Associations Between Polygenic Risk for Alzheimer’s Disease and Grey Matter Volume Are Dependent on APOE, Pathological and Diagnostic Status
Abstract
1. Introduction
2. Materials and Methods
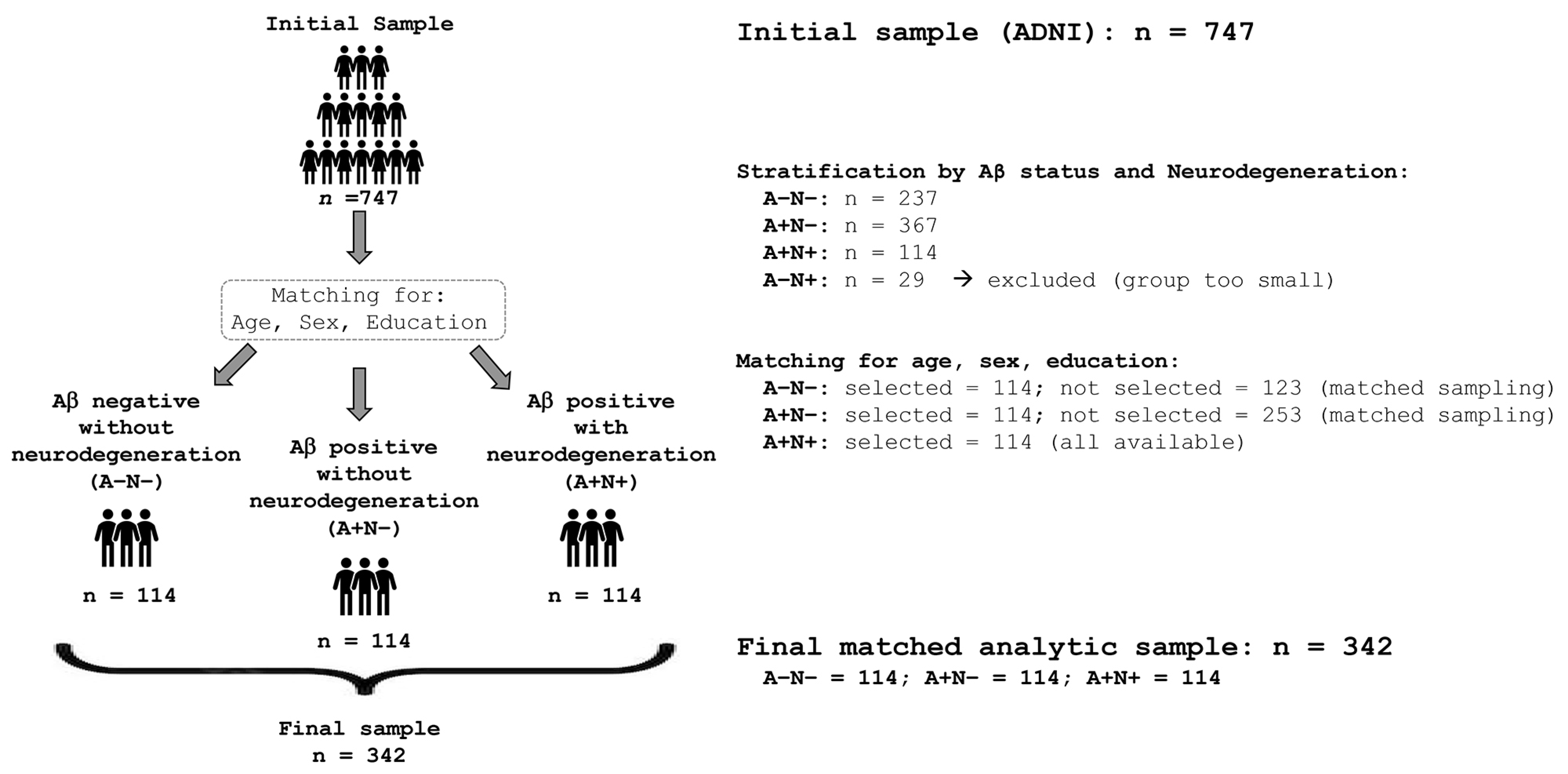
2.1. Participants
2.2. APOE Genotype
2.3. PRS Calculation
2.4. MRI Data and Pre-Processing
2.5. Clinical and Cognitive Data
2.6. Data Analysis
3. Results
3.1. PRSs Across Groups
3.2. Associations Between PRSs and Regional GM Volume Within Individual Groups
3.3. Associations Between PRSs and Regional GM Volumes Within Groups Stratified by APOE Genotype
3.4. Associations Between PRSs and Regional GM Volumes in the Whole Sample
3.5. Associations Between PRSs and Regional GM Volumes Stratified by Diagnosis and APOE Carrier Status
3.6. ROI-Wise Associations Between AD PRS and Regional Grey Matter Volume with Joint FDR Across ROIs, Groups, and PRSs
3.7. Sensitivity Analysis—PRSs Without APOE
3.8. Sensitivity Analysis—Aβ Positivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prince, M.; Wimo, A.; Guerchet, M.; Ali, G.; Wu, Y.; Prina, M. World Alzheimer Report 2015—The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends; Alzheimer’s Disease International: London, UK, 2015; 84p, Available online: http://www.alz.co.uk/research/world-report-2015 (accessed on 4 July 2025).
- Cummings, J.L. Alzheimer’s disease. N. Engl. J. Med. 2004, 351, 56–67. [Google Scholar] [CrossRef]
- Jack, C.R.; Andrews, S.J.; Beach, T.G.; Buracchio, T.; Dunn, B.; Graf, A.; Hansson, O.; Ho, C.; Jagust, W.; McDade, E.; et al. Revised criteria for the diagnosis and staging of Alzheimer’s disease. Nat. Med. 2024, 30, 2121–2124. [Google Scholar] [CrossRef]
- Tanzi, R.E.; Bertram, L. New frontiers in Alzheimer’s disease genetics. Neuron 2001, 32, 181–184. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Bellenguez, C.; Küçükali, F.; Jansen, I.E.; Kleineidam, L.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Campos-Martin, R.; Grenier-Boley, B.; Andrade, V.; et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 2022, 54, 412–436. [Google Scholar] [CrossRef]
- Jones, L.; Holmans, P.A.; Hamshere, M.L.; Harold, D.; Moskvina, V.; Ivanov, D.; Pocklington, A.; Abraham, R.; Hollingworth, P.; Sims, R.; et al. Genetic Evidence Implicates the Immune System and Cholesterol Metabolism in the Aetiology of Alzheimer’s Disease. PLoS ONE 2010, 5, e13950. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Desikan, R.S.; Fan, C.C.; Wang, Y.; Schork, A.J.; Cabral, H.J.; Cupples, L.A.; Thompson, W.K.; Besser, L.; Kukull, W.A.; Holland, D.; et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017, 14, e1002258. [Google Scholar] [CrossRef]
- Leonenko, G.; Baker, E.; Stevenson-Hoare, J.; Sierksma, A.; Fiers, M.; Williams, J.; de Strooper, B.; Escott-Price, V. Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat. Commun. 2021, 12, 4506. [Google Scholar] [CrossRef] [PubMed]
- Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013, 9, e1003348. [Google Scholar] [CrossRef]
- Escott-Price, V.; Sims, R.; Bannister, C.; Harold, D.; Vronskaya, M.; Majounie, E.; Badarinarayan, N.; Morgan, K.; Passmore, P.; Holmes, C.; et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015, 138, 3673–3684. [Google Scholar] [CrossRef]
- Tan, C.H.; Bonham, L.W.; Fan, C.C.; Mormino, E.C.; Sugrue, L.P.; Broce, I.J.; Hess, C.P.; Yokoyama, J.S.; Rabinovici, G.D.; Miller, B.L.; et al. Polygenic hazard score, amyloid deposition and Alzheimer’s neurodegeneration. Brain 2019, 142, 460–470. [Google Scholar] [CrossRef]
- Axelrud, L.K.; Santoro, M.L.; Pine, D.S.; Talarico, F.; Gadelha, A.; Manfro, G.G.; Pan, P.M.; Jackowski, A.; Picon, F.; Brietzke, E.; et al. Polygenic Risk Score for Alzheimer’s Disease: Implications for Memory Performance and Hippocampal Volumes in Early Life. Am. J. Psychiatry 2018, 175, 555–563. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.; Li, A.; Liu, S.; Qin, W.; Yu, C.; Liu, Y.; Liu, B.; Jiang, T. Polygenic risk for Alzheimer’s disease influences precuneal volume in two independent general populations. Neurobiol. Aging 2018, 64, 116–122. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wu, B.; Kuo, K.; Zhang, W.; Ma, Q.; Xiang, S.; Li, Y.; Wang, Z.; Dong, Q.; Feng, J.; et al. Association between polygenic risk for Alzheimer’s disease and brain structure in children and adults. Alzheimer’s Res. Ther. 2023, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.H.H.; de Bruijn, R.F.A.G.; Hofman, A.; Uitterlinden, A.G.; van Duijn, C.M.; Vernooij, M.W.; Koudstaal, P.J.; Ikram, M.A. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimer’s Dement. 2015, 11, 1277–1285. [Google Scholar] [CrossRef]
- Andrews, S.J.; Das, D.; Cherbuin, N.; Anstey, K.J.; Easteal, S. Association of genetic risk factors with cognitive decline: The PATH through life project. Neurobiol. Aging 2016, 41, 150–158. [Google Scholar] [CrossRef]
- Verhaaren, B.F.J.; Vernooij, M.W.; Koudstaal, P.J.; Uitterlinden, A.G.; van Duijn, C.M.; Hofman, A.; Breteler, M.M.B.; Ikram, M.A. Alzheimer’s disease genes and cognition in the nondemented general population. Biol. Psychiatry 2013, 73, 429–434. [Google Scholar] [CrossRef]
- Hansson, O.; Seibyl, J.; Stomrud, E.; Zetterberg, H.; Trojanowski, J.Q.; Bittner, T.; Lifke, V.; Corradini, V.; Eichenlaub, U.; Batrla, R.; et al. CSF biomarkers of Alzheimer’s disease concord with amyloid-β PET and predict clinical progression: A study of fully automated immunoassays in BioFINDER and ADNI cohorts. Alzheimer’s Dement. 2018, 14, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.D.; Pontecorvo, M.J.; Clark, C.M.; Carpenter, A.P.; Jennings, D.L.; Sadowsky, C.H.; Adler, L.P.; Kovnat, K.D.; Seibyl, J.P.; Arora, A.; et al. Performance characteristics of amyloid PET with florbetapir F 18 in patients with alzheimer’s disease and cognitively normal subjects. J. Nucl. Med. 2012, 53, 378–384. [Google Scholar] [CrossRef]
- Saykin, A.J.; Shen, L.; Yao, X.; Kim, S.; Nho, K.; Risacher, S.L.; Ramanan, V.K.; Foroud, T.M.; Faber, K.M.; Sarwar, N.; et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimer’s Dement. 2015, 11, 792–814. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Conomos, M.P.; Miller, M.B.; Thornton, T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015, 39, 276–293. [Google Scholar] [CrossRef]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Chen, C.; Ni, Y.; Feng, Y.A.; Smoller, J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019, 10, 1776. [Google Scholar] [CrossRef] [PubMed]
- Manca, R.; Pardiñas, A.F.; Venneri, A. The neural signatures of psychoses in Alzheimer’s disease: A neuroimaging genetics approach. Eur. Arch. Psychiatry Clin. Neurosci. 2023, 273, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; O’Reilly, P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 2019, 8, giz082. [Google Scholar] [CrossRef]
- Jack, C.R.; Bernstein, M.A.; Fox, N.C.; Thompson, P.; Alexander, G.; Harvey, D.; Borowski, B.; Britson, P.J.; L Whitwell, J.; Ward, C.; et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 2008, 27, 685–691. [Google Scholar] [CrossRef]
- Klöppel, S.; Stonnington, C.M.; Chu, C.; Draganski, B.; Scahill, R.I.; Rohrer, J.D.; Fox, N.C.; Jack, C.R.; Ashburner, J.; Frackowiak, R.S.J. Automatic classification of MR scans in Alzheimer’s disease. Brain 2008, 131, 681–689. [Google Scholar] [CrossRef]
- Marchewka, A.; Kherif, F.; Krueger, G.; Grabowska, A.; Frackowiak, R.; Draganski, B. Influence of magnetic field strength and image registration strategy on voxel-based morphometry in a study of Alzheimer’s disease. Hum. Brain Mapp. 2014, 35, 1865–1874. [Google Scholar] [CrossRef]
- Schmitter, D.; Roche, A.; Maréchal, B.; Ribes, D.; Abdulkadir, A.; Bach-Cuadra, M.; Daducci, A.; Granziera, C.; Klöppel, S.; Maeder, P.; et al. An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. Neuroimage Clin. 2015, 7, 7–17. [Google Scholar] [CrossRef]
- Ashburner, J.; Friston, K.J. Voxel-based morphometry--the methods. Neuroimage 2000, 11, 805–821. [Google Scholar] [CrossRef]
- Diaz-de-Grenu, L.Z.; Acosta-Cabronero, J.; Chong, Y.F.V.; Pereira, J.M.S.; Sajjadi, S.A.; Williams, G.B.; Nestor, P.J. A brief history of voxel-based grey matter analysis in Alzheimer’s disease. J. Alzheimer’s Dis. 2014, 38, 647–659. [Google Scholar] [CrossRef]
- Rolls, E.T.; Joliot, M.; Tzourio-Mazoyer, N. Implementation of a new parcellation of the orbitofrontal cortex in the automated anatomical labeling atlas. Neuroimage 2015, 122, 1–5. [Google Scholar] [CrossRef]
- De Marco, M.; Manca, R.; Kirby, J.; Hautbergue, G.M.; Blackburn, D.J.; Wharton, S.B.; Venneri, A.; Alzheimer’s Disease Neuroimaging Initiative. The Association between Polygenic Hazard and Markers of Alzheimer’s Disease Following Stratification for APOE Genotype. Curr. Alzheimer Res. 2020, 17, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Altmann, A.; Scelsi, M.A.; Shoai, M.; Silva, E.d.; Aksman, L.M.; Cash, D.M.; Hardy, J.; Schott, J.M.; Initiative, A.D.N. A comprehensive analysis of methods for assessing polygenic burden on Alzheimer’s disease pathology and risk beyond APOE. Brain Commun. 2019, 2, fcz047. [Google Scholar] [CrossRef]
- Manca, R.; Mitolo, M.; Bacalini, M.G.; Capellari, S.; Venneri, A. The impact of polygenic risk for Alzheimer’s disease on neurotransmitter-related grey matter atrophy in the Alzheimer continuum. Neurol. Sci. 2025, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Z.; Fan, D.; Shen, Y.; Chen, D.; Li, H.; Li, L.; Yang, H.; Liu, Y.; Bu, X.; et al. Association of Polygenic Risk Score with Age at Onset and Cerebrospinal Fluid Biomarkers of Alzheimer’s Disease in a Chinese Cohort. Neurosci. Bull. 2020, 36, 696–704. [Google Scholar] [CrossRef]
- Xicota, L.; Gyorgy, B.; Grenier-Boley, B.; Lecoeur, A.; Fontaine, G.; Danjou, F.; Gonzalez, J.S.; Colliot, O.; Amouyel, P.; Martin, G.; et al. Association of APOE-Independent Alzheimer Disease Polygenic Risk Score with Brain Amyloid Deposition in Asymptomatic Older Adults. Neurology 2022, 99, e462–e475. [Google Scholar] [CrossRef]
- Foley, S.F.; Tansey, K.E.; Caseras, X.; Lancaster, T.; Bracht, T.; Parker, G.; Hall, J.; Williams, J.; Linden, D.E.J. Multimodal Brain Imaging Reveals Structural Differences in Alzheimer’s Disease Polygenic Risk Carriers: A Study in Healthy Young Adults. Biol. Psychiatry 2017, 81, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Yu, H.; Cai, X.; Zhang, Y.; Pu, W.; Gao, B. A comparative study of posterior cingulate metabolism in patients with mild cognitive impairment due to Parkinson’s disease or Alzheimer’s disease. Sci. Rep. 2023, 13, 14241. [Google Scholar] [CrossRef] [PubMed]
- Poulin, S.P.; Dautoff, R.; Morris, J.C.; Barrett, L.F.; Dickerson, B.C. Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Res. 2011, 194, 7–13. [Google Scholar] [CrossRef]
- Rao, Y.L.; Ganaraja, B.; Murlimanju, B.V.; Joy, T.; Krishnamurthy, A.; Agrawal, A. Hippocampus and its involvement in Alzheimer’s disease: A review. 3 Biotech 2022, 12, 55. [Google Scholar] [CrossRef]
- Van Hoesen, G.W.; Augustinack, J.C.; Dierking, J.; Redman, S.J.; Thangavel, R. The parahippocampal gyrus in Alzheimer’s disease. Clinical and preclinical neuroanatomical correlates. Ann. N. Y. Acad. Sci. 2000, 911, 254–274. [Google Scholar] [CrossRef]
- Pengas, G.; Hodges, J.R.; Watson, P.; Nestor, P.J. Focal posterior cingulate atrophy in incipient Alzheimer’s disease. Neurobiol. Aging 2010, 31, 25–33. [Google Scholar] [CrossRef]
- Harrison, T.M.; Mahmood, Z.; Lau, E.P.; Karacozoff, A.M.; Burggren, A.C.; Small, G.W.; Bookheimer, S.Y. An Alzheimer’s Disease Genetic Risk Score Predicts Longitudinal Thinning of Hippocampal Complex Subregions in Healthy Older Adults. eNeuro 2016, 3, ENEURO.0098-16.2016. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, S.; Kang, S.W.; Park, M.; Baik, K.; Yoo, H.S.; Chung, S.J.; Jeong, S.H.; Jung, J.H.; Lee, P.H.; et al. Interaction of CSF α-synuclein and amyloid beta in cognition and cortical atrophy. Alzheimer’s Dement. 2021, 13, e12177. [Google Scholar] [CrossRef]
- Sabuncu, M.R.; Buckner, R.L.; Smoller, J.W.; Lee, P.H.; Fischl, B.; Sperling, R.A. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb. Cortex 2012, 22, 2653–2661. [Google Scholar] [CrossRef]
- Xiao, E.; Chen, Q.; Goldman, A.L.; Tan, H.Y.; Healy, K.; Zoltick, B.; Das, S.; Kolachana, B.; Callicott, J.H.; Dickinson, D.; et al. Late-Onset Alzheimer’s Disease Polygenic Risk Profile Score Predicts Hippocampal Function. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2017, 2, 673–679. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Lowe, V.J.; Wiste, H.J.; Senjem, M.L.; Weigand, S.D.; Therneau, T.M.; Boeve, B.F.; Josephs, K.A.; Fang, P.; Pandey, M.K.; Murray, M.E.; et al. Widespread brain tau and its association with ageing, Braak stage and Alzheimer’s dementia. Brain 2018, 141, 271–287. [Google Scholar] [CrossRef]
- Leal, S.L.; Lockhart, S.N.; Maass, A.; Bell, R.K.; Jagust, W.J. Subthreshold Amyloid Predicts Tau Deposition in Aging. J. Neurosci. 2018, 38, 4482–4489. [Google Scholar] [CrossRef]
- La Joie, R.; Visani, A.V.; Baker, S.L.; Brown, J.A.; Bourakova, V.; Cha, J.; Chaudhary, K.; Edwards, L.; Iaccarino, L.; Janabi, M.; et al. Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET. Sci. Transl. Med. 2020, 12, eaau5732. [Google Scholar] [CrossRef]
- Schäfer, A.; Chaggar, P.; Thompson, T.B.; Goriely, A.; Kuhl, E. Predicting brain atrophy from tau pathology: A summary of clinical findings and their translation into personalized models. Brain Multiphysics 2021, 2, 100039. [Google Scholar] [CrossRef]
- Convit, A.; de Asis, J.; de Leon, M.J.; Tarshish, C.Y.; De Santi, S.; Rusinek, H. Atrophy of the medial occipitotemporal, inferior, and middle temporal gyri in non-demented elderly predict decline to Alzheimer’s disease. Neurobiol. Aging 2000, 21, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Bejanin, A.; Schonhaut, D.R.; La Joie, R.; Kramer, J.H.; Baker, S.L.; Sosa, N.; Ayakta, N.; Cantwell, A.; Janabi, M.; Lauriola, M.; et al. Tau pathology and neurodegeneration contribute to cognitive impairment in Alzheimer’s disease. Brain 2017, 140, 3286–3300. [Google Scholar] [CrossRef]
- Filiou, R.; Bier, N.; Slegers, A.; Houzé, B.; Belchior, P.; Brambati, S.M. Connected speech assessment in the early detection of Alzheimer’s disease and mild cognitive impairment: A scoping review. Aphasiology 2020, 34, 723–755. [Google Scholar] [CrossRef]
- Petti, U.; Baker, S.; Korhonen, A. A systematic literature review of automatic Alzheimer’s disease detection from speech and language. J. Am. Med. Inf. Assoc. 2020, 27, 1784–1797. [Google Scholar] [CrossRef]
- Minoshima, S.; Giordani, B.; Berent, S.; Frey, K.A.; Foster, N.L.; Kuhl, D.E. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer’s disease. Ann. Neurol. 1997, 42, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Anchisi, D.; Borroni, B.; Franceschi, M.; Kerrouche, N.; Kalbe, E.; Beuthien-Beumann, B.; Cappa, S.; Lenz, O.; Ludecke, S.; Marcone, A.; et al. Heterogeneity of brain glucose metabolism in mild cognitive impairment and clinical progression to Alzheimer disease. Arch. Neurol. 2005, 62, 1728–1733. [Google Scholar] [CrossRef]
- Rane Levendovszky, S. Cross-Sectional and Longitudinal Hippocampal Atrophy, Not Cortical Thinning, Occurs in Amyloid-Negative, p-Tau-Positive, Older Adults With Non-Amyloid Pathology and Mild Cognitive Impairment. Front. Neuroimaging 2022, 1, 828767. [Google Scholar] [CrossRef]
- Josephs, K.A.; Murray, M.E.; Tosakulwong, N.; Whitwell, J.L.; Knopman, D.S.; Machulda, M.M.; Weigand, S.D.; Boeve, B.F.; Kantarci, K.; Petrucelli, L.; et al. Tau aggregation influences cognition and hippocampal atrophy in the absence of beta-amyloid: A clinico-imaging-pathological study of primary age-related tauopathy (PART). Acta Neuropathol. 2017, 133, 705–715. [Google Scholar] [CrossRef]
- Echávarri, C.; Aalten, P.; Uylings, H.B.M.; Jacobs, H.I.L.; Visser, P.J.; Gronenschild, E.H.B.M.; Verhey, F.R.J.; Burgmans, S. Atrophy in the parahippocampal gyrus as an early biomarker of Alzheimer’s disease. Brain Struct. Funct. 2011, 215, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Hamza, E.A.; Moustafa, A.A.; Tindle, R.; Karki, R.; Nalla, S.; Hamid, M.S.; El Haj, M. Effect of APOE4 Allele and Gender on the Rate of Atrophy in the Hippocampus, Entorhinal Cortex, and Fusiform Gyrus in Alzheimer’s Disease. Curr. Alzheimer Res. 2023, 19, 943–953. [Google Scholar] [CrossRef]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef]
- Mu, Y.; Gage, F.H. Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Mol. Neurodegener. 2011, 6, 85. [Google Scholar] [CrossRef]
- Killiany, R.J.; Hyman, B.T.; Gomez-Isla, T.; Moss, M.B.; Kikinis, R.; Jolesz, F.; Tanzi, R.; Jones, K.; Albert, M.S. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology 2002, 58, 1188–1196. [Google Scholar] [CrossRef]
- Scahill, R.I.; Schott, J.M.; Stevens, J.M.; Rossor, M.N.; Fox, N.C. Mapping the evolution of regional atrophy in Alzheimer’s disease: Unbiased analysis of fluid-registered serial MRI. Proc. Natl. Acad. Sci. USA 2002, 99, 4703–4707. [Google Scholar] [CrossRef]
- Mueller, S.G.; Schuff, N.; Yaffe, K.; Madison, C.; Miller, B.; Weiner, M.W. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer’s disease. Hum. Brain Mapp. 2010, 31, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, M.E.; Pike, G.B. MRI of healthy brain aging: A review. NMR Biomed. 2021, 34, e4564. [Google Scholar] [CrossRef] [PubMed]
- Capogna, E.; Manca, R.; De Marco, M.; Hall, A.; Soininen, H.; Venneri, A. Understanding the effect of cognitive/brain reserve and depression on regional atrophy in early Alzheimer’s disease. Postgrad. Med. 2019, 131, 533–538. [Google Scholar] [CrossRef] [PubMed]
| Variable | A+N+ (n = 114) | A+N− (n = 114) | A−N− (n = 114) | χ2 | p |
|---|---|---|---|---|---|
| Age (years) | 78.2 ± 6.8 | 77.6 ± 6.3 | 76.5 ± 6.2 | 4.58 | 0.101 |
| Education (years) | 16.3 ± 2.9 | 16.2 ± 2.7 | 16.3 ± 2.7 | 0.26 | 0.878 |
| Sex (m/f) | 81/33 | 84/30 | 82/32 | 0.20 | 0.903 |
| Diagnosis (CU/MCI/AD) | 64/48/60 a,b | 53/55/6 a | 25/70/19 | 100.38 | <0.001 |
| APOE (carrier/non-carrier) | 69/45 a | 60/54 a | 14/100 | 62.76 | <0.001 |
| PRS1 | PRS2 | |||||
|---|---|---|---|---|---|---|
| β | SE | p | β | SE | p | |
| A+N+ | ||||||
| Left amygdala | −0.025 | 0.010 | 0.016 | −0.025 | 0.010 | 0.017 |
| Left parahippocampal gyrus | −0.067 | 0.031 | 0.031 | −0.068 | 0.031 | 0.028 |
| A−N− | ||||||
| Right posterior cingulate cortex | −0.045 | 0.015 | 0.003 | −0.046 | 0.014 | 0.002 |
| A+N− | ||||||
| Right hippocampus | −0.061 | 0.029 | 0.039 | −0.058 | 0.029 | 0.047 |
| PRS1 | PRS2 | |||||
|---|---|---|---|---|---|---|
| β | SE | p | β | SE | p | |
| A+N+ non-carriers | ||||||
| Left amygdala | n.s. | n.s. | n.s. | −0.040 | 0.020 | 0.048 |
| A−N− non-carriers | ||||||
| Left superior temporal gyrus | −0.274 | 0.127 | 0.030 | −0.281 | 0.125 | 0.025 |
| Right superior temporal gyrus | −0.322 | 0.153 | 0.036 | −0.325 | 0.149 | 0.029 |
| Right posterior cingulate cortex | −0.107 | 0.048 | 0.025 | n.s. | n.s. | n.s. |
| A−N− carriers | ||||||
| Left amygdala | 0.030 | 0.000 | <0.001 | 0.032 | 0.000 | <0.001 |
| Right amygdala | −0.002 | 0.000 | <0.001 | −0.003 | 0.000 | <0.001 |
| Left hippocampus | 0.108 | 0.000 | <0.001 | 0.121 | 0.000 | <0.001 |
| Right hippocampus | 0.177 | 0.000 | <0.001 | 0.202 | 0.000 | <0.001 |
| Left parahippocampal gyrus | 0.136 | 0.000 | <0.001 | 0.155 | 0.000 | <0.001 |
| Right parahippocampal gyrus | 0.249 | 0.000 | <0.001 | 0.284 | 0.000 | <0.001 |
| Left middle temporal gyrus | 0.631 | 0.005 | <0.001 | 0.714 | 0.003 | <0.001 |
| Right middle temporal gyrus | 0.204 | 0.003 | <0.001 | 0.231 | 0.002 | <0.001 |
| Left superior temporal gyrus | 0.556 | 0.001 | <0.001 | 0.627 | 0.001 | <0.001 |
| Right superior temporal gyrus | 0.368 | 0.001 | <0.001 | 0.417 | 0.001 | <0.001 |
| Left fusiform gyrus | 0.364 | 0.000 | <0.001 | 0.403 | 0.000 | <0.001 |
| Right fusiform gyrus | 0.026 | 0.000 | <0.001 | 0.028 | 0.000 | <0.001 |
| Left medial prefrontal cortex | 0.032 | 0.000 | <0.001 | 0.036 | 0.000 | <0.001 |
| Right medial prefrontal cortex | 0.111 | 0.000 | <0.001 | 0.124 | 0.000 | <0.001 |
| Left posterior cingulate cortex | −0.118 | 0.000 | <0.001 | −0.132 | 0.000 | <0.001 |
| Right posterior cingulate cortex | 0.020 | 0.000 | <0.001 | 0.023 | 0.000 | <0.001 |
| A+N− non-carriers | ||||||
| Left amygdala | −0.040 | 0.017 | 0.020 | n.s. | n.s. | n.s. |
| PRS1 | PRS2 | |||||
|---|---|---|---|---|---|---|
| β | SE | p | β | SE | p | |
| Left amygdala | −0.048 | 0.022 | 0.025 | −0.050 | 0.022 | 0.025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nocella, V.; Manca, R.; Venneri, A. Associations Between Polygenic Risk for Alzheimer’s Disease and Grey Matter Volume Are Dependent on APOE, Pathological and Diagnostic Status. Genes 2025, 16, 1128. https://doi.org/10.3390/genes16101128
Nocella V, Manca R, Venneri A. Associations Between Polygenic Risk for Alzheimer’s Disease and Grey Matter Volume Are Dependent on APOE, Pathological and Diagnostic Status. Genes. 2025; 16(10):1128. https://doi.org/10.3390/genes16101128
Chicago/Turabian StyleNocella, Valerio, Riccardo Manca, and Annalena Venneri. 2025. "Associations Between Polygenic Risk for Alzheimer’s Disease and Grey Matter Volume Are Dependent on APOE, Pathological and Diagnostic Status" Genes 16, no. 10: 1128. https://doi.org/10.3390/genes16101128
APA StyleNocella, V., Manca, R., & Venneri, A. (2025). Associations Between Polygenic Risk for Alzheimer’s Disease and Grey Matter Volume Are Dependent on APOE, Pathological and Diagnostic Status. Genes, 16(10), 1128. https://doi.org/10.3390/genes16101128






