Prevalence of IMPG1 and IMPG2 Mutations Leading to Retinitis Pigmentosa or Vitelliform Macular Dystrophy in a Cohort of Patients with Inherited Retinal Dystrophies
Abstract
1. Introduction
2. Methods
2.1. Subjects and Examinations
2.2. Pathogenicity Prediction
2.3. Modeling and Evaluation of Modeled Structures
2.4. Protein Stability Prediction
2.5. Interatomic Analysis in Reference and Mutant Proteins
2.6. Physiochemical Analysis
3. Results
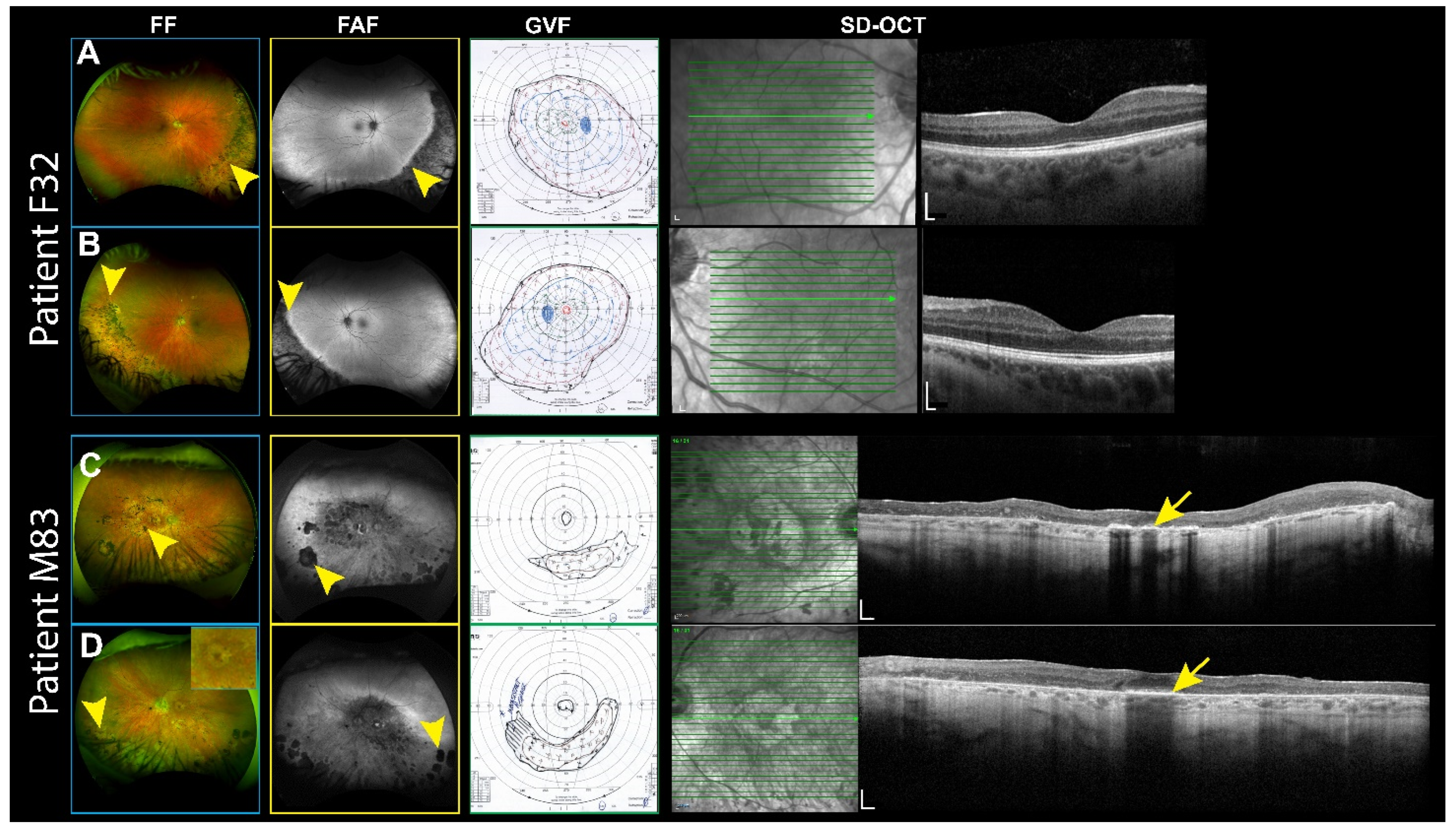
3.1. RP Patients
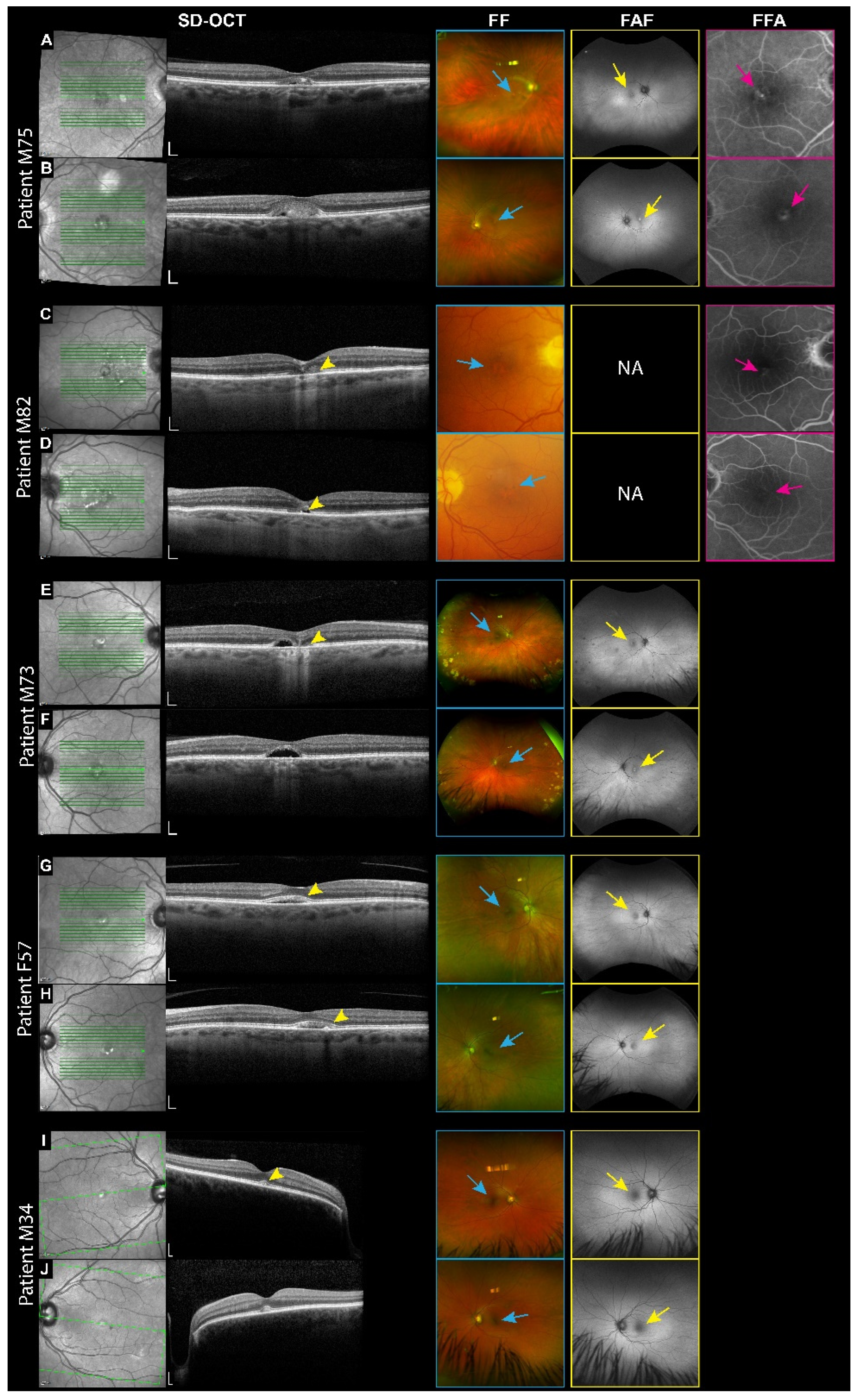
3.2. VMD Patients
3.3. Pathogenicity Prediction
3.4. Prediction of Protein Stability for Point Mutations
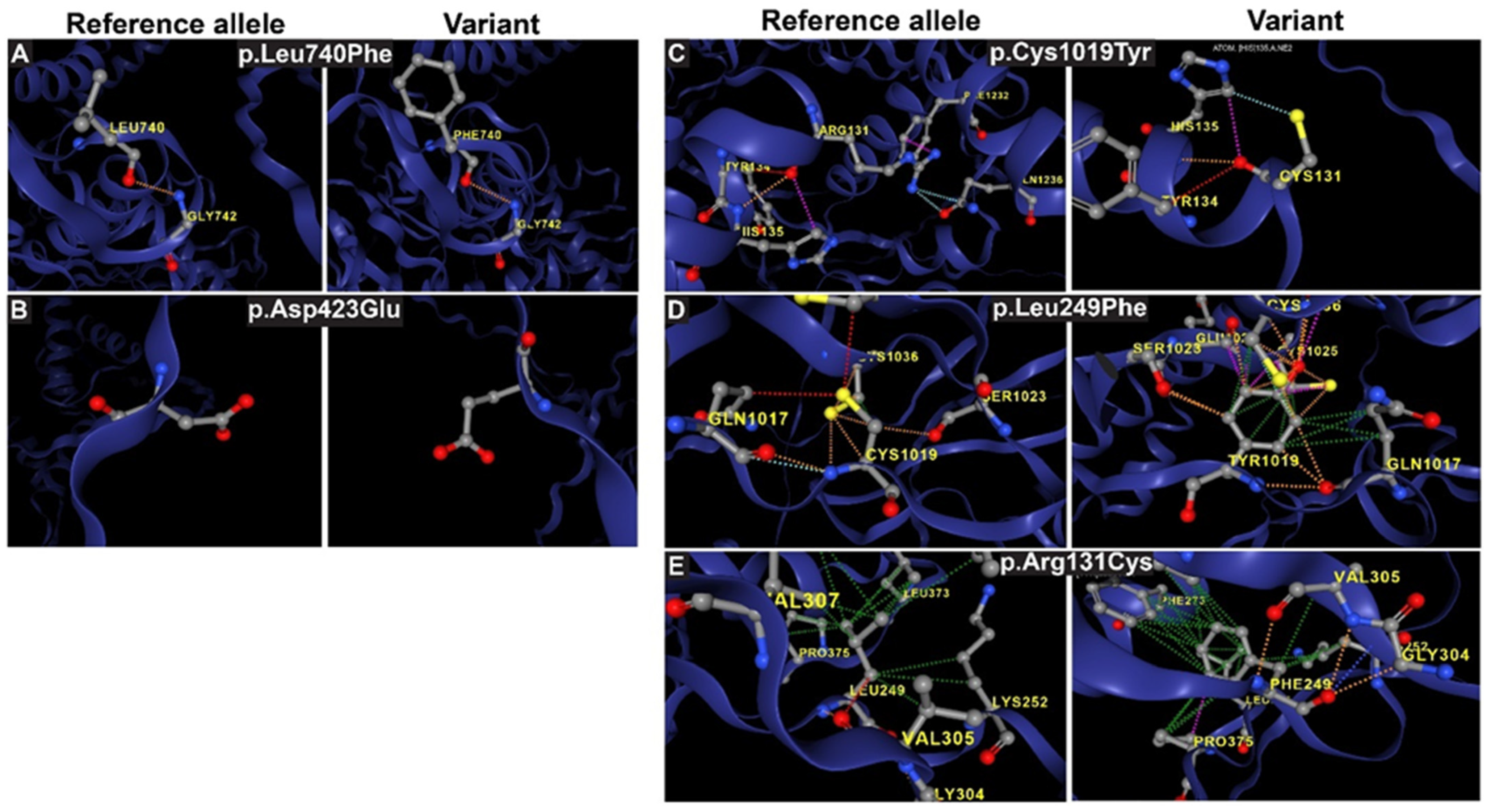
3.5. Analysis of Interatomic Interactions in Reference and Mutant IMPG1/2 Proteins
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishikawa, M.; Sawada, Y.; Yoshitomi, T. Structure and function of the interphotoreceptor matrix surrounding retinal photoreceptor cells. Exp. Eye Res. 2015, 133, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Felemban, M.; Dorgau, B.; Hunt, N.C.; Hallam, D.; Zerti, D.; Bauer, R.; Ding, Y.; Collin, J.; Steel, D.; Krasnogor, N.; et al. Extracellular matrix component expression in human pluripotent stem cell-derived retinal organoids recapitulates retinogenesis in vivo and reveals an important role for IMPG1 and CD44 in the development of photoreceptors and interphotoreceptor matrix. Acta Biomater. 2018, 74, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.B. Retina Metabolism and Metabolism in the Pigmented Epithelium: A Busy Intersection. Annu. Rev. Vis. Sci. 2021, 7, 665–692. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.B.; Lindsay, K.J.; Du, J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015, 93, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, Y.; Du, J.; Salido, E.M. Retinal Metabolic Profile on IMPG2 Deficiency Mice with Subretinal Lesions. Adv. Exp. Med. Biol. 2023, 1415, 457–463. [Google Scholar] [CrossRef]
- Acharya, S.; Foletta, V.C.; Lee, J.W.; Rayborn, M.E.; Rodriguez, I.R.; Young, W.S., 3rd; Hollyfield, J.G. SPACRCAN, a novel human interphotoreceptor matrix hyaluronan-binding proteoglycan synthesized by photoreceptors and pinealocytes. J. Biol. Chem. 2000, 275, 6945–6955. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lee, J.W.; Nishiyama, K.; Shadrach, K.G.; Rayborn, M.E.; Hollyfield, J.G. SPACRCAN in the interphotoreceptor matrix of the mouse retina: Molecular, developmental and promoter analysis. Exp. Eye Res. 2003, 76, 1–14. [Google Scholar] [CrossRef]
- Acharya, S.; Rodriguez, I.R.; Moreira, E.F.; Midura, R.J.; Misono, K.; Todres, E.; Hollyfield, J.G. SPACR, a novel interphotoreceptor matrix glycoprotein in human retina that interacts with hyaluronan. J. Biol. Chem. 1998, 273, 31599–31606. [Google Scholar] [CrossRef]
- Kuehn, M.H.; Hageman, G.S. Expression and characterization of the IPM 150 gene (IMPG1) product, a novel human photoreceptor cell-associated chondroitin-sulfate proteoglycan. Matrix Biol. 1999, 18, 509–518. [Google Scholar] [CrossRef]
- Hollyfield, J.G.; Rayborn, M.E.; Nishiyama, K.; Shadrach, K.G.; Miyagi, M.; Crabb, J.W.; Rodriguez, I.R. Interphotoreceptor matrix in the fovea and peripheral retina of the primate Macaca mulatta: Distribution and glycoforms of SPACR and SPACRCAN. Exp. Eye Res. 2001, 72, 49–61. [Google Scholar] [CrossRef]
- Acharya, S.; Rayborn, M.E.; Hollyfield, J.G. Characterization of SPACR, a sialoprotein associated with cones and rods present in the interphotoreceptor matrix of the human retina: Immunological and lectin binding analysis. Glycobiology 1998, 8, 997–1006. [Google Scholar] [CrossRef][Green Version]
- Kuehn, M.H.; Hageman, G.S. Molecular characterization and genomic mapping of human IPM 200, a second member of a novel family of proteoglycans. Mol. Cell Biol. Res. Commun. 1999, 2, 103–110. [Google Scholar] [CrossRef]
- Lee, J.W.; Chen, Q.; Rayborn, M.E.; Shadrach, K.G.; Crabb, J.W.; Rodriguez, I.R.; Hollyfield, J.G. SPACR in the interphotoreceptor matrix of the mouse retina: Molecular, biochemical and immunohistochemical characterization. Exp. Eye Res. 2000, 71, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Salido, E.M.; Ramamurthy, V. Proteoglycan IMPG2 Shapes the Interphotoreceptor Matrix and Modulates Vision. J. Neurosci. 2020, 40, 4059–4072. [Google Scholar] [CrossRef] [PubMed]
- Hollyfield, J.G.; Rayborn, M.E.; Midura, R.J.; Shadrach, K.G.; Acharya, S. Chondroitin sulfate proteoglycan core proteins in the interphotoreceptor matrix: A comparative study using biochemical and immunohistochemical analysis. Exp. Eye Res. 1999, 69, 311–322. [Google Scholar] [CrossRef]
- Mitchell, B.; Coulter, C.; Geldenhuys, W.J.; Rhodes, S.; Salido, E.M. Interphotoreceptor matrix proteoglycans IMPG1 and IMPG2 proteolyze in the SEA domain and reveal localization mutual dependency. Sci. Rep. 2022, 12, 15535. [Google Scholar] [CrossRef]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Hanany, M.; Shalom, S.; Ben-Yosef, T.; Sharon, D. Comparison of Worldwide Disease Prevalence and Genetic Prevalence of Inherited Retinal Diseases and Variant Interpretation Considerations. Cold Spring Harb. Perspect. Med. 2024, 14, a041277. [Google Scholar] [CrossRef] [PubMed]
- Daiger, S.P.; Sullivan, L.S.; Bowne, S.J. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013, 84, 132–141. [Google Scholar] [CrossRef]
- Wright, A.F.; Chakarova, C.F.; Abd El-Aziz, M.M.; Bhattacharya, S.S. Photoreceptor degeneration: Genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet. 2010, 11, 273–284. [Google Scholar] [CrossRef]
- Khan, A.O.; Al Teneiji, A.M. Homozygous and heterozygous retinal phenotypes in families harbouring IMPG2 mutations. Ophthalmic Genet. 2019, 40, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Bandah-Rozenfeld, D.; Collin, R.W.; Banin, E.; van den Born, L.I.; Coene, K.L.; Siemiatkowska, A.M.; Zelinger, L.; Khan, M.I.; Lefeber, D.J.; Erdinest, I.; et al. Mutations in IMPG2, encoding interphotoreceptor matrix proteoglycan 2, cause autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010, 87, 199–208. [Google Scholar] [CrossRef]
- van Lith-Verhoeven, J.J.; Hoyng, C.B.; van den Helm, B.; Deutman, A.F.; Brink, H.M.; Kemperman, M.H.; de Jong, W.H.; Kremer, H.; Cremers, F.P. The benign concentric annular macular dystrophy locus maps to 6p12.3-q16. Investig. Ophthalmol. Vis. Sci. 2004, 45, 30–35. [Google Scholar] [CrossRef] [PubMed]
- van Huet, R.A.; Collin, R.W.; Siemiatkowska, A.M.; Klaver, C.C.; Hoyng, C.B.; Simonelli, F.; Khan, M.I.; Qamar, R.; Banin, E.; Cremers, F.P.; et al. IMPG2-associated retinitis pigmentosa displays relatively early macular involvement. Investig. Ophthalmol. Vis. Sci. 2014, 55, 3939–3953. [Google Scholar] [CrossRef]
- Gonzalez-Gomez, A.; Romero-Trevejo, J.L.; Garcia-Ben, A.; Garcia-Campos, J.M. Bull’s eye maculopathy caused by a novel IMPG-1 mutation. Ophthalmic Genet. 2019, 40, 71–73. [Google Scholar] [CrossRef]
- Meunier, I.; Manes, G.; Bocquet, B.; Marquette, V.; Baudoin, C.; Puech, B.; Defoort-Dhellemmes, S.; Audo, I.; Verdet, R.; Arndt, C.; et al. Frequency and clinical pattern of vitelliform macular dystrophy caused by mutations of interphotoreceptor matrix IMPG1 and IMPG2 genes. Ophthalmology 2014, 121, 2406–2414. [Google Scholar] [CrossRef] [PubMed]
- Brandl, C.; Schulz, H.L.; Charbel Issa, P.; Birtel, J.; Bergholz, R.; Lange, C.; Dahlke, C.; Zobor, D.; Weber, B.H.F.; Stohr, H. Mutations in the Genes for Interphotoreceptor Matrix Proteoglycans, IMPG1 and IMPG2, in Patients with Vitelliform Macular Lesions. Genes 2017, 8, 170. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Aguilera, N.; Bower, A.J.; Li, J.; Ullah, E.; Dubra, A.; Cukras, C.; Brooks, B.P.; Jeffrey, B.G.; Hufnagel, R.B.; et al. Photoreceptor and Retinal Pigment Epithelium Relationships in Eyes With Vitelliform Macular Dystrophy Revealed by Multimodal Adaptive Optics Imaging. Investig. Opthalmology Vis. Sci. 2022, 63, 27. [Google Scholar] [CrossRef]
- Chatterjee, S.; Gupta, S.; Chaudhry, V.N.; Chaudhry, P.; Mukherjee, A.; Mutsuddi, M. Whole exome sequencing identifies a novel splice-site mutation in IMPG2 gene causing Stargardt-like juvenile macular dystrophy in a north Indian family. Gene 2022, 816, 146158. [Google Scholar] [CrossRef]
- Hamel, C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.A.; Guziewicz, K.E.; Lee, C.J.; Kalathur, R.C.; Pulido, J.S.; Marmorstein, L.Y.; Marmorstein, A.D. Bestrophin 1 and retinal disease. Progress. Retin. Eye Res. 2017, 58, 45–69. [Google Scholar] [CrossRef]
- AlAshwal, S.M.; Yassin, S.H.; Kalaw, F.G.P.; Borooah, S. PRPH2-associated Retinal Diseases: A Systematic Review of Phenotypic Findings. Am. J. Ophthalmol. 2024, 271, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Leys, M. A Family Affected by Novel C213W Mutation in PRPH2: Long-Term Follow-Up of CNV Secondary to Pattern Dystrophy. Ophthalmic Surg. Lasers Imaging Retin. 2020, 51, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Manes, G.; Meunier, I.; Avila-Fernandez, A.; Banfi, S.; Le Meur, G.; Zanlonghi, X.; Corton, M.; Simonelli, F.; Brabet, P.; Labesse, G.; et al. Mutations in IMPG1 cause vitelliform macular dystrophies. Am. J. Hum. Genet. 2013, 93, 571–578. [Google Scholar] [CrossRef]
- Gómez-Benlloch, A.; Garrell-Salat, X.; Cobos, E.; López, E.; Esteve-Garcia, A.; Ruiz, S.; Vázquez, M.; Sararols, L.; Biarnés, M. Optical Coherence Tomography in Inherited Macular Dystrophies: A Review. Diagnostics 2024, 14, 878. [Google Scholar] [CrossRef] [PubMed]
- Lindenberg, S.; Mahmoudi, A.; Oncel, D.; Corradetti, G.; Emamverdi, M.; Almidani, L.; Farahani, A.; Wakatsuki, Y.; He, Y.; Saju M, S.; et al. Acquired Vitelliform Lesions in Intermediate Age-Related Macular Degeneration: A Cross Sectional Study. Ophthalmol. Retin. 2024, 8, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Rödelsperger, C.; Schuelke, M.; Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 2010, 7, 575–576. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.A.; Edwards, K.J.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017, 45, W229–W235. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Randall, A.; Baldi, P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct. Funct. Bioinform. 2006, 62, 1125–1132. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Olivier, G.; Corton, M.; Intartaglia, D.; Verbakel, S.K.; Sergouniotis, P.I.; Le Meur, G.; Dhaenens, C.M.; Naacke, H.; Avila-Fernandez, A.; Hoyng, C.B.; et al. Pathogenic variants in IMPG1 cause autosomal dominant and autosomal recessive retinitis pigmentosa. J. Med. Genet. 2021, 58, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Verbakel, S.K.; Van Huet, R.A.C.; Boon, C.J.F.; Den Hollander, A.I.; Collin, R.W.J.; Klaver, C.C.W.; Hoyng, C.B.; Roepman, R.; Klevering, B.J. Non-syndromic retinitis pigmentosa. Progress. Retin. Eye Res. 2018, 66, 157–186. [Google Scholar] [CrossRef]
- Paez-Escamilla, M.; Scanga, H.L.; Liasis, A.; Nischal, K.K. Macular atrophy in JAG1- related Alagille syndrome: A case series. Ophthalmic Genet. 2022, 43, 230–234. [Google Scholar] [CrossRef]
- Ruiz-Chavolla, D.; Barragán-Arévalo, T.; Cortes-Muñoz, D.; Sánchez-Ruiz, J.; Zenteno, J.C.; Ledesma-Gil, G. Macular atrophy and focal choroidal excavation in a patient with JAG1-related alagille syndrome. Ophthalmic Genet. 2024, 45, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, X.; Xu, H.; Huang, L.; Zhang, S.; Liu, W.; Yang, Y.; Fei, P.; Li, S.; Yang, M.; et al. Exome sequencing revealed Notch ligand JAG1 as a novel candidate gene for familial exudative vitreoretinopathy. Genet. Med. 2020, 22, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Da Palma, M.M.; Igelman, A.D.; Ku, C.; Burr, A.; You, J.Y.; Place, E.M.; Wang, N.-K.; Oh, J.K.; Branham, K.E.; Zhang, X.; et al. Characterization of the Spectrum of Ophthalmic Changes in Patients With Alagille Syndrome. Investig. Opthalmol. Vis. Sci. 2021, 62, 27. [Google Scholar] [CrossRef] [PubMed]
- Habibi, I.; Falfoul, Y.; Tran, H.V.; El Matri, K.; Chebil, A.; El Matri, L.; Schorderet, D.F. Different Phenotypes in Pseudodominant Inherited Retinal Dystrophies. Front. Cell Dev. Biol. 2021, 9, 625560. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.N.; Chana, R.; Ali, N.; Wright, G.; Webster, A.R.; Moore, A.T.; Michaelides, M. Advanced diagnostic genetic testing in inherited retinal disease: Experience from a single tertiary referral centre in the UK National Health Service. Clin. Genet. 2017, 91, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.; Grewal, P.S.; Narayan, A.; Alser, M.; Ali, N.; Fujinami, K.; Webster, A.R.; Michaelides, M. Sector Retinitis Pigmentosa: Extending the Molecular Genetics Basis and Elucidating the Natural History. Am. J. Ophthalmol. 2021, 221, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Brecher, R.; Bird, A.C. Adult vitelliform macular dystrophy. Eye 1990, 4, 210–215. [Google Scholar] [CrossRef]
- Birtel, J.; Caswell, R.; De Silva, S.R.; Herrmann, P.; Rehman, S.; Lotery, A.J.; Mahroo, O.A.; Michaelides, M.; Webster, A.R.; MacLaren, R.E.; et al. IMPG2-Related Maculopathy. Am. J. Ophthalmol. 2024, 258, 32–42. [Google Scholar] [CrossRef]
- 58th Annual Symposium of the International Society for Clinical Electrophysiology of Vision (ISCEV 2020). Doc. Ophthalmol. 2020, 141, 1–37. [CrossRef] [PubMed]
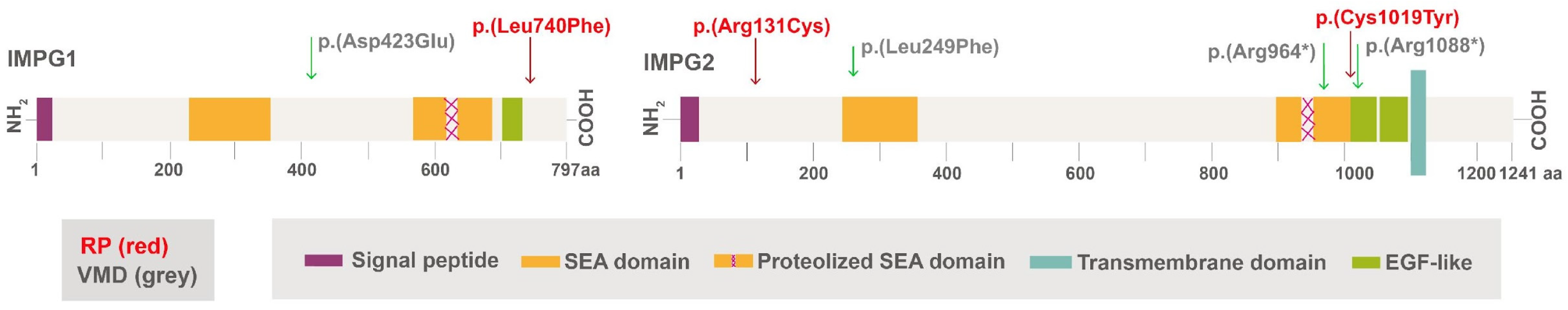
| Patient ID#: Sex and Age (Y) | Gene Involved | cDNA | Protein | Zygosity and Inheritance | Type of Mutation | Presentation of Disease | OD–OS VA |
|---|---|---|---|---|---|---|---|
| F32 | IMGP1 | c.2218C>T | p.Leu740Phe | Het; D | Missense | Sectoral RP | 20/25–20/25 |
| M83 | IMPG2 | c.3056G>A & c.391C>T | p.Cys1019Tyr & p.Arg131Cys | Het for both genes; R | Missense | RP | 20/70–20/30 |
| M75 | IMPG1 | c.1269C>A | p.Asp423Glu | Het; D | Missense | VMD | 20/40–20/40 |
| M82 | IMPG2 | c.745C>T | p.Leu249Phe | Het; D | Missense | VMD | 20/30–20/25 |
| M73 | IMPG2 | c.2890C>T | p.Arg964* | Het; D | Nonsense | VMD | 20/100–20/50 |
| F57 | IMPG2 | c.2890C>T | p.Arg964* | Het; D | Nonsense | VMD | 20/20–20/20 |
| M34 | IMPG2 | c.3262C>T | p.Arg1088* | Het; D | Nonsense | VMD | 20/20–20/20 |
| Gene | Variant | SIFT Score | PolyPhen Score | CADD Score | FATHMM Coding | MutationTaster Score | Prediction | Existing Variant ID (ClinVar) |
|---|---|---|---|---|---|---|---|---|
| IMPG1 | p.Leu740Phe | 0.09 | 0.575 | 3.6 | - | 22 | Likely Benign | rs15048636 |
| IMPG1 | p.Asp423Glu | 0.02 | 0.98 | 2.5 | - | 45 | Pathogenic | Novel |
| IMPG2 | p.Cys1019Tyr | 0 | 0.996 | 25.8 | 0.0376 | 46 | Pathogenic | rs2107208112 |
| IMPG2 | p.Arg131Cys | 0 | 0.645 | 21.7 | 0.0577 | 91 | Pathogenic | rs150344327 |
| IMPG2 | p.Leu249Phe | 0 | 0.998 | 22.6 | 0.0076 | 78 | Pathogenic | rs376443291 |
| IMPG2 | p.Arg964* | - | - | 11.2 | 0.011 | 58 | Pathogenic | rs267606975 |
| IMPG2 | p.Arg1088* | - | - | 13.4 | 0.0513 | 29 | Pathogenic | rs199867882 |
| Variant | Gene | mCSM AAG | SDM AAG | DUET AAG | iMutant AAG | MuProAAG |
|---|---|---|---|---|---|---|
| p.Leu740Phe | IMPG1 | −0.588 (DSTB) | −0.33 (DSTB) | −0.591 (DSTB) | −0.85 (DSTB) | −0.930 (DecStab) |
| p.Asp423Glu | IMPG1 | −0.653 (DSTB) | −0.52 (DSTB) | −0.138 (DSTB) | −0.76 (DecStab) | −0.478 (DecStab) |
| p.Cys1019Tyr | IMPG2 | −0.826 (DSTB) | −0.78 (DSTB) | −0.85 (DSTB) | −0.14 (DSTB) | −1.266 (DSTB) |
| p.Arg131Cys | IMPG2 | −0.007 (DSTB) | −0.22 (DSTB) | −0.018 (DSTB) | −1.13 (DSTB) | −0.627 (DSTB) |
| p.Leu249Phe | IMPG2 | −1.743 (DSTB) | −0.66 (DSTB) | −1.77 (DSTB) | −0.55 (DSTB) | −1.525 (DSTB) |
| Physiochemical Properties | Reference Allele | p.Asp423Glu | p.Leu740Phe |
|---|---|---|---|
| Molecular weight | 89,387 | 89,419 | 89,421 |
| Theoretical pI | 4.79 | 4.81 | 4.79 |
| Total number of atoms | 12,450 | 12,457 | 12,451 |
| Instability Index | 53.57 | 53.76 | 53.16 |
| Aliphatic Index | 75.63 | 75.63 | 75.14 |
| GRAVY | −0.482 | −0.474 | −0.483 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, M.; Chatterjee, S.; Leys, M.; Odom, J.V.; Salido, E.M. Prevalence of IMPG1 and IMPG2 Mutations Leading to Retinitis Pigmentosa or Vitelliform Macular Dystrophy in a Cohort of Patients with Inherited Retinal Dystrophies. Genes 2025, 16, 43. https://doi.org/10.3390/genes16010043
Yuan M, Chatterjee S, Leys M, Odom JV, Salido EM. Prevalence of IMPG1 and IMPG2 Mutations Leading to Retinitis Pigmentosa or Vitelliform Macular Dystrophy in a Cohort of Patients with Inherited Retinal Dystrophies. Genes. 2025; 16(1):43. https://doi.org/10.3390/genes16010043
Chicago/Turabian StyleYuan, Ming, Souradip Chatterjee, Monique Leys, J. Vernon Odom, and Ezequiel M. Salido. 2025. "Prevalence of IMPG1 and IMPG2 Mutations Leading to Retinitis Pigmentosa or Vitelliform Macular Dystrophy in a Cohort of Patients with Inherited Retinal Dystrophies" Genes 16, no. 1: 43. https://doi.org/10.3390/genes16010043
APA StyleYuan, M., Chatterjee, S., Leys, M., Odom, J. V., & Salido, E. M. (2025). Prevalence of IMPG1 and IMPG2 Mutations Leading to Retinitis Pigmentosa or Vitelliform Macular Dystrophy in a Cohort of Patients with Inherited Retinal Dystrophies. Genes, 16(1), 43. https://doi.org/10.3390/genes16010043





