Abstract
Background: As a key rice breeding resource, aromatic rice is widely cultivated in agriculture due to its unique aroma. Badh2 mutations cause function loss, enabling rice’s characteristic aroma. Methods: In this study, we analyzed several badh2 mutation types across 8 japonica and 16 indica aromatic rice lines. Based on the 7 bp deletion in badh2-E2 identified in japonica aromatic lines, we developed a multiplex-ready PCR assay for badh2 genotyping. Additionally, leveraging the deletion mutation in badh2-E7 from the indica aromatic line Yexiang, we designed a KASP marker. Results: All 8 japonica aromatic lines carried a 7 bp deletion in badh2-E2, while 12 indica aromatic lines harbored an 8 bp deletion in badh2-E7, and 4 additional indica aromatic lines exhibited an 8 bp deletion in badh2-E2. The multiplex-ready PCR assay was used to screen 200 individual plants from the aromatic rice line Jia 58: 199 plants showed the expected results, while the remaining 1 exhibited two fluorescent signal peaks—suggesting that it may be a heterozygous individual. Using the KASP marker, we performed genotyping analysis on F7 progeny individuals derived from the cross between Yexiang (aromatic line) and Yuenongsimiao (non-aromatic line). Combined with phenotypic observations, we successfully screened out an elite aromatic line named Zhexiangzhenhe, which not only possesses aroma but also maintains superior agronomic traits. Conclusions: The multiplex-ready PCR assay and KASP markers facilitate high-throughput genotyping in large-scale breeding populations, providing breeders with a rapid and efficient selection tool to accelerate aromatic trait improvement in rice.
1. Introduction
Rice is the most important stamp crop in the world, especially in China and South-east Asia. Rice grains are a main source of income for farmers. Due to its special aroma and good market value, the demand for aromatic rice has been increasing in recent years. In order to improve the breeding efficiency of aromatic rice varieties, identifying an inexpensive, simple, and high-throughput marker for aromatic rice is becoming increasingly important for rice farmers. The farmers can use the marker to quickly select the ideal individual plant that not only has high yield and good resistance to stress but also aromatic smell.
Cultivating an aromatic rice variety is the most valuable aspect of rice breeding, as farmers can earn more from planting aromatic rice. According to previous studies, 2-acetyl-1-pyrroline (2-AP) levels play a crucial role in the grains of aromatic rice, despite detecting one hundred compounds in them [1,2,3,4]. The 2-AP concentration in aromatic rice grains is higher than non-aromatic ones and can be detected by human olfactory organs [5,6]. Presently, there are two methods of detecting flavor or aroma in rice grains: a subjective method and an objective method. Discriminating between the aromatic and non-aromatic varieties using the subjective method mainly depends on the analysts’ taste of individual grains or the smell of the leaf tissue or grain that has been heated in water or reacted with KOH or I2/KI solutions [7]. Objective detection uses gas chromatography to measure the quantity of rice grains in rice and distinguish the rice varieties [8,9]. However, the subjective method needs a large amount of man power. Subjective detection accuracy is still not very reliable. The objective method requires expensive instruments and equipment and has high test costs. Given the limitation of these methods and the development, identification, and cloning of rice aromatic genes, marker-assisted selection appears to have more advantages than subjective and objective detection in rice breeding. Marker-assisted selection allows for the rapid selection of individual plants with aromatic traits by segregating generations, and these traits can be quickly stabilized [10,11]. Moreover, it is not affected by environmental conditions, making it a simple and efficient selection method [12].
In a previous study, badh2, a single recessive gene, which is located on chromosome 8 of rice, was reported to contain 14 introns and 15 exons, and it is the main gene that encodes betaine aldehyde dehydrogenase (Badh2), which is associated with rice aroma. Fragment deletion on exons 2 and 7 will lead to the function loss of badh2 and inhibits 2-acetyl-1-pyrroline (2-AP) synthesis, which produces aroma in rice grains [13,14,15,16,17]. A small fragment deletion can be used as an SNP marker in molecular marker-assisted breeding programs for rice. SNP markers have been successfully developed and used in aromatic rice breeding [18]. In addition, several PCR-based molecular markers have been developed to detect the badh2 gene in rice. With the continuous discovery and identification of badh2 gene haplotypes, molecular markers based on the characteristics of these haplotypes have been continuously developed and applied in marker-assisted breeding, enabling breeders to rapidly improve the aromatic traits of rice [14,19,20,21,22]. Presently, different kinds of methods for SNP detection were used in molecular breeding, including allele-specific PCR, cleaved amplified polymorphism sequences (CAPSs), derived CAPSs (dCAPSs), high-resolution melting (HRM), temperature switch PCR (TS-PCR), and Kompetitive allele-specific PCR (KASP) [23]. As KASP has many advantages, including high throughput, high speed and accuracy, good stability, and low cost, it has been widely used in marker-assistant selection, bulked segregation analysis, and single nucleotide polymorphism (SNP) genotyping [23]. As a new method for SNP genotyping, multiplex-ready PCR can be widely applied in marker-assisted breeding, and it is particularly effective in identifying and utilizing small-fragment insertion–deletion (Indel) polymorphisms. In particular, for cases where common PCR amplification methods cannot be used to design primers for genotyping analysis due to the high GC content of DNA fragments near SNP loci, primers can be designed using DNA regions with low GC content upstream and downstream of SNPs, and their polymorphisms can be identified through PCR amplification [24,25,26,27].
In this study, we designed specific primers for amplifying badh2-E2 and badh2-E7 in different types of aromatic rice varieties and identified the characteristics of badh2 gene mutations using Sanger sequencing. Based on the sequencing results of the badh2 gene, multiplex-ready PCR and KASP molecular markers were designed for genotyping analysis and marker-assisted selection of the badh2 gene, respectively. A recombinant inbred line (RILs) population was constructed by crossing the aromatic rice line Yexiang with the conventional non-aromatic rice line Yunongsimiao. Using KASP molecular markers, 98 homozygous aromatic lines were detected among 384 RILs. Through the pedigree method combined with 2-AP detection, an aromatic rice line, Zhexiangzhenhe, with aroma and excellent agronomic traits was bred.
2. Materials and Methods
2.1. Plant Materials
To identify the mutation types in the badh2-E2 and badh2-E7 loci among aromatic rice varieties currently promoted in production, this study selected eight aromatic japonica rice lines, sixteen aromatic indica rice lines, and two conventional non-aromatic rice lines (Table 1). All rice materials were planted in the summer season of 2022 in a paddy field at the Yangdu Experimental Base of the Zhejiang Academy of Agricultural Sciences, Haining County, China. Yexiang is an aromatic indica rice line with excellent taste and cooking quality, while Yuenongsimiao is a non-aromatic indica line characterized by high yield and strong adaptability to diverse environmental conditions. The F1 plants were generated from a cross between Yexiang (as the female parent) and Yuenongsimiao (as the male parent). Approximately 500 F2 seeds from this cross were sown. In each subsequent generation, around 50 plants with desirable agronomic traits were selected and self-pollinated until the F7 generation. A total of 384 plants were then genotyped using a functional KASP marker targeting the badh2 gene. Ninety-eight F8 RILs with homozygous badh2 genotypes were planted in the summer season of 2022 for phenotypic and yield-related observations. An elite line named Zhexiangzhenhe was selected to measure the 2-AP content, and phenotyping was conducted to determine the yield trait.

Table 1.
Twenty-four aromatic and two non-aromatic rice lines were selected in this study.
The procedures are presented in Figure 1 in this study.
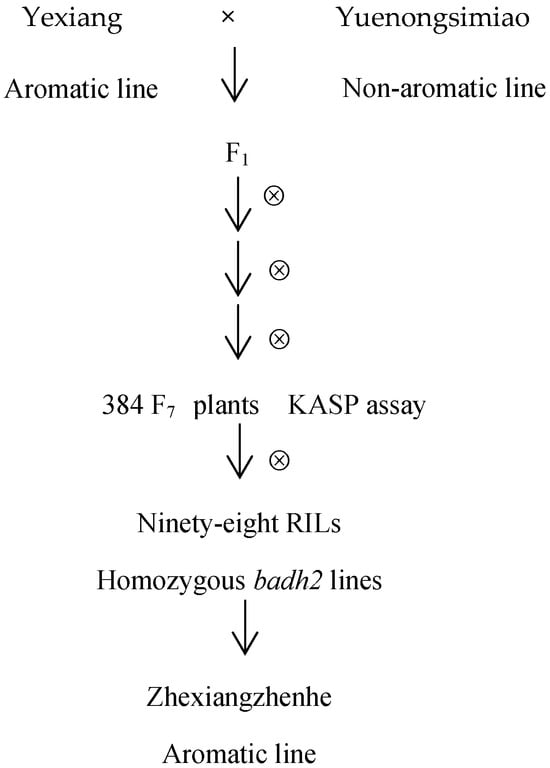
Figure 1.
Selection scheme of the aromatic line of ZhexiangZhenhe via KASP.
2.2. DNA Isolation and Marker Development
Genomic DNA was extracted from the leaf with a length of 1–2 cm using the CTAB buffer at the booting stage [28]. The DNA concentration was measured using a Nanodrop onec-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and adjusted to a working concentration of 100 ng/μL. To identify the deletion mutations at the badh2-E2 and badh2-E7 loci of the aromatic rice lines, specific primers were designed at positions 200–300 bp upstream and downstream of the deletion sites, respectively (Table 2). Based on the DNA sequence of the rice Oryza sativa cv. Nipponbare, a 554 bp fragment at the badh2-E2 locus and a 442 bp fragment at the badh2-E2 locus were amplified for Sanger sequencing analysis, respectively. According to the mutation type of eight aromatic japonica rice lines, a specific primer was designed for multiplex-ready PCR (Table 2). A 234 bp fragment for the aromatic line and a 241 bp fragment for the non-aromatic line were amplified for multiplex-ready PCR analysis. PCR reactions were prepared following the standard protocol for AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA, USA), with primer concentrations optimized for target amplification. The PCR amplification conditions were as follows: an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 45 s, and extension at 72 °C for 45 s. This was then followed by 8 additional cycles of 94 °C for 30 s (denaturation), 53 °C for 45 s (annealing), and 72 °C for 45 s (extension); finally, a final extension step was performed at 72 °C for 10 min. Subsequently, 1 µL of the PCR product was mixed with 22 µL of formamide and 0.5 µL of ROX standard (ABI), and the mixture was analyzed using an ABI 3730XL Prism Genetic Analyzer. The assay method for multiplex-ready PCR analysis was performed as previously described [27]. To design KASP markers for genotyping, approximately 200 bp flanking sequences (both upstream and downstream) surrounding the SNP locus of exon 7 of the badh2 gene were analyzed. KASP primers were then designed following standard KASP guidelines and synthesized by LGC Limited (Teddington, Middlesex, UK). The standard FAM (5′-GAAGGTGACCAAGTTCATGCT-3′) and HEX (5′-GAAGGTCGGAGTCAACGGATT-3′) fluorescent tags were incorporated and linked to allele-specific forward primers, each targeting the SNP at their respective 3′ ends. A common reverse primer was designed using the Primer 3 software [29], with the resulting amplification length restricted to less than 100 bp. The KASP assay method was performed as previously described [23].

Table 2.
Primers for sequence analysis and marker development in this study.
2.3. Experiment Design and Measuring 2-AP Content
To observe phenotypic and agronomic traits, ninety-eight F8 RIls homozygous positive for badh2 genes were planted in plots during the summer season in 2022. Each line was planted in 6 rows with 6 plants per row, maintaining a plant spacing and row spacing of 20 cm × 20 cm. Agronomic traits, including plant height, panicle length, number of filled grains per panicle, seed setting rate, and thousand-grain weight, were measured from 10 plants per line. Yuenongsimiao, an elite non-aromatic rice line, was included as a standard variety for comparison. Each agronomic trait measurement was conducted with three biological replicates, and each replicate included 10 plants of the same line to ensure the reliability of the data.
For the determination of 2-AP content, a volatile aromatic compound, 30 g of mature seeds was collected from the plants. After dehusking, the brown rice was ground into rice flour. Using 2,4,6-trimethylpyridine as the internal standard (concentration: 229.25 ng·ml-1), 400 mg of the rice flour sample was weighed in triplicate. The samples were placed in 10 mL narrow-mouth glass vials, and 0.8 mL of an ethanol extraction reagent containing the internal standard was added. The vials were incubated in an oven at 80 °C for 3 h for extraction. After extraction, the samples were taken out and allowed to cool to room temperature, then filtered through a 0.22 μm disposable syringe filter. A 150 μL aliquot of the filtrate was transferred into a liner tube, which was then placed in a 2 mL sample vial. The determination was performed using a GC-MS (gas chromatography-mass spectrometry) instrument (Shimadzu Corporation, Kyoto, Japan) [30]. The detection data were analyzed using the Excel 2010 and SPSS 17.0 software. The measurement of plant 2-AP content was performed at Huazhong Agricultural University, Wuhan, China.
2.4. Statistical Analysis
Statistical analyses were performed using SPSS 17.0 (IBM Corp., Armonk, NY, USA). Phenotypic differences between Zhexiangzhenhe and Yuenongsimiao were compared via one-way analysis of variance (ANOVA), followed by Tukey’s honestly significant difference (HSD) test for multiple comparisons.
3. Results
3.1. Identification of Mutation Type in Aromatic Lines
To clarify badh2 mutation characteristics in currently cultivated aromatic rice, Sanger sequencing was performed on 24 aromatic lines targeting badh2-E2 and badh2-E7. PCR amplification was performed on sixteen aromatic indica rice lines, eight aromatic japonica rice lines, and two non-aromatic lines using gene-specific primers. Sanger sequencing results revealed that the sixteen aromatic indica rice lines and eight aromatic japonica rice lines all harbor three distinct types of mutations in the badh2-E2 and badh2-E7 loci compared to two non-aromatic control lines. Among them, all eight japonica rice varieties exhibit a 7 bp (5′-GGCGCCG-3′) deletion in the badh2-E2 locus, with consistent mutations across all aromatic japonica rice lines. However, sixteen aromatic indica rice lines have two mutation types in the badh2 gene. Of these, twelve lines, including Nongxiang18, Nongxiang42, Yuzhenxiang, Chaungxiang5, Meixiangzhan2, Xiangyaxiangzhan, Yexiang, Zhexiangsimiao, 99xiang, Taixiang8, 19xiang, and Yingxiangsimiao, exhibit an 8 bp (5′-GATTATGG-3′) deletion in badh2-E7. Four lines, including Zhuxianglisi, Meixiangxinzhan, Junhexiangzhan, and Guangliangxiang3, exhibit an 8 bp (5′-ACCCCCAC-3′) deletion in badh2-E2 (Figure 2). These results indicate that badh2 mutations are subspecies-specific—7 bp deletion in badh2-E2 for japonica, and two distinct deletions (8 bp in badh2-E7 or badh2-E2) for indica.

Figure 2.
Mutation-type comparison of badh2 alleles in different aromatic lines and non-aromatic lines. The blue-bottomed horizontal line indicates different base deletion fragments: 7 bp (5′-GGCGCCG-3′) deletion in badh2-E2 of japonica aromatic lines, 8 bp (5′-GATTATGG-3′) deletion in badh2-E7 of 12 indica aromatic lines, and 8 bp (5′-ACCCCCAC-3′) deletion in badh2-E2 of 4 indica aromatic lines.
3.2. Multiplex-Ready PCR Assay
Based on the Sanger sequencing results of the badh2 gene for eight japonica aromatic lines, we developed the marker for multiplex-ready PCR. Based on the fluorescence intensities from ABI 3730XL, a single fluorescent signal was detected in all eight japonica aromatic rice varieties, with a molecular size of 234 bp. To ensure the purity of aromatic rice varieties, 200 individual plants of the Jia 58 aromatic line were randomly selected, and multiplex-ready PCR analysis was performed on their badh2 genes. The results showed that 199 individual plants had only one fluorescent signal peak, while the remaining plant had two fluorescent signal peaks, indicating that it might be a heterozygous plant, which was confirmed via phenotypic analysis (Figure 3). The multiplex-ready PCR assay exhibited high accuracy (99.5%) in genotyping, confirming its suitability for purification of aromatic rice varieties.
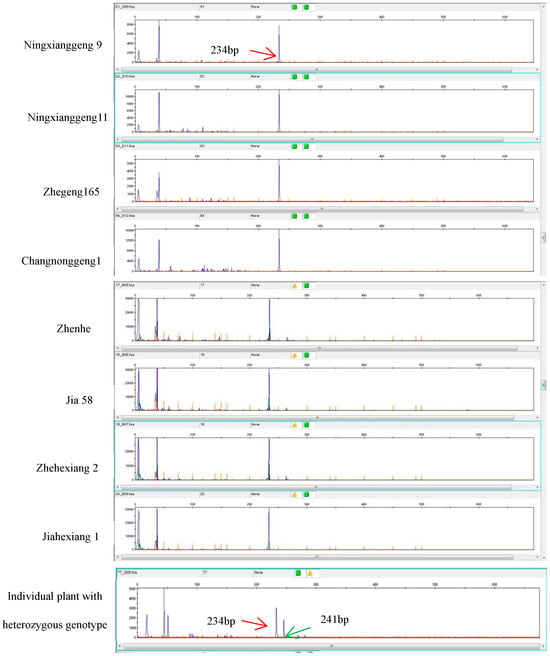
Figure 3.
ABI 3730XL electrotraces showing multiplex-ready PCR amplified from eight aromatic japonica lines and individual plants with a heterozygous genotype. Individual plants with a homozygous badh2 gene show only one fluorescent signal peak with a size of 234 bp, while individual plants with a heterozygous badh2 gene exhibit two fluorescent signal peaks with sizes of 234 bp and 241 bp, respectively.
3.3. KASP Assay for the badh2 Gene
To generate lines that possess both aromatic genes and excellent agronomic traits, genotyping analysis of the badh2 gene was conducted on 384 F7 lines from the cross of Yexiang (aromatic) × Yuenongsimiao (non-aromatic) using KASP markers. The KASP assay for the badh2 gene showed that ninety-eight individual plants were of the homozygous genotype with a blue signal, similar to Yexiang, while 238 individual plants were of the homozygous genotype with a red signal, similar to Yuenongsimiao, and thirty-nine individual plants were of the heterozygous genotype with a green signal. Unfortunately, the genotype of thirty-nine individual plants could not be determined with a pink signal (Figure 4).
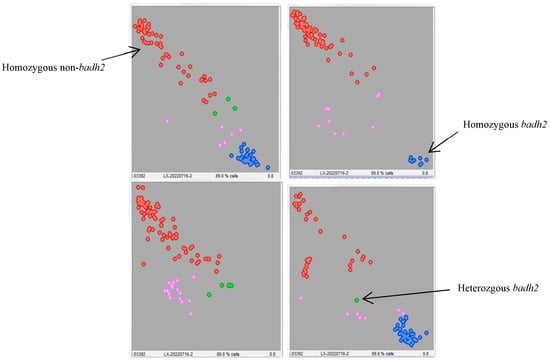
Figure 4.
KASP assay of individual plants for the badh2 gene in 384 F7 lines. The blue signal represents the homozygous genotype associated with aromatic traits; the red signal represents the homozygous genotype associated with non-aromatic traits; the green signal represents the heterozygous genotype, which also corresponds to non-aromatic traits; and the pink signal indicates genotypes that could not be determined.
3.4. Measurement of 2-AP Content and Agronomic Characteristic
To determine the 2-AP content, mature seeds from Zhexiangzhenhe which possess homozygous badh2 and Yuenongsimiao (non-aromatic) were selected and harvested. After dehusking, the seeds were ground into brown rice flour, and the 2-AP content was measured using a gas chromatography-mass spectrometry (GC-MS) system. The 2-AP content detection results showed that the average content in Zhexiangzhenhe was 0.345 μg·g−1, which was significantly higher than that in Yuenongsimiao, whose average content was only 0.032 μg·g−1 (p < 0.01) (Figure 5).
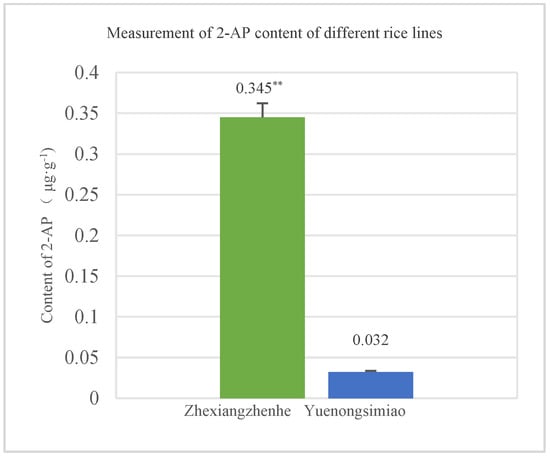
Figure 5.
Measurement of 2-AP between Zhexiangzhenhe (aromatic line) and Yuenongsimiao (non-aromatic line). ** indicated that there are significant differences between different lines at the 0.01 level.
To assess differences in agronomic traits between Zhexiangzhenhe and Yuenongsimiao, statistical analyses were conducted on agronomic traits, including plant height, panicle length, effective panicles, total number of grains per panicle, number of filled grains per panicle, seed-setting rate, and 1000-grain weight (Table 3). The results revealed no significant differences between the two lines, indicating that Zhexiangzhenhe retained Yuenongsimiao’s agronomic traits while incorporating the aromatic gene badh2. This enables it to exhibit both aromatic rice quality and high-yield characteristics.

Table 3.
Agronomic traits comparison between Zhexiangzhenhe and Yuenongsimiao.
4. Discussion
As a staple food in regions such as East Asia, Southeast Asia, and South Asia, rice plays a crucial role in agricultural production. Aromatic rice, which is favored by consumers for its unique aroma, has higher commercial value compared to ordinary rice of the same category. Therefore, breeders have introduced aromatic genes to enable rice varieties to produce aromatic grains while retaining excellent agronomic traits. This not only increases agricultural benefits but also facilitates the promotion of aromatic rice varieties. 2-AP is the main component responsible for the aroma in aromatic rice. It can be detected in all tissues of aromatic rice varieties except roots [2]. A mutation in the badh2 gene on rice chromosome 8 results in premature termination of aldehyde dehydrogenase translation. Consequently, the functionally conserved domain is not translated, rendering the enzyme incapable of catalyzing the dehydrogenation reaction. This leads to 2-AP loss, which is responsible for the characteristic aroma [3,13].
Different mutations in the badh2 gene have been identified in some specific aromatic rice varieties. For example, Amarawathi et al. reported a new mutation in the badh2 gene which has a 7 bp insertion in exon 8 in addition to an 8 bp deletion in badh2-E7 [19]. Shao et al. reported an 803 bp deletion between exons 4 and 5, which was found in the aromatic variety Zaimiaoxiangnuo [31]. Shi et al. reported that in the aromatic rice variety Nankai 138, a 3 bp deletion was identified in the 5′ untranslated region of the badh2 gene, accompanied by an 8 bp insertion in its promoter region [16]. However, the 8 bp deletion mutation in badh2-E7 and the 7 bp deletion mutation in badh2-E2 are mutation types widely found in most aromatic rice varieties [3,4,15,17]. In this study, the sequencing results of twenty-four aromatic rice lines showed that eight japonica aromatic rice lines had an 8 bp deletion mutation in the badh2-E2 locus, which was highly conserved. Among the sixteen indica aromatic rice lines, twelve had a 7 bp deletion mutation in the badh2-E7 locus, which was the same as the mutation in aromatic rice such as Basmati [3,17]. However, an 8 bp deletion mutation was identified in the badh2-E2 among the other four indica aromatic rice lines, and this mutation differs from those found in aromatic rice varieties [3,16,17,19,31]. Meanwhile, no other mutations were detected in badh2-E2 or other positions of badh2-E7, suggesting that this could potentially be a new type of mutation. This new mutation enriches the genetic variation library of the badh2 gene and provides a new molecular marker for the selection of aromatic traits in indica rice.
As an important molecular breeding technology, molecular marker-assisted selection has played a significant role in the research on improving the aromatic traits of rice. Specifically, the molecular marker techniques developed based on PCR and restriction enzyme digestion have been widely used in aromatic quality improvement [14,16,18,20,21,22]. However, with the continuous development and optimization of fluorescent labeling technology, multiplex-ready PCR and KASP technologies, which were developed based on PCR and fluorescent labeling, have demonstrated advantages such as high throughput and speed in molecular breeding and trait improvement, leading to their further application [23,31,32,33]. In this study, based on the 7 bp deletion in badh2-E2 in eight japonica aromatic rice varieties, we developed a multiplex-ready PCR technology to perform genotyping analysis of the badh2 gene in these lines. Additionally, we screened 200 individual plants from the Jia 58 aromatic rice line and successfully removed one heterozygous plant, providing technical support for the purification and rejuvenation of aromatic rice varieties. In this study, based on the 8 bp deletion in badh2-E7 identified in the Yexiang aromatic line, we developed KASP markers. These markers were used to screen 384 F7 plants derived from the cross between Yexiang and Yuenongsimiao. Through genotyping, ninety-eight F8 plants with homozygous badh2 were obtained. Combined with phenotypic observations, an elite line named Zhexiangzhenhe was selected, which exhibited excellent agronomic traits and retained the aromatic phenotype. Analyzing the 2-AP content revealed that Zhexiangzhenhe exhibited significantly higher 2-AP levels than the non-aromatic line Yuenongsimiao, while its agronomic traits showed no significant differences compared to Yuenongsimiao. Thus, KASP markers enable accurate target gene selection and facilitate high-throughput genotyping in large-scale breeding populations, thereby providing breeders with a rapid and efficient molecular marker-assisted selection tool to accelerate target trait improvement.
This study developed multiplex-ready PCR and KASP markers for the badh2 gene, which have been verified in specific rice materials, but there are still some limitations. First, the transferability of the developed markers across different breeding backgrounds needs to be further verified. At present, the markers are mainly validated in japonica and indica rice in China. Whether they can be effectively applied to other rice subspecies (such as aromatic rice in South Asia) remains to be tested, as differences in genetic background may affect the amplification efficiency and genotyping accuracy of the markers. Secondly, the current study only focused on the mutation of the badh2 gene, but the aroma of rice is a complex quantitative trait, which may be regulated by other genes and environmental factors [34]. Therefore, further research on the interaction between badh2 and other genes, as well as the influence of environmental factors on 2-AP synthesis, is needed to comprehensively understand the genetic mechanism of rice aroma.
5. Conclusions
This study identified three subspecies-specific badh2 mutation types in 24 aromatic rice lines: a 7 bp deletion in badh2-E2 (japonica), an 8 bp deletion in badh2-E7 (75% of indica), and a potential novel 8 bp deletion in badh2-E2 (25% of indica). Two high-throughput detection markers were developed: multiplex-ready PCR for accurate badh2-E2 genotyping (suitable for variety purification) and KASP for high-throughput badh2-E7 screening (suitable for large breeding populations). Both markers were validated to be reliable—enabling efficient selection of aromatic genotypes and overcoming the limitations of traditional aroma detection methods. Using KASP markers, we successfully bred the elite aromatic line Zhexiangzhenhe, which combines high 2-AP content (0.345 μg·g−1) and superior agronomic traits, providing a valuable germplasm resource for aromatic rice breeding. The marker system and genetic insights from this study lay a foundation for accelerating aroma trait improvement in rice.
Author Contributions
H.F. and H.H., writing—original draft, investigation, methodology, and data collection. L.Y. and L.W., methodology and data collection. J.L., funding acquisition and writing—review. Y.Q., funding acquisition, conceptualization, supervision, and writing—original draft, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Project of the Science and Technology Department of Zhejiang Province, China (Grant Number 2021C02063-2), and the Project of Hangzhou Science and Technology Development Plan, China (Grant Number 2020ZDSJ0241).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors would like to thank Researcher Duo Wang at the Hunan Golden Fenghua Seed Industry Technology Co., Ltd. for growing the rice plants in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 2-AP | 2-acetyl-1-pyrroline |
| CAPS | Cleaved amplified polymorphism sequence |
| dCAPS | Derived CAPS |
| HRM | High-resolution melting |
| TS-PCR | Temperature switch PCR |
| KASP | Kompetitive allele-specific PCR |
| badh2-E2 | The second exon of badh2 |
| badh2-E7 | The seventh exon of badh2 |
| SNP | Single nucleotide polymorphism |
| RILs | Recombinant inbred lines |
| PH | Plant height |
| PL | Panicle length |
| EP | Effective panicles 104/ha |
| TNGP | Total number of grains per panicle |
| NFGP | Number of filled grains per panicle |
| SSR | Seed sating rat |
| TGW | Thousand-grain weight (g) |
References
- Yajima, I.; Yanai, T.; Nakamura, M.; Sakakibar, H.; Habu, T. Volatile flavor components of cooked rice kaorimai (scented rice, O. sativa japonica). Agric. Biol. Chem. 1979, 43, 2425–2429. [Google Scholar]
- Buttery, R.G.; Ling, L.C.; Juliano, B.O.; Turnbaugh, J.G. Cooked rice aroma and 2-acetyl-1-pyrroline. J. Agric. Food Chem. 1983, 31, 823–826. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L.E. The gene for fragrance in rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Gillies, S.A.; Brushett, D.J.; Waters, D.L.; Henry, R.J. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 2008, 68, 439–449. [Google Scholar] [CrossRef]
- Grosch, W.; Schieberle, P. Flavor of cereal products—A review. Cereal Chem. 1997, 74, 91–97. [Google Scholar] [CrossRef]
- Wilkie, K.; Wootton, M.; Paton, J.E. Sensory testing of Australian fragrant, imported fragrant, and non-fragrant rice aroma. Int. J. Food Prop. 2004, 7, 27–36. [Google Scholar] [CrossRef]
- Sood, B.C.; Sidiq, E.A. A rapid technique for scent determination in rice. Indian J. Genet. Plant Breed. 1978, 38, 268–271. [Google Scholar]
- Widjaja, R.; Craske, J.; Wotton, D. Comparative studies on volatile components of non-fragrant and fragrant rices. J. Sci. Food Agric. 1996, 70, 151–161. [Google Scholar] [CrossRef]
- Sriseadka, T.; Wongpornchai, S.; Kitsawatpaiboon, P. Rapid method for quantitative analysis of the aroma impact compound, 2-acetyl-1-pyrroline, in fragrant rice using automated headspace gas chromatography. J. Agric. Food Chem. 2006, 54, 8183–8189. [Google Scholar] [CrossRef]
- Singh, A.; Gopalakrishnan, S.; Singh, V.P.; Prabhu, K.V.; Mohapatra, T.; Singh, S.N.; Sharma, T.R.; Nagarajan, M.; Vinod, K.K.; Singh, U.D.; et al. Marker assisted selection: A paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 2011, 71, 120–128. [Google Scholar]
- Kiani, G. Marker aided selection for aroma in F2 populations of rice. Afr. J. Biotechnol. 2013, 10, 15845–15848. [Google Scholar] [CrossRef]
- Collard, B.; Jahufer, M.; Brouwer, J.; Pang, E. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Chen, S.; Yang, Y.; Shi, W.; Ji, Q.; He, F.; Zhang, Z.; Cheng, Z.; Liu, X.; Xu, M. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 2008, 20, 1850–1861. [Google Scholar] [CrossRef]
- Sakthivel, K.; Shobha Rani, N.; Pandey, M.K.; Sivaranjani, A.K.; Neeraja, C.N.; Balachandran, S.M.; Sheshu Madhav, M.; Viraktamath, B.C.; Prasad, G.S.V.; Sundaram, R.M. Development of a simple functional marker for fragrance in rice and its validation in Indian basmati and non-basmati fragrant rice varieties. Mol. Breed. 2009, 24, 185–190. [Google Scholar] [CrossRef]
- Shao, G.; Tang, S.; Chen, M.; Wei, X.; He, J.; Luo, J.; Jiao, G.; Hu, Y.; Xie, L.; Hu, P. Haplotype variation at Badh2, the gene determining fragrance in rice. Genomics 2013, 101, 157–162. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, G.; Xu, X.; Li, J. Discovery of a new fragrance allele and development of functional markers for identifying diverse fragrant genotypes in rice. Mol. Breed. 2014, 33, 701–708. [Google Scholar] [CrossRef]
- Yu, Y.; Shao, G.; Sheng, Z.; Jiang, H.; He, J.; Sun, Y.; Cai, Y.; Hu, P.; Tang, S. Genetic diversity of global aromatic rice varieties. Plant Divers. Resour. 2015, 37, 871–880. [Google Scholar]
- Jin, Q.; Waters, D.; Cordeiro, G.M.; Henry, R.J.; Reinke, R.F. A single nucleotide polymorphism (SNP) marker linked to the fragrance gene in rice (Oryza sativa L.). Plant Sci. 2003, 165, 359–364. [Google Scholar] [CrossRef]
- Amarawathi, Y.; Singh, R.; Singh, A.K.; Singh, V.P.; Mohapatra, T.; Sharma, T.R.; Singh, N.K. Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 2008, 21, 49–65. [Google Scholar] [CrossRef]
- Shi, W.; Yang, Y.; Chen, S.; Xu, M. Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 2008, 22, 185–192. [Google Scholar] [CrossRef]
- Xu, X.L.; Zhao, G.C.; Li, J.Y. Development of molecular markers used to identify two types of fragrant rice and analysis of mutation sites od Badh2 gene in 24 varieties of fragrant rice. Plant Divers. Resour. 2011, 33, 667–673. [Google Scholar]
- Hashemi, F.S.G.; Rafii, M.Y.; Ismail, M.R.; Mohamed, M.T.; Rahim, H.A.; Latif, M.A.; Aslani, F. The genetic and molecular origin of natural variation for the fragrance trait in an elite Malaysian aromatic rice through quantitative trait loci mapping using SSR and gene-based markers. Gene 2015, 555, 101–107. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, L.; Song, J.; Ma, G.; Wang, J. Development and utilization of the functional co-dominant KASP marker for thermo-sensitive genic male sterility in rice (Oryza sativa L). Genet. Resour. Crop Evol. 2022, 69, 635–643. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Hayden, M.J.; Nguyen, T.M.; Waterman, A.; Chalmers, K.J. Multiplex-ready PCR: A new method for multiplexed SSR and SNP genotyping. BMC Genom. 2008, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Kim, Y.Y.; Ka, K.H.; Lee, H.S.; Bak, W.C.; Jeong, S.J.; Seong, J.Y.; Suh, D.S. Microsatellite markers for population-genetic studies of shiitake (Lentinula edodes) strains. Genes Genom. 2009, 31, 403–411. [Google Scholar] [CrossRef]
- Bonneau, J.; Hayden, M. Multiplex-Ready Technology for mid-throughput genotyping of molecular markers. Methods Mol. Biol. 2014, 1145, 47–57. [Google Scholar] [PubMed]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Grimm, C.C.; Bergman, C.; Delgado, J.T.; Bryant, R. Screening for 2-acetyl-1-pyrroline in the headspace of rice using SPME/GC-MS. J. Agric. Food Chem. 2001, 49, 245–249. [Google Scholar] [CrossRef]
- Shao, G.N.; Tang, A.; Tang, S.Q.; Luo, J.; Jiao, G.A.; Wu, J.L.; Hu, P.S. A new deletion mutation of fragrant gene and the development of three molecular markers for fragrance in rice. Plant Breed. 2011, 130, 172–176. [Google Scholar] [CrossRef]
- Myint, K.M.; Arikit, S.; Wanchana, S.; Yoshihashi, T.; Choowongkomon, K.; Vanavichit, A. A PCR-based marker for a locus conferring the aroma in Myanmar rice (Oryza sativa L.). Theor. Appl. Genet. 2012, 125, 887–896. [Google Scholar] [CrossRef]
- Zhong, Q.; Jia, Q.; Yin, W.; Wang, Y.; Rao, Y.; Mao, Y. Advances in cloning functional genes for rice yield traits and molecular design breeding in China. Front. Plant Sci. 2023, 14, 1206165. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.A.; Sackville Hamilton, N.R.; Calingacion, M.N.; Verhoeven, H.A.; Butardo, V.M. Is there a second fragrance gene in rice? Plant Biotechnol. J. 2008, 6, 416–423. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).