The Identification of Auxin Response Factors and Expression Analyses of Different Floral Development Stages in Roses
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of ARF Genes in Roses
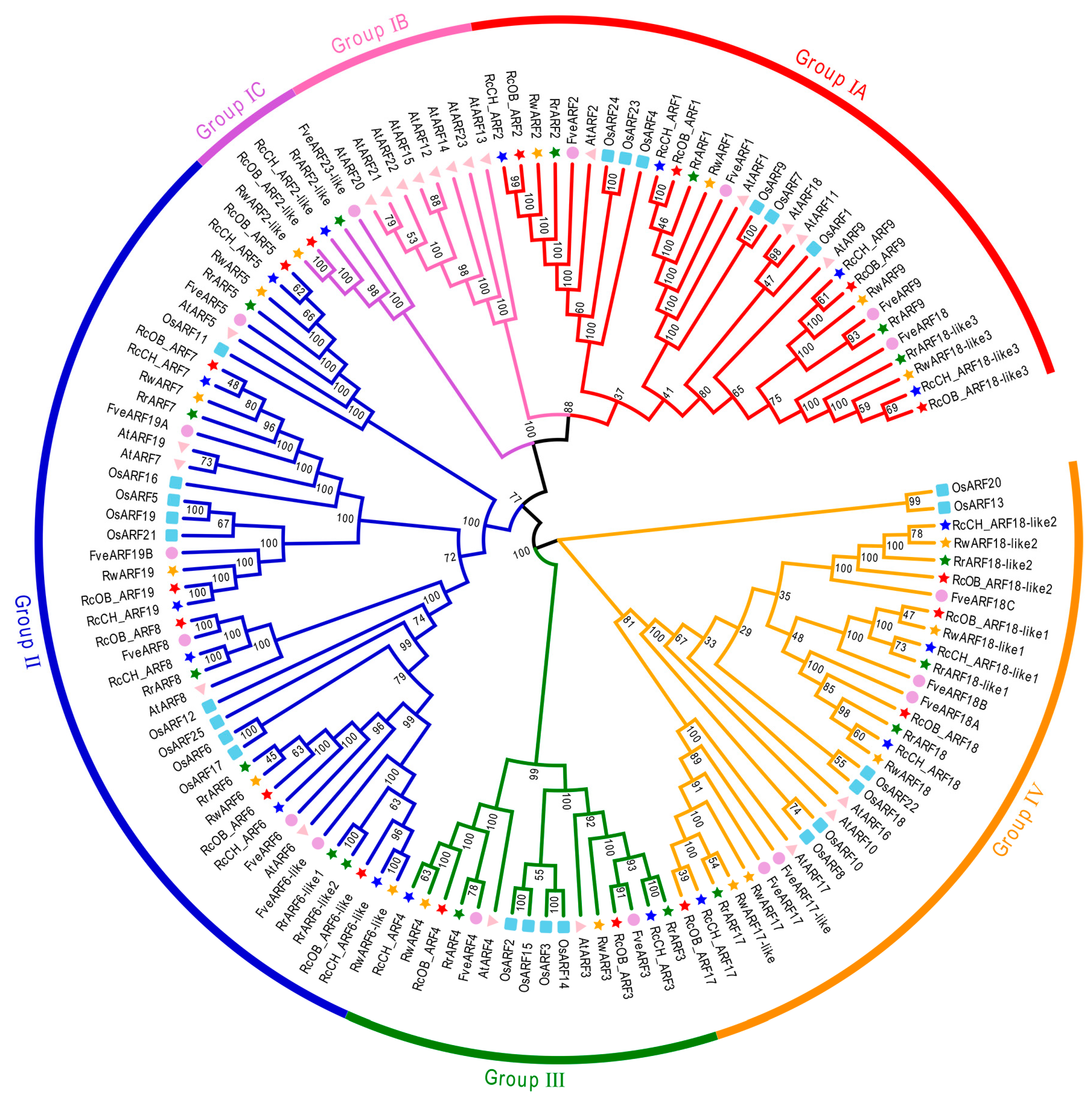
2.2. Phylogenetic Analysis of ARFs in Roses
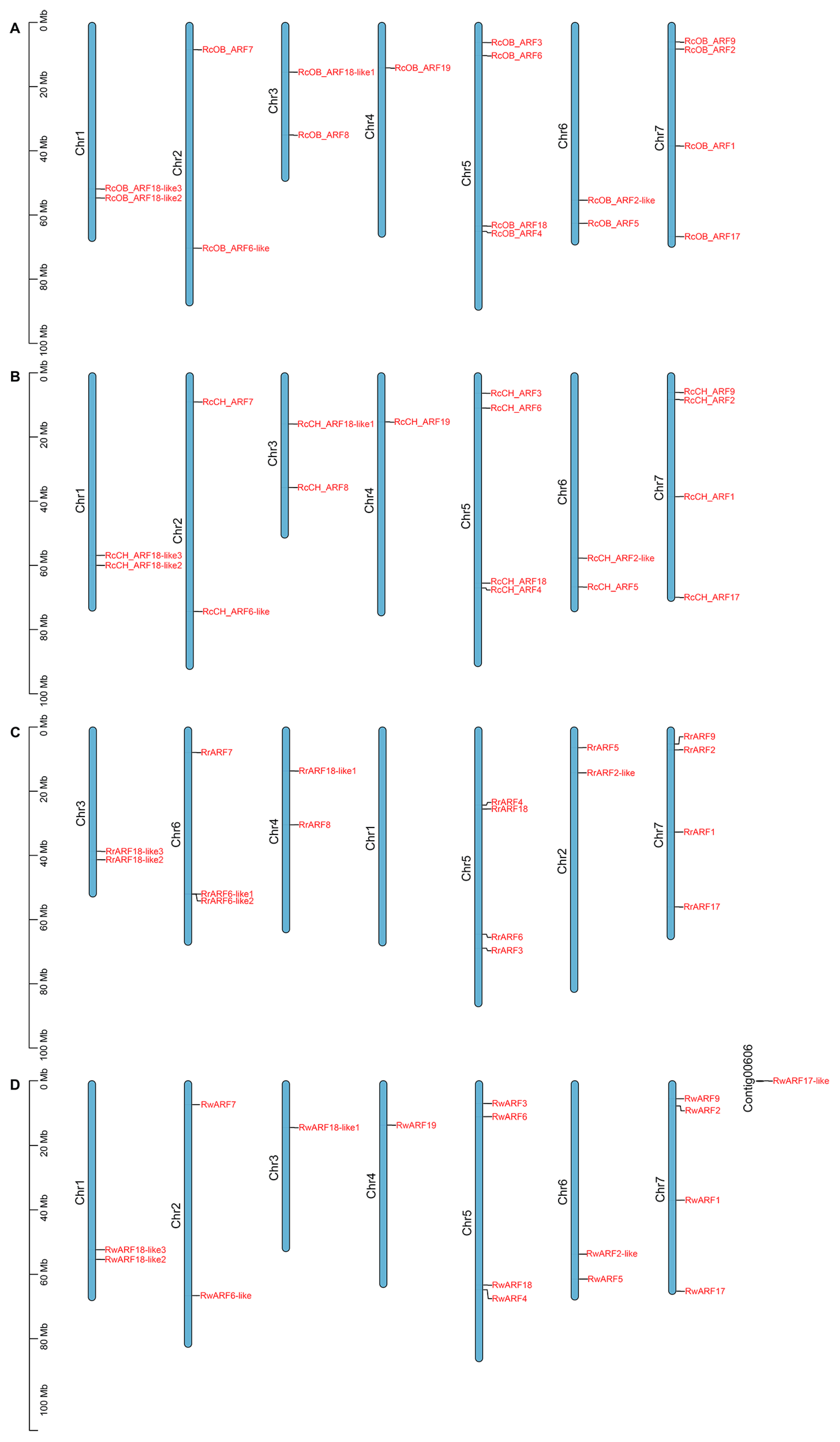
2.3. Chromosomal Location Analysis of ARFs
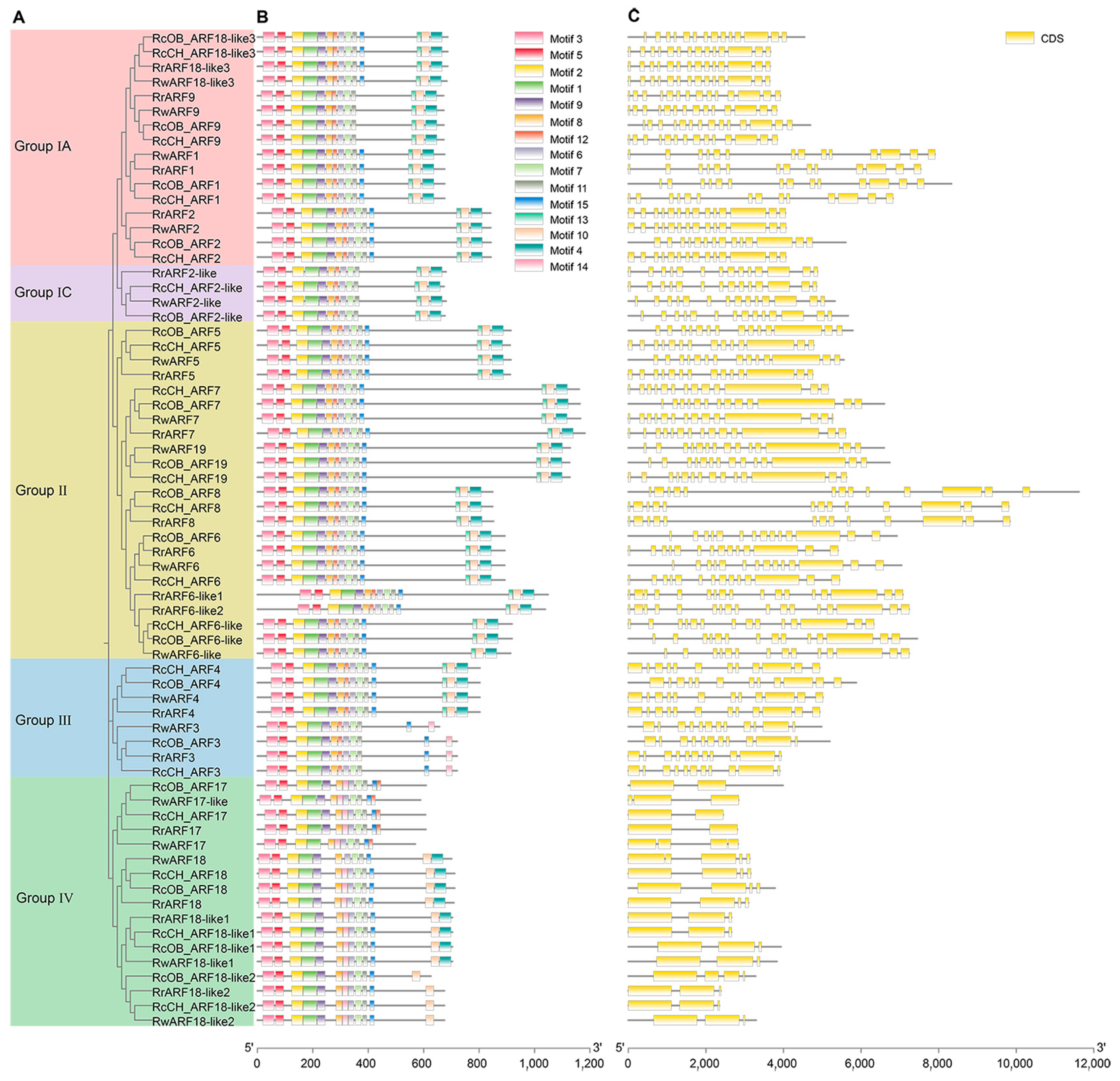
2.4. Motifs and Gene Structure Analysis of ARFs
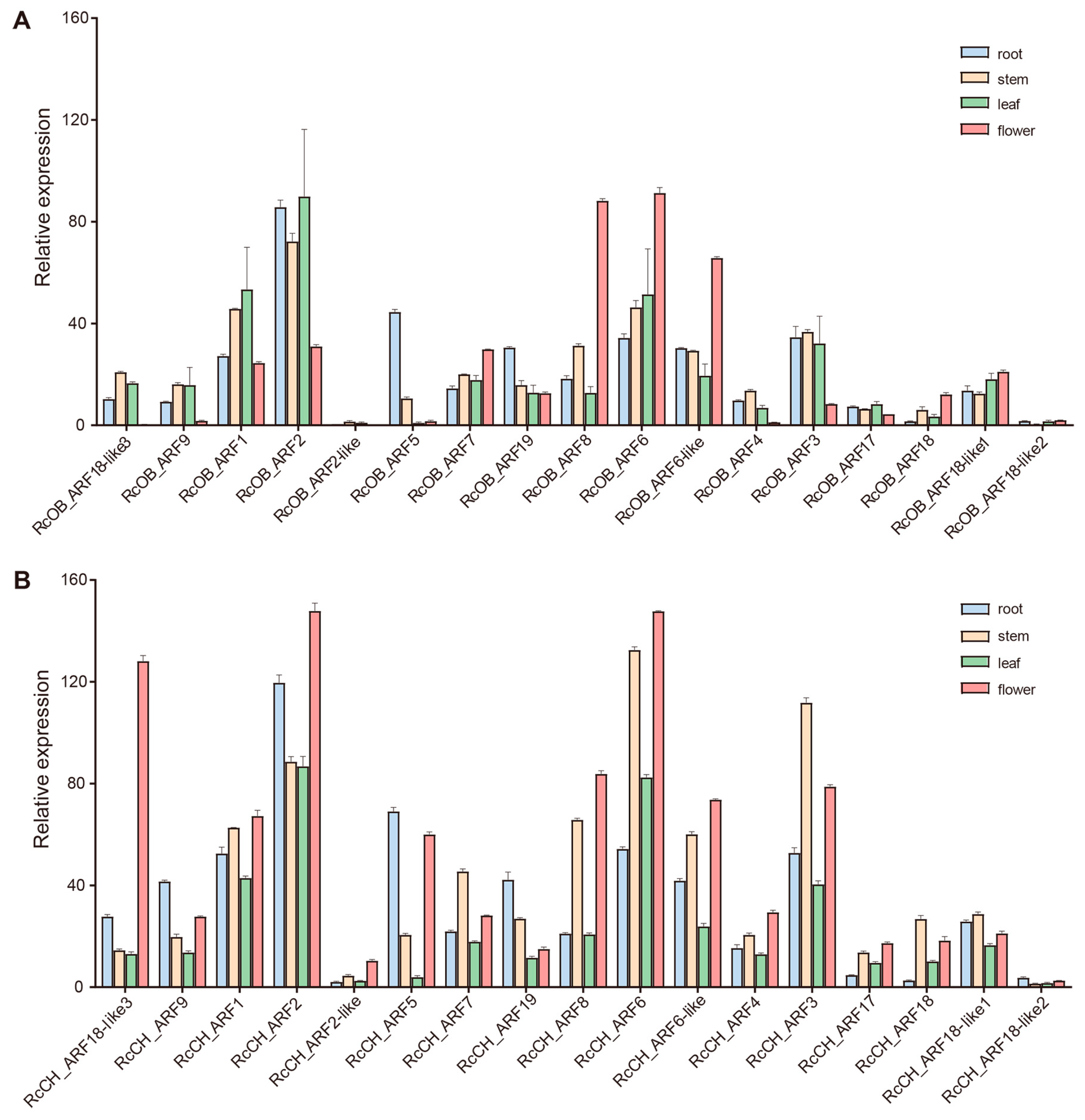
2.5. Transcriptome Analysis of ARFs
3. Results
3.1. Identification of ARF Genes in Four Rosa Cultivars
3.2. Phylogenetic Analysis
3.3. Chromosomal Locations of ARFs in Rosa
3.4. Motifs and Gene Structure Analysis of the Four Rosa Cultivars
3.5. Expression of ARFs in Roses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mockaitis, K.; Estelle, M. Auxin Receptors and Plant Development: A New Signaling Paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yahaya, B.S.; Li, J.; Wu, F. Enigmatic role of auxin response factors in plant growth and stress tolerance. Front. Plant Sci. 2024, 15, 1398818. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Nishihama, R.; Weijers, D.; Kohchi, T. Evolution of nuclear auxin signaling: Lessons from genetic studies with basal land plants. J. Exp. Bot. 2018, 69, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, S.; Qi, Y. Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 2022, 65, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Benkova, E.; Jäger, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jürgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005, 24, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Boer, D.R.; et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc. Nat. Acad. Sci. USA 2020, 117, 24557–24566. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, H.; Pan, Y.; Niu, Y.; Guo, L.; Ma, Y.; Tian, S.; Wei, J.; Wang, C.; Yang, X.; et al. Cell- and noncell-autonomous AUXIN RESPONSE FACTOR3 controls meristem proliferation and phyllotactic patterns. Plant Physiol. 2022, 190, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef]
- Liu, Z.; Miao, L.; Huo, R.; Song, X.; Johnson, C.; Kong, L.; Sundaresan, V.; Yu, X. ARF2–ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant Cell Physiol. 2018, 59, 179–189. [Google Scholar] [CrossRef]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.; Zouine, M.; Fu, Y.; Chateigner-Boutin, A.-L.; Mila, I.; Frasse, P.; Wang, H.; Audran, C.; Roustan, J.-P.; Bouzayen, M. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE 2014, 9, e84203. [Google Scholar] [CrossRef]
- Die, J.V.; Gil, J.; Millan, T. Genome-wide identification of the auxin response factor gene family in Cicer arietinum. BMC Genom. 2018, 19, 301. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, S.; Wu, F.; Wei, M.; Li, J.; Yang, F. Genome-wide identification and functional characterization of auxin response factor (ARF) genes in Eggplant. Int. J. Mol. Sci. 2022, 23, 6219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, W.; Shen, H.; Yang, L. Genome-wide identification and expression analysis of ARF gene family in embryonic development of Korean pine (Pinus koraiensis). BMC Plant Biol. 2024, 24, 267. [Google Scholar] [CrossRef] [PubMed]
- Si, C.; Zeng, D.; da Silva, J.A.T.; Qiu, S.; Duan, J.; Bai, S.; He, C. Genome-wide identification of Aux/IAA and ARF gene families reveal their potential roles in flower opening of Dendrobium officinale. BMC Genom. 2023, 24, 199. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, C.; Li, X.; Xu, H.; Liang, Y.; Ma, N.; Fei, Z.; Gao, J.; Jiang, C.Z.; Ma, C. Transcriptome Profiling of Petal Abscission Zone and Functional Analysis of an Aux/IAA Family Gene RhIAA16 Involved in Petal Shedding in Rose. Front. Plant Sci. 2016, 7, 1375. [Google Scholar] [CrossRef]
- Jing, W.; Zhang, S.; Fan, Y.; Deng, Y.; Wang, C.; Lu, J.; Sun, X.; Ma, N.; Shahid, M.O.; Li, Y.; et al. Molecular Evidences for the Interactions of Auxin, Gibberellin, and Cytokinin in Bent Peduncle Phenomenon in Rose (Rosa sp.). Int. J. Mol. Sci. 2020, 21, 1360. [Google Scholar] [CrossRef]
- Khan, R.U.; Khan, M.S.; Rashid, A.; Farooq, A. Effect of exogenous indole-3-acetic acid and naphthalene acetic acid on regeneration of damask rose cuttings in three growing media. Pak. J. Biol. Sci. 2007, 10, 3626–3631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Otiende, M.A.; Fricke, K.; Nyabundi, J.O.; Ngamau, K.; Hajirezaei, M.R.; Druege, U. Involvement of the auxin-cytokinin homeostasis in adventitious root formation of rose cuttings as affected by their nodal position in the stock plant. Planta 2021, 254, 65. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Bharti, N.; Singh, A.P.; Tripathi, S.K.; Pandey, S.P.; Chauhan, A.S.; Kulkarni, A.; Sane, A.P. Petal abscission in fragrant roses is associated with large scale differential regulation of the abscission zone transcriptome. Sci. Rep. 2020, 10, 17196. [Google Scholar] [CrossRef]
- Liang, Y.; Jiang, C.; Liu, Y.; Gao, Y.; Lu, J.; Aiwaili, P.; Fei, Z.; Jiang, C.Z.; Hong, B.; Ma, C.; et al. Auxin Regulates Sucrose Transport to Repress Petal Abscission in Rose (Rosa hybrida). Plant Cell 2020, 32, 3485–3499. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Li, Y.; Li, Y.; Wang, Y.; Jiang, C.; Choisy, P.; Xu, T.; Cai, Y.; Pei, D.; et al. AUXIN RESPONSE FACTOR 18-HISTONE DEACETYLASE 6 module regulates floral organ identity in rose (Rosa hybrida). Plant Physiol. 2021, 186, 1074–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hussain, N.; Ma, Y.; Zuo, L.; Jiang, Y.; Sun, X.; Gao, J. The ARF2-MYB6 module mediates auxin-regulated petal expansion in rose. J. Exp. Bot. 2023, 74, 4489–4502. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, Q.; Lan, L.; Peng, D.; Guan, H.; Luo, K.; Bao, M.; Bendahmane, M.; Fu, X.; Wu, Z. Haplotype-resolved genome assembly of the diploid Rosa chinensis provides insight into the mechanisms underlying key ornamental traits. Mol. Hortic. 2024, 4, 14. [Google Scholar] [CrossRef]
- Chen, F.; Su, L.; Hu, S.; Xue, J.Y.; Liu, H.; Liu, G.; Jiang, Y.; Du, J.; Qiao, Y.; Fan, Y.; et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic. Res. 2021, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.C.; Jiang, X.D.; Yang, G.Q.; Cui, W.H.; Suo, Z.Q.; Wang, W.J.; Sun, Y.B.; Wang, D.; Cheng, X.C.; Li, X.M.; et al. Rose without prickle: Genomic insights linked to moisture adaptation. Natl. Sci. Rev. 2021, 8, nwab092. [Google Scholar] [CrossRef]
- Luo, H.; Li, T.; Guan, Y.; Zhang, Z.; Zhang, Z.; Zhang, Z.; Li, H. FvemiR160-FveARF18A-FveAP1/FveFUL module regulates flowering time in woodland strawberry. Plant J. 2023, 117, 1130–1147. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yu, J.; Zhao, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Dissecting the genome-wide evolution and function of R2R3-MYB transcription factor family in Rosa chinensis. Genes 2019, 10, 823. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xiong, Y.; Kong, X.; Huang, G. Roles of auxin response factors in rice development and stress responses. Plant Cell Environ. 2022, 46, 1075–1086. [Google Scholar] [CrossRef]
- Dong, X.; Guan, Y.; Zhang, Z.; Li, H. miR390-tasiRNA3-ARF4 pathway is involved in regulating flowering time in woodland strawberry. Plant Cell Rep. 2022, 41, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhao, Y. A role for auxin in flower development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Goldental-Cohen, S.; Israeli, A.; Ori, N.; Yasuor, H. Auxin response dynamics during wild-type and entire flower development in tomato. Plant Cell Physiol. 2017, 58, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gong, M.; Xu, X.; Li, H.; Deng, W. Roles of auxin in the growth, development, and stress tolerance of horticultural plants. Cells 2022, 11, 2761. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Csukasi, F.; Catalá, C. Mechanisms regulating auxin action during fruit development. Physiol. Plantarum 2014, 151, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, J.; Xu, X.; Wang, R.; Liu, Y.; Huang, S.; Wei, H.; Wei, Z. Molecular Mechanisms of Diverse Auxin Responses during Plant Growth and Development. Int. J. Mol. Sci. 2022, 23, 12495. [Google Scholar] [CrossRef]
- Yi, S.N.; Mao, J.X.; Zhang, X.Y.; Li, X.M.; Zhang, Z.H.; Li, H. FveARF2 negatively regulates fruit ripening and quality in strawberry. Front. Plant Sci. 2022, 13, 1023739. [Google Scholar] [CrossRef] [PubMed]
- Li, B.J.; Shi, Y.N.; Xiao, Y.N.; Jia, H.R.; Yang, X.F.; Dai, Z.R.; Sun, Y.F.; Shou, J.H.; Jiang, G.H.; Grierson, D.; et al. AUXIN RESPONSE FACTOR 2 mediates repression of strawberry receptacle ripening via auxin-ABA interplay. Plant Physiol. 2024, 196, 2638–2653. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Ping, Y.; Bao, C.; Liu, C.; Tahir, M.M.; Li, X.; Song, Y.; Xu, W.; Ma, F.; Guan, Q. Mdm-miR160-MdARF17-MdWRKY33 module mediates freezing tolerance in apple. Plant J. 2023, 114, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Lu, Q.; Liu, Z.; Lv, T.; Li, X.; Bu, H.; Liu, W.; Xu, Y.; Yuan, H.; Wang, A. Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol. 2020, 226, 1781–1795. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; He, J.; Ping, Y.; Guo, J.; Hou, N.; Cao, F.; Li, X.; Geng, D.; Wang, S.; Chen, P.; et al. The positive feedback regulatory loop of miR160-Auxin Response Factor 17-HYPONASTIC LEAVES 1 mediates drought tolerance in apple trees. Plant Physiol. 2022, 188, 1686–1708. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.; Zhang, X.; Zheng, B.; Han, Y.; Zhang, R.X. PpARF6 acts as an integrator of auxin and ethylene signaling to promote fruit ripening in peach. Hortic. Res. 2023, 10, uhad158. [Google Scholar] [CrossRef]
- Chen, G.; Yue, Y.; Li, L.; Li, Y.; Li, H.; Ding, W.; Shi, T.; Yang, X.; Wang, L. Genome-wide identification of the auxin response factor (ARF) gene family and their expression analysis during flower development of Osmanthus fragrans. Forests 2020, 11, 245. [Google Scholar] [CrossRef]
- Liu, R.; Guo, Z.; Lu, S. Genome-wide identification and expression analysis of the Aux/IAA and auxin response factor gene family in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 10494. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, L.; Li, W.; Dang, Z.; Xie, Y.; Zhao, W.; Zhao, L.; Li, W.; Yang, C.; Xu, C.; et al. Genome-wide identification and expression analysis of auxin response factor gene family in Linum usitatissimum. Int. J. Mol. Sci. 2023, 24, 11006. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, K.; Guo, L.; Liu, X.; Zhang, Z. AUXIN RESPONSE FACTOR3 plays distinct role during early flower development. Plant Signal. Behav. 2018, 13, e1467690. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Vayssières, A.; Ó’Maoiléidigh, D.S.; Yang, X.; Vincent, C.; Bertran Garcia de Olalla, E.; Cerise, M.; Franzen, R.; Coupland, G. MicroRNA172 controls inflorescence meristem size through regulation of APETALA2 in Arabidopsis. New Phytol. 2022, 235, 356–371. [Google Scholar] [CrossRef] [PubMed]

| Species | Gene Name | Gene ID |
|---|---|---|
| R. chinensis ‘OB’ | RcOB_ARF18-like3 | RcHm_v2.0_Chr1g0360041 |
| RcOB_ARF9 | RcHm_v2.0_Chr7g0186081 | |
| RcOB_ARF1 | RcHm_v2.0_Chr7g0219771 | |
| RcOB_ARF2 | RcHm_v2.0_Chr7g0188911 | |
| RcOB_ARF2-like | RcHm_v2.0_Chr6g0292551 | |
| RcOB_ARF5 | RcHm_v2.0_Chr6g0302551 | |
| RcOB_ARF7 | RcHm_v2.0_Chr2g0095551 | |
| RcOB_ARF19 | RcHm_v2.0_Chr4g0397771 | |
| RcOB_ARF8 | RcHm_v2.0_Chr3g0487771 | |
| RcOB_ARF6 | RcHm_v2.0_Chr5g0014961 | |
| RcOB_ARF6-like | RcHm_v2.0_Chr2g0152951 | |
| RcOB_ARF4 | RcHm_v2.0_Chr5g0060011 | |
| RcOB_ARF3 | RcHm_v2.0_Chr5g0009381 | |
| RcOB_ARF17 | RcHm_v2.0_Chr7g0240691 | |
| RcOB_ARF18 | RcHm_v2.0_Chr5g0058761 | |
| RcOB_ARF18-like1 | RcHm_v2.0_Chr3g0469661 | |
| RcOB_ARF18-like2 | RcHm_v2.0_Chr1g0363241 | |
| R. chinensis ‘CH’ | RcCH_ARF18-like3 | evm.model.hB_v1.0_chr1.2384 |
| RcCH_ARF9 | evm.model.hB_v1.0_chr7.760 | |
| RcCH_ARF1 | evm.model.hB_v1.0_chr7.3082 | |
| RcCH_ARF2 | evm.model.hB_v1.0_chr7.966 | |
| RcCH_ARF2-like | evm.model.hB_v1.0_chr6.2903 | |
| RcCH_ARF5 | evm.model.hB_v1.0_chr6.3683 | |
| RcCH_ARF7 | evm.model.hB_v1.0_chr2.914 | |
| RcCH_ARF19 | evm.model.hB_v1.0_chr4.708 | |
| RcCH_ARF8 | evm.model.hB_v1.0_chr3.2842 | |
| RcCH_ARF6 | evm.model.hB_v1.0_chr5.1066 | |
| RcCH_ARF6-like | evm.model.hB_v1.0_chr2.4774 | |
| RcCH_ARF4 | evm.model.hB_v1.0_chr5.3774 | |
| RcCH_ARF3 | evm.model.hB_v1.0_chr5.662 | |
| RcCH_ARF17 | evm.model.hB_v1.0_chr7.4543 | |
| RcCH_ARF18 | evm.model.hB_v1.0_chr5.3710 | |
| RcCH_ARF18-like1 | evm.model.hB_v1.0_chr3.1581 | |
| RcCH_ARF18-like2 | evm.model.hB_v1.0_chr1.2612 | |
| R. rugosa | RrARF18-like3 | evm.model.Chr3.3276 |
| RrARF9 | evm.model.Chr7.749 | |
| RrARF1 | evm.model.Chr7.3607 | |
| RrARF2 | evm.model.Chr7.969 | |
| RrARF2-like | evm.model.Chr2.1948 | |
| RrARF5 | evm.model.Chr2.923 | |
| RrARF7 | evm.model.Chr6.1049 | |
| RrARF8 | evm.model.Chr4.3418 | |
| RrARF6 | evm.model.Chr5.5782 | |
| RrARF6-like1 | evm.model.Chr6.5248 | |
| RrARF6-like2 | evm.model.Chr6.5260 | |
| RrARF4 | evm.model.Chr5.2304 | |
| RrARF3 | evm.model.Chr5.6284 | |
| RrARF17 | evm.model.Chr7.5673 | |
| RrARF18 | evm.model.Chr5.2402 | |
| RrARF18-like1 | evm.model.Chr4.1801 | |
| RrARF18-like2 | evm.model.Chr3.3560 | |
| R. wichurana | RwARF18-like3 | Rw1G025590.1 |
| RwARF9 | Rw7G006530.1 | |
| RwARF1 | Rw7G027920.1 | |
| RwARF2 | Rw7G008560.1 | |
| RwARF2-like | Rw6G029710.1 | |
| RwARF5 | Rw6G037050.1 | |
| RwARF7 | Rw2G007600.1 | |
| RwARF19 | Rw4G006600.1 | |
| RwARF6 | Rw5G010120.1 | |
| RwARF6-like | Rw2G040900.1 | |
| RwARF4 | Rw5G036880.1 | |
| RwARF3 | Rw5G006830.1 | |
| RwARF17-like | Rw0G013480.1 | |
| RwARF17 | Rw7G041690.1 | |
| RwARF18 | Rw5G036140.1 | |
| RwARF18-like1 | Rw3G014400.1 | |
| RwARF18-like2 | Rw1G027880.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, R.; Zhang, X.; Luo, K.; Tembrock, L.R.; Li, S.; Wu, Z. The Identification of Auxin Response Factors and Expression Analyses of Different Floral Development Stages in Roses. Genes 2025, 16, 41. https://doi.org/10.3390/genes16010041
Huang R, Zhang X, Luo K, Tembrock LR, Li S, Wu Z. The Identification of Auxin Response Factors and Expression Analyses of Different Floral Development Stages in Roses. Genes. 2025; 16(1):41. https://doi.org/10.3390/genes16010041
Chicago/Turabian StyleHuang, Rui, Xiaoni Zhang, Kaiqing Luo, Luke R. Tembrock, Sen Li, and Zhiqiang Wu. 2025. "The Identification of Auxin Response Factors and Expression Analyses of Different Floral Development Stages in Roses" Genes 16, no. 1: 41. https://doi.org/10.3390/genes16010041
APA StyleHuang, R., Zhang, X., Luo, K., Tembrock, L. R., Li, S., & Wu, Z. (2025). The Identification of Auxin Response Factors and Expression Analyses of Different Floral Development Stages in Roses. Genes, 16(1), 41. https://doi.org/10.3390/genes16010041







