189, GM120, GM015, GM159, GM134, GM185, GM109, GM053, GM041, GM037, GM058, GM083 | ||
| Cluster IV | 40 | GM025, GM186, GM176, GM174, GM162, GM139, GM045, GM017, GM057, GM107, GM031, GM101, GM155, GM090, GM092 |
| GM055, GM064, GM195, GM081, GM029, GM019, GM099, GM080, GM023, GM148, GM094, GM030, GM049, GM032, GM082 | ||
| GM077, GM144, GM085, GM026, GM018 | ||
| GM028, GM114, GM074, GM033, GM010 |
| Number | GM-019 | GM-020 | GM-021 | GM-022 | GM-023 |
|---|---|---|---|---|---|
| GM-010 | 0.454 | ||||
| GM-011 | 0.561 | 0.471 | |||
| GM-012 | 0.522 | 0.502 | 0.411 | ||
| GM-013 | 0.509 | 0.571 | 0.308 | 0.702 | |
| GM-014 | 0.421 | 0.405 | 0.476 | 0.559 | 0.629 |
| Number | GM-182 | GM-183 | GM-184 | GM-185 | GM-186 |
|---|---|---|---|---|---|
| GM-174 | 0.475 | 0.661 | 0.713 | 0.622 | 0.578 |
| GM-175 | 0.562 | 0.572 | 0.477 | 0.532 | 0.607 |
| GM-176 | 0.595 | 0.829 | 0.902 | 0.658 | 0.798 |
| GM-177 | 0.696 | 0.611 | 0.489 | 0.797 | 0.574 |
| GM-178 | 0.498 | 0.597 | 0.632 | 0.512 | 0.482 |
| Number | Primer Name | Chromosomal Location | Number of Polymorphic Bands | Effective Number of Alleles | Gene Diversity | Polymorphism Information Content |
|---|---|---|---|---|---|---|
| 1 | bnlg439g1 | 1.03 | 8 | 6.25 | 0.84 | 0.73 |
| 2 | umc1335g2 | 1.06 | 4 | 2.85 | 0.63 | 0.31 |
| 3 | umc1147g3 | 1.07 | 7 | 5.01 | 0.82 | 0.68 |
| 4 | bnlg1671g4 | 1.1 | 6 | 3.99 | 0.74 | 0.61 |
| 5 | phi96100g3 | 2.01 | 7 | 4.89 | 0.79 | 0.72 |
| 6 | umc2007g2 | 2.04 | 5 | 4.03 | 0.62 | 0.58 |
| 7 | umc1536g4 | 2.07 | 3 | 3.19 | 0.75 | 0.69 |
| 8 | bnlg1940g1 | 2.08 | 8 | 6.99 | 0.88 | 0.79 |
| 9 | bnlg1520g4 | 2.09 | 4 | 4.72 | 0.81 | 0.73 |
| 10 | umc2105g1 | 3 | 9 | 7.02 | 0.86 | 0.82 |
| 11 | phi053g2 | 3.05 | 7 | 5.18 | 0.77 | 0.7 |
| 12 | umc1489g3 | 3.07 | 6 | 3.98 | 0.78 | 0.76 |
| 13 | phi072g1 | 4 | 4 | 3.76 | 0.71 | 0.64 |
| 14 | bnlg490g3 | 4.04 | 5 | 3.99 | 0.81 | 0.74 |
| 15 | bnlg2291g2 | 4.07 | 4 | 3.04 | 0.67 | 0.56 |
| 16 | umc1999g3 | 4.09 | 7 | 5.94 | 0.77 | 0.74 |
| 17 | umc2115g4 | 5.02 | 5 | 5.01 | 0.75 | 0.71 |
| 18 | umc1705g2 | 5.03 | 7 | 6.96 | 0.82 | 0.79 |
| 19 | umc1429g3 | 5.03 | 8 | 7.92 | 0.84 | 0.81 |
| 20 | bnlg2305g1 | 5.07 | 9 | 6.32 | 0.86 | 0.83 |
| 21 | bnlg161g2 | 6 | 5 | 4.99 | 0.78 | 0.75 |
| 22 | bnlg249g4 | 6.01 | 7 | 6.38 | 0.84 | 0.8 |
| 23 | bnlg1702g1 | 6.05 | 6 | 5.63 | 0.78 | 0.73 |
| 24 | phi299852g3 | 6.07 | 5 | 4.97 | 0.76 | 0.72 |
| 25 | umc1545g1 | 7 | 6 | 5.73 | 0.75 | 0.71 |
| 26 | umc2160g4 | 7.01 | 5 | 3.95 | 0.75 | 0.71 |
| 27 | umc1936g4 | 7.03 | 6 | 5.28 | 0.76 | 0.74 |
| 28 | umc1125g2 | 7.04 | 5 | 4.76 | 0.77 | 0.74 |
| 29 | bnlg2235g4 | 8.02 | 7 | 6.46 | 0.81 | 0.79 |
| 30 | bnlg240g1 | 8.06 | 8 | 7.51 | 0.83 | 0.81 |
| 31 | phi080g2 | 8.08 | 6 | 5.39 | 0.79 | 0.76 |
| 32 | phi233376g3 | 8.09 | 5 | 4.65 | 0.74 | 0.72 |
| 33 | umc2084g3 | 9.01 | 6 | 5.51 | 0.78 | 0.75 |
| 34 | phi065g1 | 9.03 | 4 | 3.74 | 0.73 | 0.7 |
| 35 | umc1492g2 | 9.04 | 3 | 1.75 | 0.42 | 0.3 |
| 36 | umc1231g4 | 9.05 | 6 | 4.86 | 0.78 | 0.73 |
| 37 | phi041g4 | 10 | 2 | 1.71 | 0.47 | 0.42 |
| 38 | umc1432g1 | 10.02 | 3 | 2.21 | 0.62 | 0.58 |
| 39 | umc2163g3 | 10.04 | 5 | 4.68 | 0.76 | 0.71 |
| 40 | umc1506g2 | 10.05 | 7 | 6.01 | 0.82 | 0.79 |
| Forms | Quantities | Serial Number |
|---|---|---|
| Cluster I | 53 | GM001, GM003, GM005, GM017, GM023, GM024, GM030, GM036, GM044, GM050, GM052, GM053, GM055, GM058, GM066, GM072, GM075, GM080, GM085, GM086, GM087, GM088, GM090, GM091, GM095, GM097, GM099, GM100, GM106, GM109, GM111, GM112, GM115, GM122, GM125, GM130, GM133, GM140, GM143, GM147, GM148, GM152, GM153, GM161, GM165, GM168, GM175, GM177, GM178, GM181, GM188, GM190, GM192 |
| Cluster II | 102 | GM002, GM004, GM006, GM008, GM009, GM011, GM013, GM014, GM015, GM016, GM018, GM019, GM020, GM021, GM022, GM026, GM027, GM028, GM029, GM031, GM033, GM034, GM035, GM038, GM039, GM041, GM042, GM046, GM047, GM049, GM054, GM056, GM057, GM060, GM061, GM062, GM063, GM065, GM067, GM069, GM071, GM074, GM077, GM078, GM079, GM082, GM083, GM084, GM094, GM096, GM098, GM102, GM103, GM104, GM110, GM113, GM117, GM118, GM119, GM120, GM121, GM123, GM124, GM126, GM127, GM128, GM131, GM134, GM135, GM136, GM137, GM138, GM144, GM145, GM146, GM151, GM156, GM159, GM162, GM163, GM164, GM166, GM167, GM169, GM172, GM176, GM179, GM180, GM182, GM183, GM185, GM187, GM189, GM191, GM193, GM194, GM195, GM196, GM197, GM198, GM199, GM200 |
| Cluster III | 28 | GM007, GM010, GM012, GM025, GM032, GM037, GM040, GM045, GM048, GM051, GM059, GM068, GM070, GM073, GM076, GM089, GM092, GM093, GM101, GM105, GM107, GM108, GM114, GM116, GM132, GM149, GM150, GM160 |
| Cluster IV | 17 | GM043, GM064, GM081, GM129, GM139, GM141, GM142, GM154, GM155, GM157, GM158, GM170, GM171, GM173, GM174, GM184, GM186 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Zheng, X.; Wang, S.; Ai, Y.; Zhao, R.; Lan, J. Genetic Diversity of Fresh Maize Germplasm Revealed by Morphological Traits and SSR Markers. Genes 2025, 16, 1138. https://doi.org/10.3390/genes16101138
Guo S, Zheng X, Wang S, Ai Y, Zhao R, Lan J. Genetic Diversity of Fresh Maize Germplasm Revealed by Morphological Traits and SSR Markers. Genes. 2025; 16(10):1138. https://doi.org/10.3390/genes16101138
Chicago/Turabian StyleGuo, Suying, Xin Zheng, Shuaiyi Wang, Yuran Ai, Rengui Zhao, and Jinhao Lan. 2025. "Genetic Diversity of Fresh Maize Germplasm Revealed by Morphological Traits and SSR Markers" Genes 16, no. 10: 1138. https://doi.org/10.3390/genes16101138
APA StyleGuo, S., Zheng, X., Wang, S., Ai, Y., Zhao, R., & Lan, J. (2025). Genetic Diversity of Fresh Maize Germplasm Revealed by Morphological Traits and SSR Markers. Genes, 16(10), 1138. https://doi.org/10.3390/genes16101138





