Integrative Multi-Omics Characterization and Structural Insights into the Poorly Annotated Integrin ITGA6 X1X2 Isoform in Mammals
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval, Alignment, and Phylogenetic Analysis
2.2. Genome Annotation Assessment and Splicing Prediction
2.3. Expression, Variant, and Mutation Analysis
2.4. Structural Modeling and Contact Analysis
3. Results
3.1. Annotation Quality of α Integrins Across Mammals
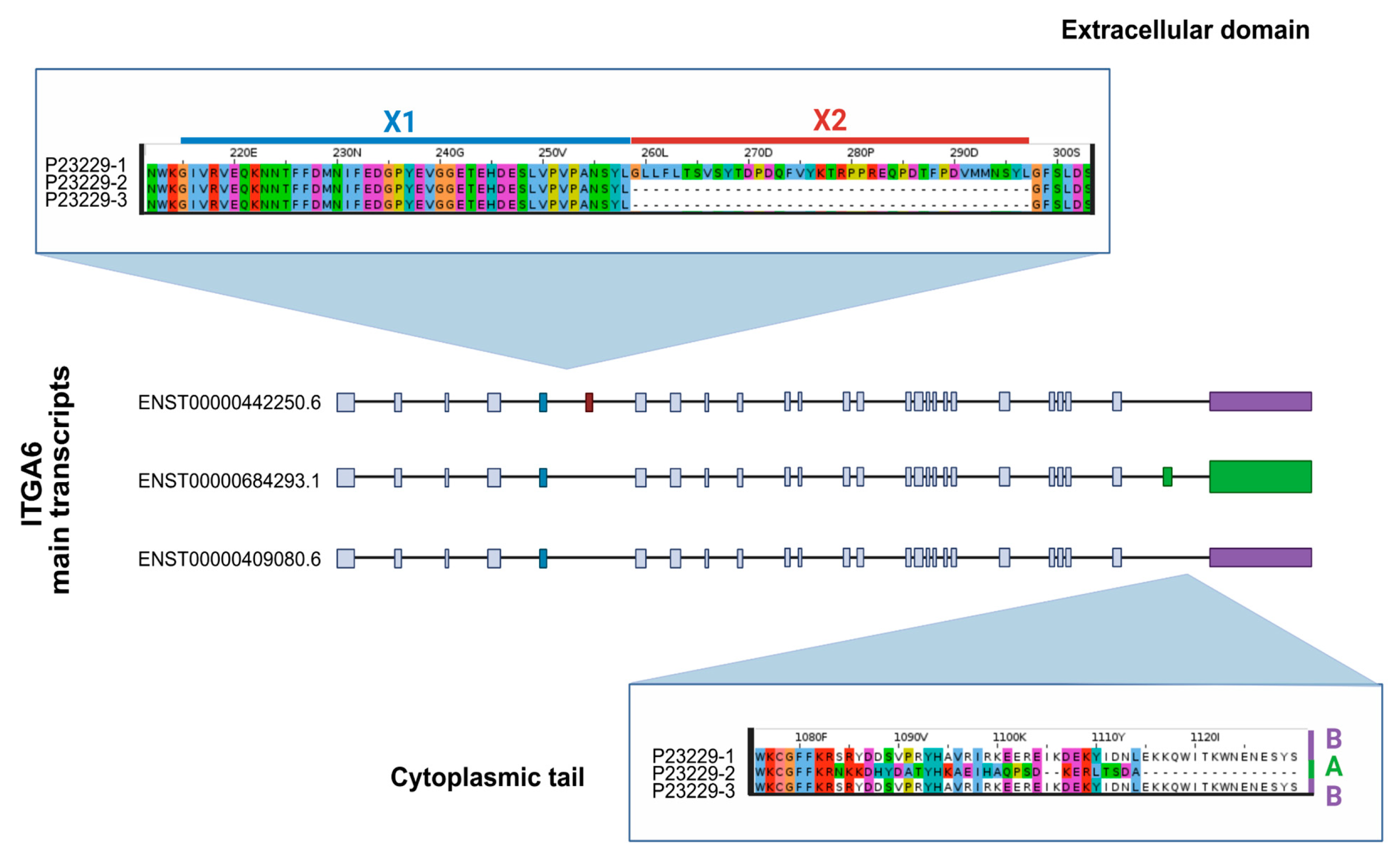
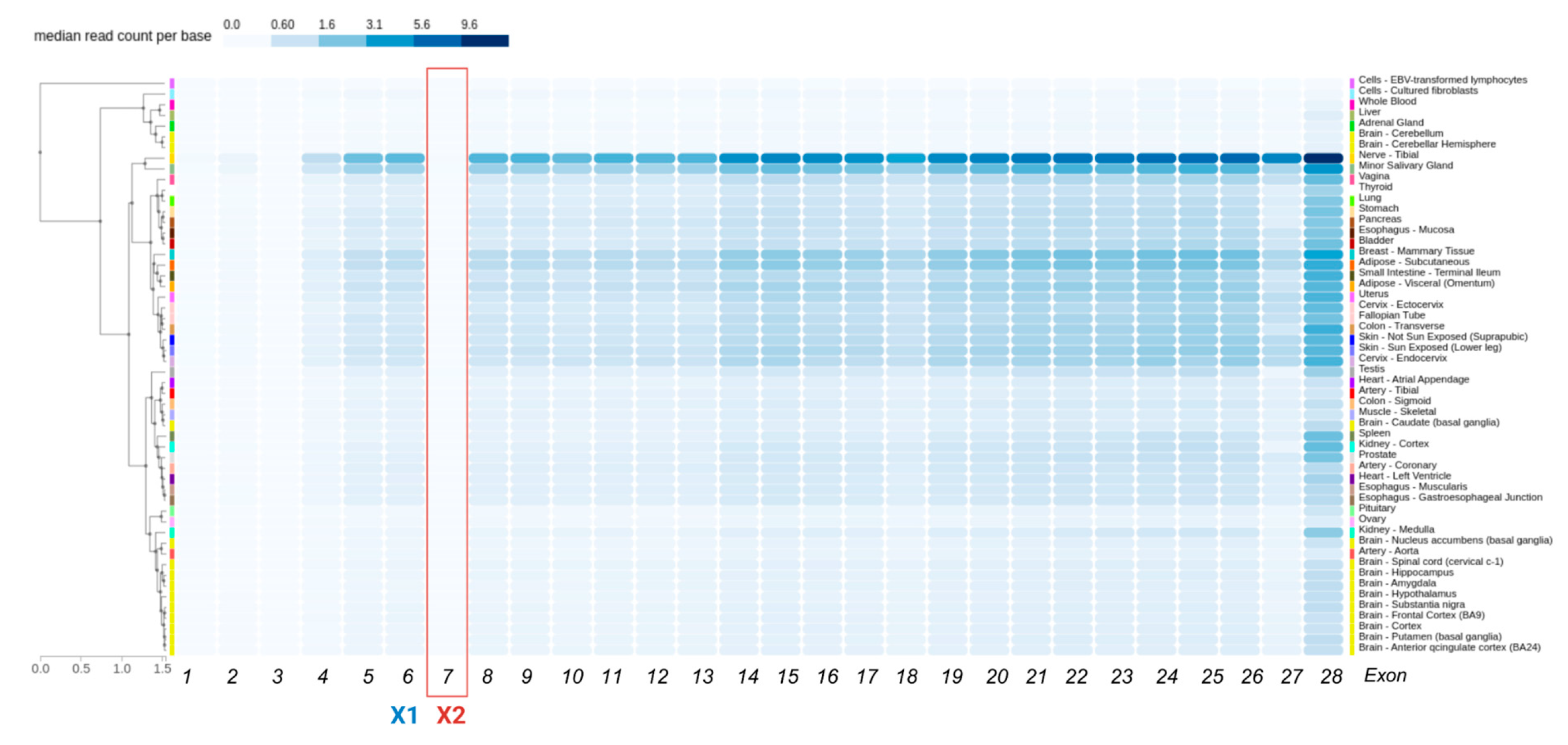
3.2. Presence of the ITGA6X1X2 Isoform Across Species
| Experiment | Read Name | Reference Span |
|---|---|---|
| ERX2613198 | ERR2596913.20999958 | NC_041765.1:59,585,017–59,585,117 |
| ERX2613229 | ERR2596944.16351931 | NC_041765.1:59,585,019–59,585,119 |
| ERX2613087 | ERR2596802.8153246 | NC_041765.1:59,585,062–59,585,162 |
| ERX2613214 | ERR2596929.22893645 | NC_041765.1:59,585,134–59,585,234 |
| ERX2613201 | ERR2596916.15064411 | NC_041765.1:59,585,177–59,585,277 |
| ERX2613119 | ERR2596834.21730323 | NC_041765.1:59,585,181–59,585,281 |
| ERX2613236 | ERR2596951.7152511 | NC_041765.1:59,583,397–59,585,162 |
| ERX2613177 | ERR2596892.11271980 | NC_041765.1:59,585,192–59,585,290 |
| ERX2613199 | ERR2596914.34156819 | NC_041765.1:59,585,063–59,585,161 |
| ERX2613116 | ERR2596831.72595928 | NC_041765.1:59,585,068–59,585,167 |
| ERX2613191 | ERR2596906.34031824 | NC_041765.1:59,585,111–59,585,210 |
| ERX2613229 | ERR2596944.50503340 | NC_041765.1:59,585,113–59,585,213 |
| ERX2613153 | ERR2596868.17044807 | NC_041765.1:59,585,069–59,585,169 |
| ERX2613217 | ERR2596932.28822930 | NC_041765.1:59,585,074–59,585,174 |
| ERX2613171 | ERR2596886.27577682 | NC_041765.1:59,585,077–59,585,176 |
| ERX2613159 | ERR2596874.8771744 | NC_041765.1:59,583,353–59,585,118 |
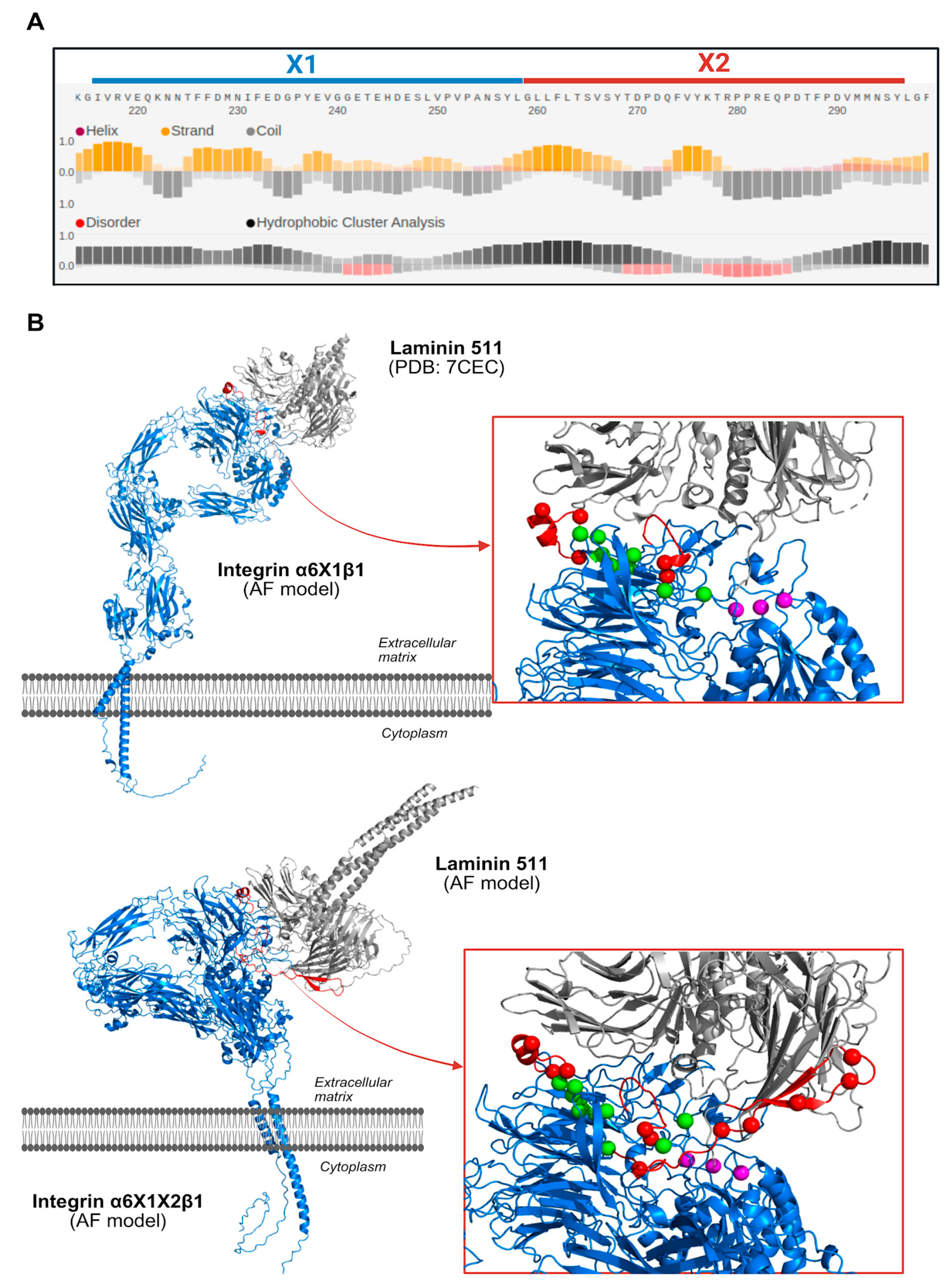
3.3. Splicing Signal Analysis for Exons X1 and X2
3.4. Transcript and Protein Evidence in Humans
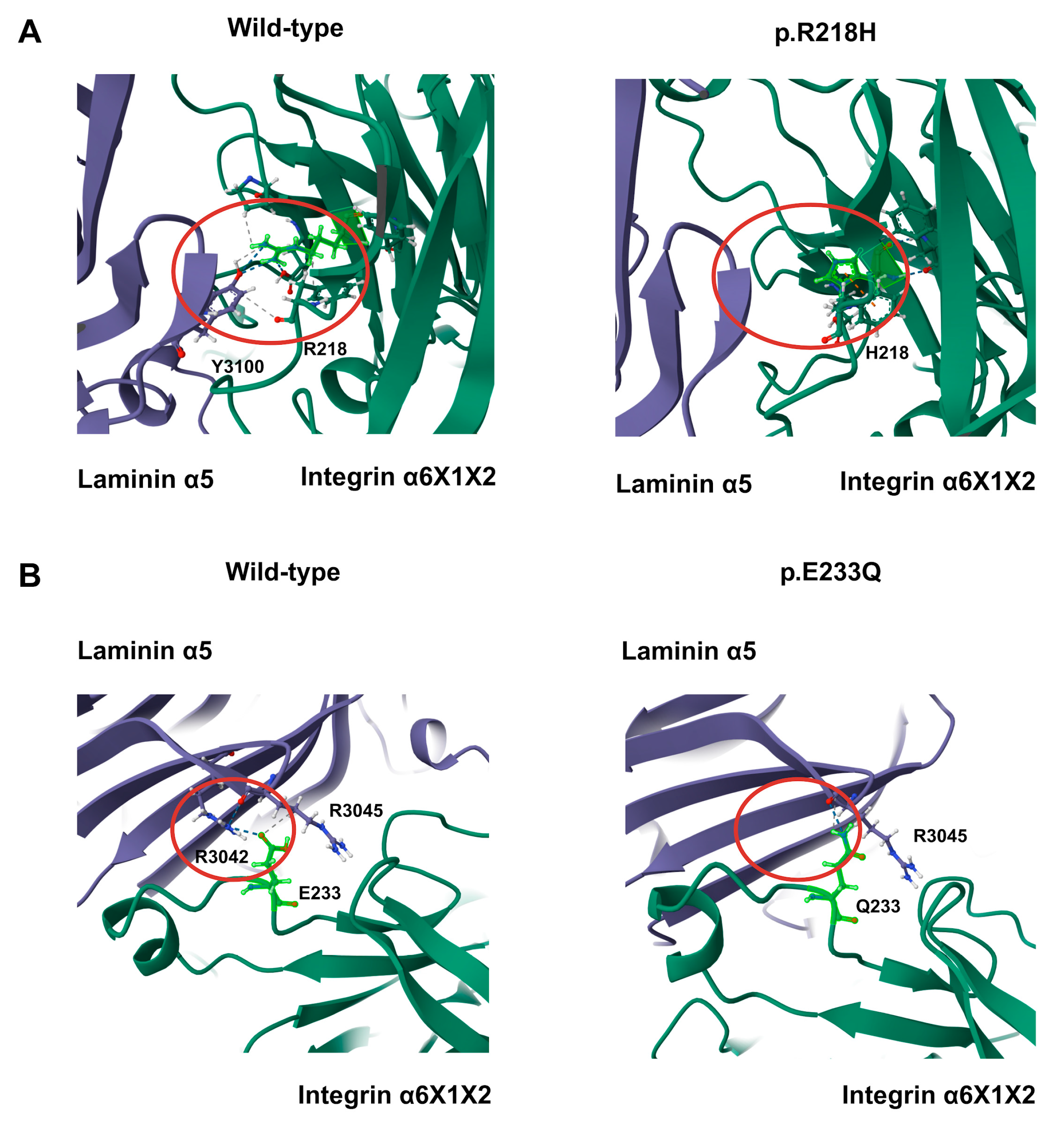
3.5. Variant Distribution and Mutations in Human Exons X1 and X2
3.6. Structural Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The Sequence of the Human Genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten Years of Next-Generation Sequencing Technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database Resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2025, 53, D20–D29. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.; Carbonell-Sala, S.; De La Vega, F.M.; Faial, T.; Frankish, A.; Gingeras, T.; Guigo, R.; Harrow, J.L.; Hatzigeorgiou, A.G.; Johnson, R.; et al. The Status of the Human Gene Catalogue. Nature 2023, 622, 41–47. [Google Scholar] [CrossRef]
- Deutekom, E.S.; Vosseberg, J.; van Dam, T.J.P.; Snel, B. Measuring the Impact of Gene Prediction on Gene Loss Estimates in Eukaryotes by Quantifying Falsely Inferred Absences. PLoS Comput. Biol. 2019, 15, e1007301. [Google Scholar] [CrossRef]
- Mudge, J.M.; Harrow, J. The State of Play in Higher Eukaryote Gene Annotation. Nat. Rev. Genet. 2016, 17, 758–772. [Google Scholar] [CrossRef]
- Marasco, L.E.; Kornblihtt, A.R. The Physiology of Alternative Splicing. Nat. Rev. Mol. Cell Biol. 2023, 24, 242–254. [Google Scholar] [CrossRef]
- Stastna, M.; Van Eyk, J.E. Analysis of Protein Isoforms: Can We Do It Better? Proteomics 2012, 12, 2937–2948. [Google Scholar] [CrossRef]
- Sulakhe, D.; D’Souza, M.; Wang, S.; Balasubramanian, S.; Athri, P.; Xie, B.; Canzar, S.; Agam, G.; Gilliam, T.C.; Maltsev, N. Exploring the Functional Impact of Alternative Splicing on Human Protein Isoforms Using Available Annotation Sources. Brief. Bioinform. 2019, 20, 1754–1768. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, Allosteric Signaling Machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Johnson, M.S.; Lu, N.; Denessiouk, K.; Heino, J.; Gullberg, D. Integrins during Evolution: Evolutionary Trees and Model Organisms. Biochim. Biophys. Acta BBA-Biomembr. 2009, 1788, 779–789. [Google Scholar] [CrossRef]
- Nishiuchi, R.; Takagi, J.; Hayashi, M.; Ido, H.; Yagi, Y.; Sanzen, N.; Tsuji, T.; Yamada, M.; Sekiguchi, K. Ligand-Binding Specificities of Laminin-Binding Integrins: A Comprehensive Survey of Laminin–Integrin Interactions Using Recombinant A3β1, A6β1, A7β1 and A6β4 Integrins. Matrix Biol. 2006, 25, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Sekiguchi, K. Chapter Six-Molecular Basis of Laminin–Integrin Interactions. In Current Topics in Membranes; Miner, J.H., Ed.; Basement Membranes; Academic Press Inc.: San Diego, CA, USA, 2015; Volume 76, pp. 197–229. [Google Scholar]
- Li, S.; Qi, Y.; McKee, K.; Liu, J.; Hsu, J.; Yurchenco, P.D. Integrin and Dystroglycan Compensate Each Other to Mediate Laminin-Dependent Basement Membrane Assembly and Epiblast Polarization. Matrix Biol. 2017, 57–58, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Manninen, A. Epithelial Polarity—Generating and Integrating Signals from the ECM with Integrins. Exp. Cell Res. 2015, 334, 337–349. [Google Scholar] [CrossRef]
- Xu, H.; LaFlamme, S.E. Contribution of Endothelial Laminin-Binding Integrins to Cellular Processes Associated with Angiogenesis. Cells 2022, 11, 816. [Google Scholar] [CrossRef] [PubMed]
- Yazlovitskaya, E.M.; Viquez, O.M.; Tu, T.; De Arcangelis, A.; Georges-Labouesse, E.; Sonnenberg, A.; Pozzi, A.; Zent, R. The Laminin Binding A3 and A6 Integrins Cooperate to Promote Epithelial Cell Adhesion and Growth. Matrix Biol. 2019, 77, 101–116. [Google Scholar] [CrossRef]
- Masunaga, T.; Ogawa, J.; Akiyama, M.; Nishikawa, T.; Shimizu, H.; Ishiko, A. Compound Heterozygosity for Novel Splice Site Mutations of ITGA6 in Lethal Junctional Epidermolysis Bullosa with Pyloric Atresia. J. Dermatol. 2017, 44, 160–166. [Google Scholar] [CrossRef]
- Xia, W.; Ni, Z.; Zhang, Z.; Sang, H.; Liu, H.; Chen, Z.; Jiang, L.; Yin, C.; Huang, J.; Li, L.; et al. Case Report: A Boy From a Consanguineous Family Diagnosed With Congenital Muscular Dystrophy Caused by Integrin Alpha 7 (ITGA7) Mutation. Front. Genet. 2021, 12, 706823. [Google Scholar] [CrossRef]
- Ramovs, V.; te Molder, L.; Sonnenberg, A. The Opposing Roles of Laminin-Binding Integrins in Cancer. Matrix Biol. 2017, 57–58, 213–243. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.L.; O’Connor, K.L. Clinical Significance of the Integrin α6β4 in Human Malignancies. Lab. Investig. 2015, 95, 976–986. [Google Scholar] [CrossRef]
- Raab-Westphal, S.; Marshall, J.F.; Goodman, S.L. Integrins as Therapeutic Targets: Successes and Cancers. Cancers 2017, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- de Melker, A.A.; Sonnenberg, A. Integrins: Alternative Splicing as a Mechanism to Regulate Ligand Binding and Integrin Signaling Events. BioEssays 1999, 21, 499–509. [Google Scholar] [CrossRef]
- de Melker, A.A.; Sterk, L.M.; Delwel, G.O.; Fles, D.L.; Daams, H.; Weening, J.J.; Sonnenberg, A. The A and B Variants of the Alpha 3 Integrin Subunit: Tissue Distribution and Functional Characterization. Lab. Investig. A J. Tech. Methods Pathol. 1997, 76, 547–563. [Google Scholar]
- Delwel, G.O.; Kuikman, I.; Sonnenberg, A. An Alternatively Spliced Exon in the Extracellular Domain of the Human A6 Integrin Subunit-Functional Analysis of the A6 Integrin Variants. Cell Adhes. Commun. 1995, 3, 143–161. [Google Scholar] [CrossRef]
- Hogervorst, F.; Admiraal, L.G.; Niessen, C.; Kuikman, I.; Janssen, H.; Daams, H.; Sonnenberg, A. Biochemical Characterization and Tissue Distribution of the A and B Variants of the Integrin Alpha 6 Subunit. J. Cell Biol. 1993, 121, 179–191. [Google Scholar] [CrossRef]
- Ziober, B.L.; Vu, M.P.; Waleh, N.; Crawford, J.; Lin, C.S.; Kramer, R.H. Alternative Extracellular and Cytoplasmic Domains of the Integrin Alpha 7 Subunit Are Differentially Expressed during Development. J. Biol. Chem. 1993, 268, 26773–26783. [Google Scholar] [CrossRef]
- Cattelino, A.; Longhi, R.; Curtis, I. de Differential Distribution of Two Cytoplasmic Variants of the A6β1 Integrin Laminin Receptor in the Ventral Plasma Membrane of Embryonic Fibroblasts. J. Cell Sci. 1995, 108, 3067–3078. [Google Scholar] [CrossRef]
- Thorsteinsdóttir, S.; Roelen, B.A.J.; Freund, E.; Gaspar, A.C.; Sonnenberg, A.; Mummery, C.L. Expression Patterns of Laminin Receptor Splice Variants α6Aβ1 and α6Bβ1 Suggest Different Roles in Mouse Development. Dev. Dyn. 1995, 204, 240–258. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, T.; Kodali, V.K.; Pujar, S.; Brover, V.; Robbertse, B.; Farrell, C.M.; Oh, D.-H.; Astashyn, A.; Ermolaeva, O.; Haddad, D.; et al. NCBI RefSeq: Reference Sequence Standards through 25 Years of Curation and Annotation. Nucleic Acids Res. 2025, 53, D243–D257. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Gremme, G.; Brendel, V.; Sparks, M.E.; Kurtz, S. Engineering a Software Tool for Gene Structure Prediction in Higher Organisms. Inf. Softw. Technol. 2005, 47, 965–978. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an Accurate RNA-Seq Aligner for Long and Short Reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Desmet, F.-O.; Hamroun, D.; Lalande, M.; Collod-Béroud, G.; Claustres, M.; Béroud, C. Human Splicing Finder: An Online Bioinformatics Tool to Predict Splicing Signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef]
- Zuallaert, J.; Godin, F.; Kim, M.; Soete, A.; Saeys, Y.; De Neve, W. SpliceRover: Interpretable Convolutional Neural Networks for Improved Splice Site Prediction. Bioinformatics 2018, 34, 4180–4188. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Lautenbacher, L.; Samaras, P.; Muller, J.; Grafberger, A.; Shraideh, M.; Rank, J.; Fuchs, S.T.; Schmidt, T.K.; The, M.; Dallago, C.; et al. Proteomics DB: Toward a FAIR Open-Source Resource for Life-Science Research. Nucleic Acids Res. 2022, 50, D1541–D1552. [Google Scholar] [CrossRef]
- Desiere, F.; Deutsch, E.W.; King, N.L.; Nesvizhskii, A.I.; Mallick, P.; Eng, J.; Chen, S.; Eddes, J.; Loevenich, S.N.; Aebersold, R. The PeptideAtlas Project. Nucleic Acids Res. 2006, 34, D655–D658. [Google Scholar] [CrossRef]
- Sondka, Z.; Dhir, N.B.; Carvalho-Silva, D.; Jupe, S.; Madhumita; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; et al. COSMIC: A Curated Database of Somatic Variants and Clinical Data for Cancer. Nucleic Acids Res. 2024, 52, D1210–D1217. [Google Scholar] [CrossRef]
- Gudmundsson, S.; Singer-Berk, M.; Watts, N.A.; Phu, W.; Goodrich, J.K.; Solomonson, M.; Consortium, G.A.D.; Rehm, H.L.; MacArthur, D.G.; O’Donnell-Luria, A. Variant Interpretation Using Population Databases: Lessons from gnomAD. Hum. Mutat. 2022, 43, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Chitipiralla, S.; Kaur, K.; Brown, G.; Chen, C.; Hart, J.; Hoffman, D.; Jang, W.; Liu, C.; Maddipatla, Z.; et al. ClinVar: Updates to Support Classifications of Both Germline and Somatic Variants. Nucleic Acids Res. 2025, 53, D1313–D1321. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, D.; Walsh, I.; Minervini, G.; Tosatto, S.C.E. FELLS: Fast Estimator of Latent Local Structure. Bioinformatics 2017, 33, 1889–1891. [Google Scholar] [CrossRef] [PubMed]
- Arimori, T.; Miyazaki, N.; Mihara, E.; Takizawa, M.; Taniguchi, Y.; Cabañas, C.; Sekiguchi, K.; Takagi, J. Structural Mechanism of Laminin Recognition by Integrin. Nat. Commun. 2021, 12, 4012. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System; Version 2.5; Schrödinger LLC: New York, NY, USA, 2015.
- Del Conte, A.; Camagni, G.F.; Clementel, D.; Minervini, G.; Monzon, A.M.; Ferrari, C.; Piovesan, D.; Tosatto, S.C.E. RING 4.0: Faster Residue Interaction Networks with Novel Interaction Types across over 35,000 Different Chemical Structures. Nucleic Acids Res. 2024, 52, W306–W312. [Google Scholar] [CrossRef]
- Bányai, L.; Patthy, L. Putative Extremely High Rate of Proteome Innovation in Lancelets Might Be Explained by High Rate of Gene Prediction Errors. Sci. Rep. 2016, 6, 30700. [Google Scholar] [CrossRef]
- Denton, J.F.; Lugo-Martinez, J.; Tucker, A.E.; Schrider, D.R.; Warren, W.C.; Hahn, M.W. Extensive Error in the Number of Genes Inferred from Draft Genome Assemblies. PLoS Comput. Biol. 2014, 10, e1003998. [Google Scholar] [CrossRef]
- Nasiri, J.; Naghavi, M.; Rad, S.N.; Yolmeh, T.; Shirazi, M.; Naderi, R.; Nasiri, M.; Ahmadi, S. Gene Identification Programs in Bread Wheat: A Comparison Study. Nucleosides Nucleotides Nucleic Acids 2013, 32, 529–554. [Google Scholar] [CrossRef]
- Uriostegui-Arcos, M.; Mick, S.T.; Shi, Z.; Rahman, R.; Fiszbein, A. Splicing Activates Transcription from Weak Promoters Upstream of Alternative Exons. Nat. Commun. 2023, 14, 3435. [Google Scholar] [CrossRef] [PubMed]
- Blakeley, P.; Siepen, J.A.; Lawless, C.; Hubbard, S.J. Investigating Protein Isoforms via Proteomics: A Feasibility Study. Proteomics 2010, 10, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Capovilla, G.; Pajoro, A.; Immink, R.G.; Schmid, M. Role of Alternative Pre-mRNA Splicing in Temperature Signaling. Curr. Opin. Plant Biol. 2015, 27, 97–103. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Le, K.; Prabhakar, B.S.; Hong, W.; Li, L. Alternative Splicing as a Biomarker and Potential Target for Drug Discovery. Acta Pharmacol. Sin. 2015, 36, 1212–1218. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative Splicing and Related RNA Binding Proteins in Human Health and Disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Kääriäinen, M.; Nissinen, L.; Kaufman, S.; Sonnenberg, A.; Järvinen, M.; Heino, J.; Kalimo, H. Expression of A7β1 Integrin Splicing Variants during Skeletal Muscle Regeneration. Am. J. Pathol. 2002, 161, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- von der Mark, H.; Williams, I.; Wendler, O.; Sorokin, L.; von der Mark, K.; Pöschl, E. Alternative Splice Variants of α7β1Integrin Selectively Recognize Different Laminin Isoforms*. J. Biol. Chem. 2002, 277, 6012–6016. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Chalancon, G.; Eustermann, S.; Wagner, G.P.; Fuxreiter, M.; Bateman, A.; Babu, M.M. Tissue-Specific Splicing of Disordered Segments That Embed Binding Motifs Rewires Protein Interaction Networks. Mol. Cell 2012, 46, 871–883. [Google Scholar] [CrossRef]
- Colak, R.; Kim, T.; Michaut, M.; Sun, M.; Irimia, M.; Bellay, J.; Myers, C.L.; Blencowe, B.J.; Kim, P.M. Distinct Types of Disorder in the Human Proteome: Functional Implications for Alternative Splicing. PLoS Comput. Biol. 2013, 9, e1003030. [Google Scholar] [CrossRef]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative Splicing in Concert with Protein Intrinsic Disorder Enables Increased Functional Diversity in Multicellular Organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro Naser, X.A.; Cestaro, A.; Tosatto, S.C.E.; Leonardi, E. Integrative Multi-Omics Characterization and Structural Insights into the Poorly Annotated Integrin ITGA6 X1X2 Isoform in Mammals. Genes 2025, 16, 1134. https://doi.org/10.3390/genes16101134
Castro Naser XA, Cestaro A, Tosatto SCE, Leonardi E. Integrative Multi-Omics Characterization and Structural Insights into the Poorly Annotated Integrin ITGA6 X1X2 Isoform in Mammals. Genes. 2025; 16(10):1134. https://doi.org/10.3390/genes16101134
Chicago/Turabian StyleCastro Naser, Ximena Aixa, Alessandro Cestaro, Silvio C. E. Tosatto, and Emanuela Leonardi. 2025. "Integrative Multi-Omics Characterization and Structural Insights into the Poorly Annotated Integrin ITGA6 X1X2 Isoform in Mammals" Genes 16, no. 10: 1134. https://doi.org/10.3390/genes16101134
APA StyleCastro Naser, X. A., Cestaro, A., Tosatto, S. C. E., & Leonardi, E. (2025). Integrative Multi-Omics Characterization and Structural Insights into the Poorly Annotated Integrin ITGA6 X1X2 Isoform in Mammals. Genes, 16(10), 1134. https://doi.org/10.3390/genes16101134







