Genome-Wide Identification and Characterization of HSP70 Gene Family in Tausch’s Goatgrass (Aegilops tauschii)
Abstract
:1. Introduction
2. Materials and Methods
2.1. A. tauschii Characteristics
2.2. Genome-Wide Identification of Hsp70 Family Members
2.3. Physicochemical Properties Analysis of Hsp70 Family Members
2.4. Multiple Alignments and Phylogenetic Analysis of Hsp70 Proteins
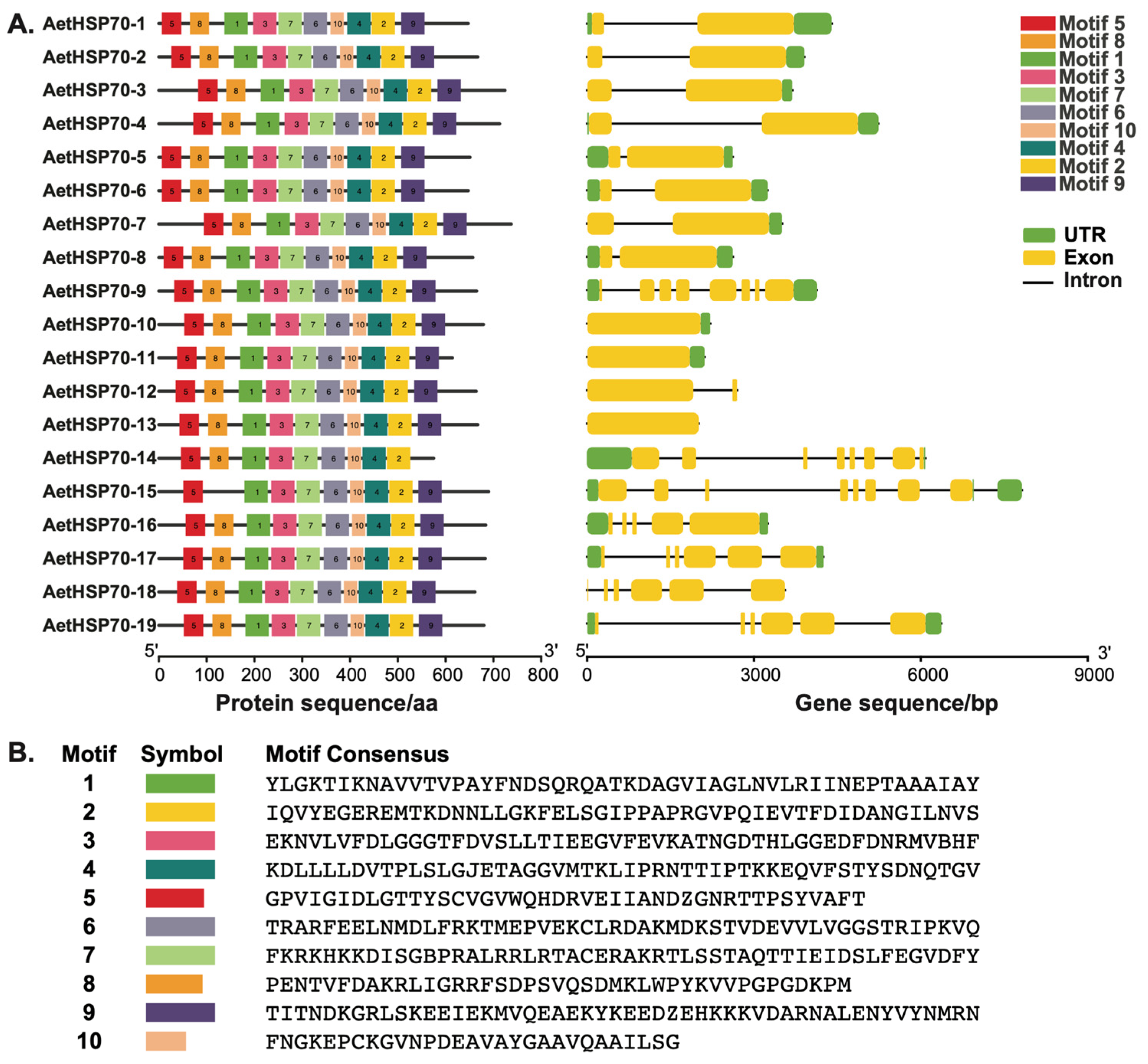
2.5. Gene Structure Analysis and Identification of Conserved Motifs
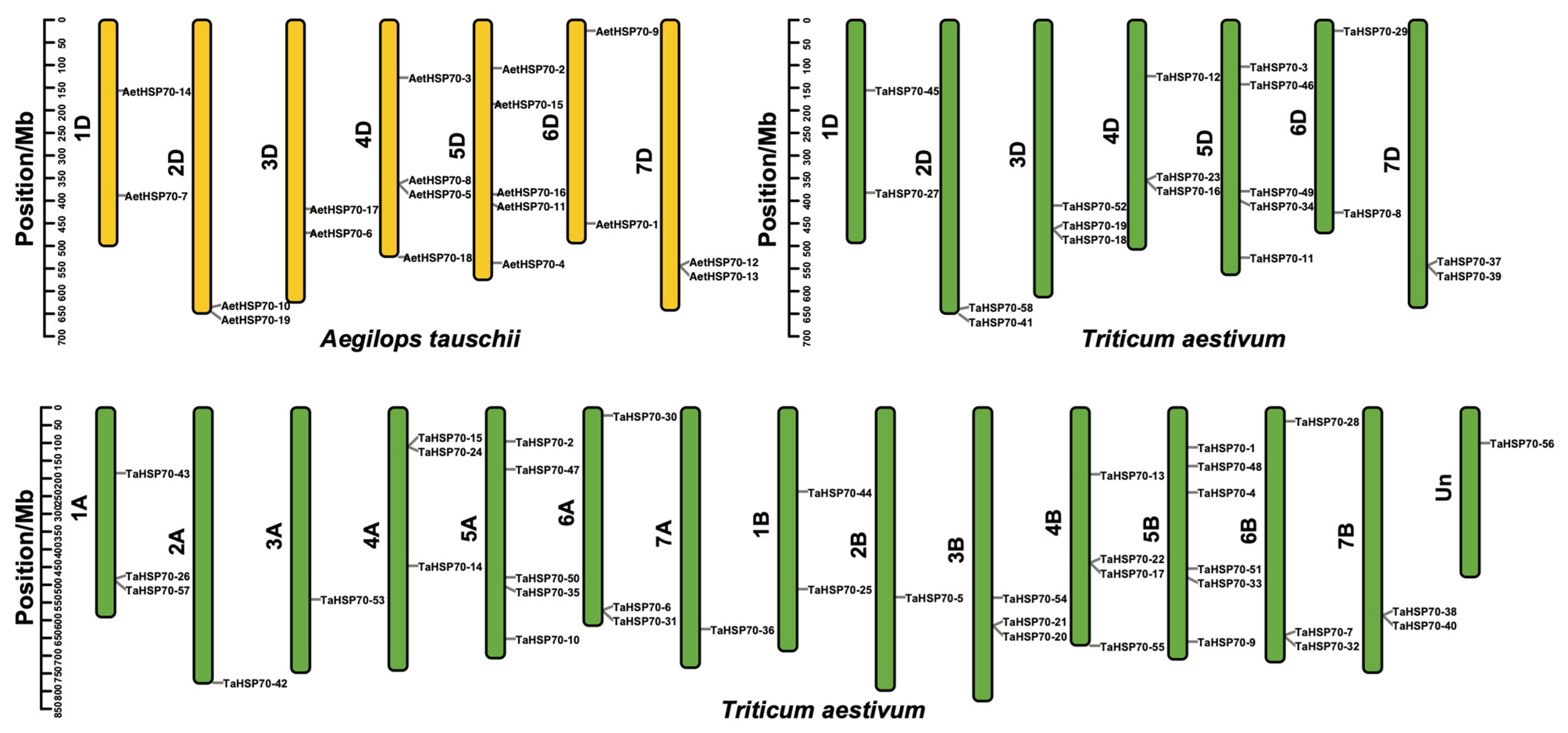
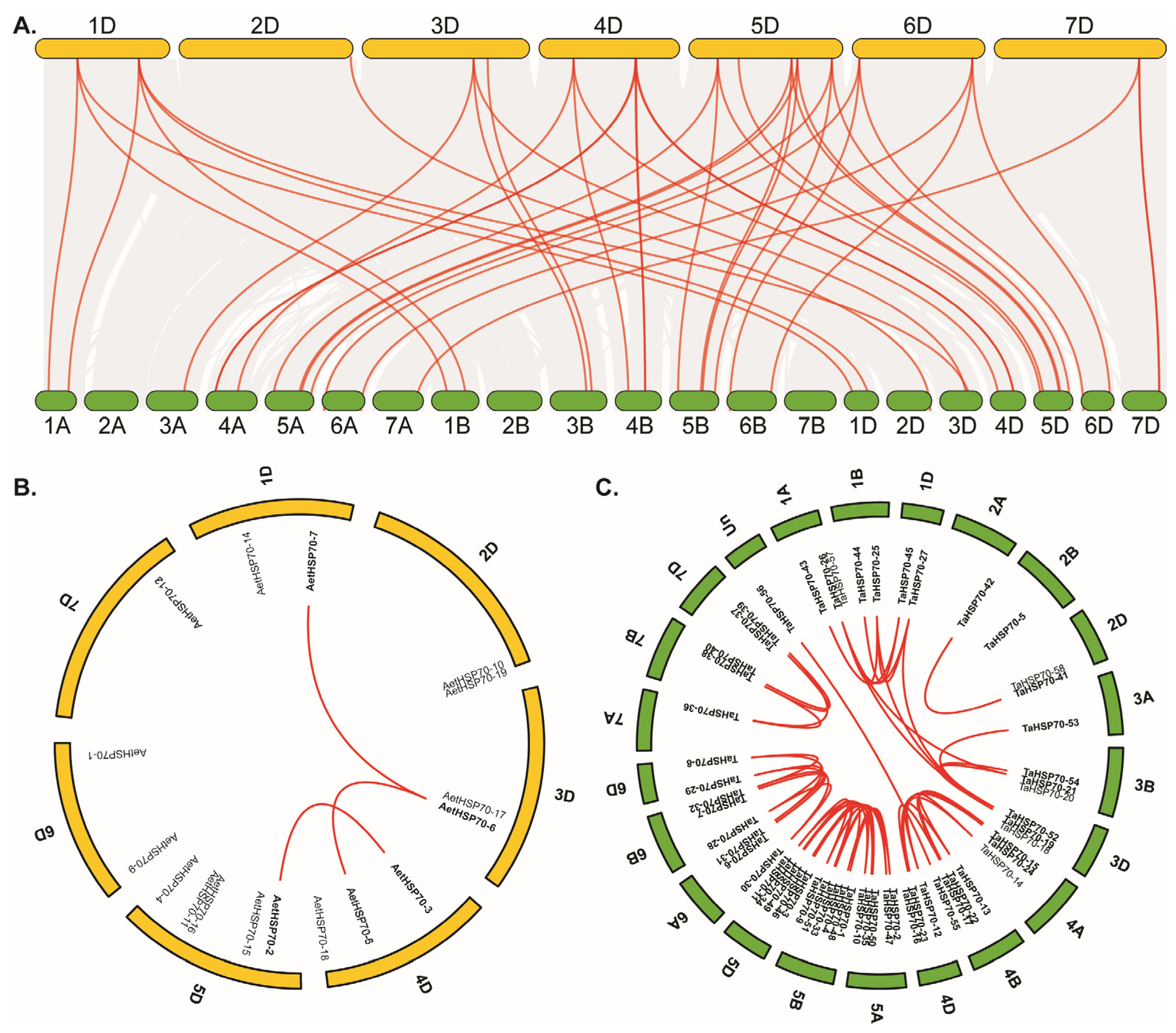
2.6. Chromosomal Location and Syntenic Analysis
2.7. Cis-Acting Element Prediction in Promoters
3. Results
3.1. Identification of Hsp70 Family Members in A. tauschii
3.2. Phylogenetic Analysis of the Hsp70 Family Proteins
3.3. Structure of AetHSP70 Genes and Conserved Motifs of AetHsp70 Proteins
3.4. Chromosomal Distribution and Syntenic Analysis of HSP70 Genes
3.5. Cis-Acting Elements of the HSP70 Gene Promoter in A. tauschii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Y.C.; Liu, C.C.; Li, Y.J.; Liao, C.M.; Vivek, S.; Chuo, G.L.; Tseng, C.Y.; Wu, Z.Q.; Shimada, T.; Suetsugu, N.; et al. Multifaceted roles of Arabidopsis heat shock factor binding protein in plant growth, development, and heat shock response. Environ. Exp. Bot. 2024, 226, 105878. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, H.K.; Dong, Q.L.; Zhang, Y.Y.; Wang, Y.M.; Li, H.Y.; Xing, G.J.; Li, Q.Y.; Dong, Y.S. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef]
- Shi, G.C.; Dong, X.H.; Chen, G.; Tan, B.P.; Yang, Q.H.; Chi, S.Y.; Liu, H.Y. Physiological responses and HSP 70 m RNA expression of GIFT strain of Nile tilapia (Oreochromis niloticus) under cold stress. Aquac. Res. 2015, 46, 658–668. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17. 0 and OsHsp23. 7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef]
- Kim, B.M.; Rhee, J.S.; Jeong, C.B.; Seod, J.S.; Parke, G.S.; Leef, Y.M.; Leea, J.S. Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2014, 166, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Srivastava, P. Heat-Shock Proteins. Curr. Protoc. Immunol. 2003, 58, A.1T.1–A.1T.6. [Google Scholar] [CrossRef]
- Garrido, C.; Schmitt, E.; Candé, C.; Vahsen, N.; Parcellier, A.; Kroemer, G. HSP27 and HSP70: Potentially oncogenic apoptosis inhibitors. Cell Cycle 2003, 2, 579–584. [Google Scholar] [CrossRef]
- Voisine, C.; Orton, K.; Morimoto, R.I. Protein misfolding, chaperone networks, and the heat shock response in the nervous system. In Molecular Neurology; Elsevier Academic Press: Cambridge, MA, USA, 2007; pp. 59–76. [Google Scholar] [CrossRef]
- Duan, Y.; Guo, J.; Ding, K.; Wang, S.J.; Zhang, H.; Dai, X.W.; Chen, Y.Y.; Govers, F.; Huang, L.L.; Kang, Z.S. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Mol. Biol. Rep. 2011, 38, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Tkáčová, J.; Angelovičová, M. Heat shock proteins (HSPs): A review. Sci. Pap. Anim. Sci. Biotechnol. 2012, 45, 349. [Google Scholar]
- Chen, Y.J.; Cheng, S.Y.; Liu, C.H.; Tsai, W.C.; Wu, H.H.; Huang, M.D. Exploration of the truncated cytosolic Hsp70 in plants—Unveiling the diverse T1 lineage and the conserved T2 lineage. Front. Plant Sci. 2023, 14, 1279540. [Google Scholar] [CrossRef]
- Wruck, F.; Avellaneda, M.J.; Koers, E.J.; Minde, D.P.; Mayer, M.P.; Kramer, G.; Mashaghi, A.; Tans, S.J. Protein folding mediated by trigger factor and Hsp70: New insights from single-molecule approaches. J. Mol. Biol. 2018, 430, 438–449. [Google Scholar] [CrossRef]
- Rasheed, A.; Ogbonnaya, F.C.; Lagudah, E.; Appels, R.; He, Z. The goat grass genome’s role in wheat improvement. Nat. Plants 2018, 4, 56–58. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhao, S.; Kong, X.; Li, Y.R.; Zhao, G.Y.; He, W.M.; Appels, R.; Pfeifer, M.; Tao, Y.; Zhang, X.Y.; et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 2013, 496, 91–95. [Google Scholar] [CrossRef]
- Lai, D.L.; Yan, J.; Fan, Y.; Li, Y.; Ruan, J.J.; Wang, J.Z.; Cheng, J.P. Genome-wide identification and phylogenetic relationships of the Hsp70 gene family of Aegilops tauschii, wild emmer wheat (Triticum dicoccoides) and bread wheat (Triticum aestivum). Biotech 2021, 11, 301. [Google Scholar] [CrossRef]
- Dvorak, J.; Luo, M.C.; Yang, Z.L.; Zhang, H.B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670. [Google Scholar] [CrossRef]
- Takumi, S.; Nishioka, E.; Morihiro, H.; Kawahara, T.; Matsuoka, Y. Natural variation of morphological traits in wild wheat progenitor Aegilops tauschii Coss. Breed. Sci. 2009, 59, 579–588. [Google Scholar] [CrossRef]
- Huang, Z.; Sui, B.; Zhang, C.; Huang, H.; Wei, S. The basis of resistance mechanism to mesosulfuron-methyl in Tausch’s goatgrass (Aegilops tauschii Coss.). Pestic. Biochem. Physiol. 2019, 155, 126–131. [Google Scholar] [CrossRef]
- Wang, H.Z.; Zhao, K.P.; Li, X.J.; Chen, X.T.; Liu, W.T.; Wang, J.X. Factors affecting seed germination and emergence of Aegilops tauschii. Weed Res. 2020, 60, 171–181. [Google Scholar] [CrossRef]
- Nazari, M.; Moosavi, S.S.; Maleki, M. Morpho-physiological and proteomic responses of Aegilops tauschii to imposed moisture stress. Plant Physiol. Biochem. 2018, 132, 445–452. [Google Scholar] [CrossRef]
- Wang, N.; Chen, H. Effects of Soil Drought on Competitiveness of the Invasive Weed Aegilops tauschii. Russ. J. Plant Physiol. 2024, 71, 114. [Google Scholar] [CrossRef]
- Liu, M.; Bian, Z.Y.; Shao, M.; Feng, Y.Q.; Ma, W.F.; Liang, G.P.; Mao, J. Expression analysis of the apple HSP70 gene family in abiotic stress and phytohormones and expression validation of candidate MdHSP70 genes. Sci. Rep. 2024, 14, 23975. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Xiao, L.D.; Zhao, J.D.; Hu, Z.Y.; Zhou, Y.L.; Liu, S.Q.; Zhou, Y. Comprehensive Genomic Analysis and Expression Profile of HSP70 Gene Family Related to Abiotic and Biotic Stress in Cucumber. Horticulturae 2023, 9, 1057. [Google Scholar] [CrossRef]
- Song, J.H.; Weng, Q.Y.; Ma, H.L.; Yuan, J.C.; Wang, L.Y.; Liu, Y.H. Cloning and expression analysis of the HSP70 gene ZmERD2 in Zea mays. Biotechnol. Biotechnol. Equip. 2016, 30, 219–226. [Google Scholar] [CrossRef]
- Su, Y.Z.; Zou, M.W.; Zhu, Y.M.; Han, X.; Li, Y.G.; Zhang, D.L.; Li, S.P. Analysis of population structure and origin in Aegilops tauschii Coss. from China through SNP markers. Genet. Resour. Crop Evol. 2020, 67, 923–934. [Google Scholar] [CrossRef]
- Li, Y.; He, L.; Li, J.; Chen, J.; Liu, C. Genome-wide Identification, Characterization and Expression Profiling of the Legume BZR Transcription Factor Gene Family. Front. Plant Sci. 2018, 9, 1332. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Kaae Sønderby, C.; Kaae Sønderby, S.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.H.; O’Reilly, M.M.; Holland, B. A subfunctionalisation model of gene family evolution predicts balanced tree shapes. Mol. Phylogenetics Evol. 2022, 176, 107566. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Liu, J.Y.; Shively, C.A.; Mitra, R.D. Quantitative analysis of transcription factor binding and expression using calling cards reporter arrays. Nucleic Acids Res. 2020, 48, 50. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Beck, E.H.; Fettig, S.; Knake, C.; Hartig, K.; Bhattarai, T. Specific and unspecific responses of plants to cold and drought stress. J. Biosci. 2007, 32, 501–510. [Google Scholar] [CrossRef]
- Lin, B.L.; Wang, J.S.; Liu, H.C.; Chen, R.W.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis HSP70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]
- Leng, L.; Liang, Q.Q.; Jiang, J.J.; Zhang, C.; Hao, Y.H.; Wang, X.L.; Su, W. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. J. Plant Res. 2017, 130, 349–363. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kundnani, P.; Grover, A. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 2013, 18, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Yu, A.M.; Li, P.; Yang, H.; Liu, G.J.; Liu, L. Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses. Sci. Rep. 2016, 6, 33650. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.; Ma, X.; Zhai, Y.F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the HSP70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef]
- Su, H.; Xing, M.; Liu, X.; Fang, Z.Y.; Yang, L.M.; Zhuang, M.; Zhang, Y.Y.; Wang, Y.; Lv, H.H. Genome-wide analysis of HSP70 family genes in cabbage (Brassica oleracea var. capitata) reveals their involvement in floral development. BMC Genom. 2019, 20, 369. [Google Scholar] [CrossRef]
- Song, Z.P.; Pan, F.L.; Lou, X.P.; Wang, D.B.; Yang, C.; Zhang, B.Q.; Zhang, H.Y. Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum. Mol. Biol. Rep. 2019, 46, 1941–1954. [Google Scholar] [CrossRef]
- Rehman, A.; Atif, R.M.; Qayyume, A.; Du, X.M.; Hinzef, L.; Azhar, M.T. Genome-wide identification and characterization of HSP70 gene family in four species of cotton. Genomics 2020, 112, 4442–4453. [Google Scholar] [CrossRef]
- Huang, Y.X.; Chen, M.M.; Chen, D.M.; Chen, H.M.; Xie, Z.H.; Dai, S.F. Enhanced HSP70 binding to m6A-methylated RNAs facilitates cold stress adaptation in mango seedlings. BMC Plant Biol. 2024, 24, 1114. [Google Scholar] [CrossRef]
- Huang, D.L.; Tian, C.; Sun, Z.J.; Niu, J.F.; Wang, G.C. Potential synergistic regulation of hsp70 and antioxidant enzyme genes in Pyropia yezoensis under high temperature stress. Algal Res. 2024, 78, 103375. [Google Scholar] [CrossRef]
- Xu, K.; Wang, P. Transcriptome-wide identification of the HSP70 gene family in Pugionium cornutum and functional analysis of PcHsp70-5 under drought stress. Planta 2024, 260, 84. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhou, H.; Feng, J.; Guo, M.Y.; Huang, H.M.; Yang, P.; Zhou, J. Involvement of HSP70 in BAG9-mediated thermotolerance in Solanum lycopersicum. Plant Physiol. Biochem. 2024, 207, 108353. [Google Scholar] [CrossRef] [PubMed]
- Vu, N.T.; Nguyen NB, T.; Ha, H.H.; Nguyen, L.N.; Luu, L.H.; Dao, H.Q.; Vu, T.T.; Huynh, H.T.T.; Le, H.T.T. Evolutionary analysis and expression profiling of the HSP70 gene family in response to abiotic stresses in tomato (Solanum lycopersicum). Sci. Prog. 2023, 106, 368504221148843. [Google Scholar] [CrossRef]
- Wang, X.S.; Jin, Z.; Ding, Y.; Guo, M. Characterization of HSP70 family in watermelon (Citrullus lanatus): Identification, structure, evolution, and potential function in response to ABA, cold and drought stress. Genet 2023, 14, 1201535. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.N.; Xie, B.Y.; Wan, F.H.; Guo, J.Y. Molecular cloning, characterization, and heterologous expression analysis of heat shock protein genes (hsp70 and hsp90) of the invasive alien weed, Ageratina adenophora (Asteraceae). Weed Biol. Manag. 2010, 10, 91–101. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Carmel, L.; Csuros, M.; Koonin, E.V. Origin and evolution of spliceosomal introns. Biol. Direct 2012, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Miurier, T.; Penny, D. The biology of intron gain and loss. Trends Genet. 2006, 22, 16–22. [Google Scholar] [CrossRef]
- Deshmukh, R.K.; Sonah, H.; Singh, N.K. Intron gain, a dominant evolutionary process supporting high levels of gene expression in rice. J. Plant Biochem. Biotechnol. 2016, 25, 142–146. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Zhao, P.; Zhang, A.; Wang, J.; Ha, M. Genome-wide analysis of HSP70s in hexaploid wheat: Tandem duplication, heat response, and regulation. Cells 2022, 11, 818. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Fang, L.; Wang, X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef]
- Sémon, M.; Wolfe, K.H. Consequences of genome duplication. Curr. Opin. Genet. Dev. 2007, 17, 505–512. [Google Scholar] [CrossRef]
- Liu, J.; Pang, X.; Cheng, Y.; Yin, C.H.; Zhang, Q.; Su, W.B.; Hu, B.; Guo, Q.W.; Ha, S.; Zhang, J.P.; et al. The HSP70 gene Family in Solanum tuberosum: Genome-Wide Identification, Phylogeny, and Expression Patterns. Sci. Rep. 2018, 8, 16628. [Google Scholar] [CrossRef]
- Jakhu, P.; Sharma, P.; Yadav, I.S.; Kaur, P.; Kaur, S.; Chhuneja, P.; Singh, K. Cloning, expression analysis and In silico characterization of HSP101: A potential player conferring heat stress in Aegilops speltoides (Tausch) Gren. Physiol. Mol. Biol. Plants 2021, 27, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.Y.; Parkin, I.A.P.; Tang, H.B.; Wang, X.Y.; Chiquet, J.; Belcram, H.; Tong, C.B.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950. [Google Scholar] [CrossRef]
- Liang, Z.; Li, M.; Liu, Z.; Wang, J. Genome-wide identification and characterization of the HSP70 gene family in allopolyploid rapeseed (Brassica napus L.) compared with its diploid progenitors. PeerJ 2019, 7, e7511. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.M.; Wang, R.C.; Ma, C.; Shi, X.; Liu, Z.S.; Wang, Z.H.; Sun, Q.X.; Cao, J.; Xu, S.B. Massive expansion and differential evolution of small heat shock proteins with wheat (Triticum aestivum L.) polyploidization. Sci. Rep. 2017, 7, 2581. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Amino Acids | MW/kD | pI | GRAVY | Localization |
|---|---|---|---|---|---|---|
| AetHSP70-1 | AET6Gv20830600 | 647 | 71.13 | 4.8 | −0.41 | Cytoplasm |
| AetHSP70-2 | AET5Gv20225300 | 667 | 73.43 | 4.93 | −0.41 | ER |
| AetHSP70-3 | AET4Gv20300300 | 724 | 78.93 | 5.19 | −0.45 | Plastid |
| AetHSP70-4 | AET5Gv21108900 | 713 | 78.12 | 5.35 | −0.39 | Plastid |
| AetHSP70-5 | AET4Gv20528300 | 651 | 71.32 | 4.83 | −0.45 | Cytoplasm |
| AetHSP70-6 | AET3Gv20805300 | 647 | 70.64 | 4.91 | −0.37 | Cytoplasm |
| AetHSP70-7 | AET1Gv20682100 | 737 | 81.13 | 7.05 | −0.54 | Plastid |
| AetHSP70-8 | AET4Gv20525900 | 657 | 71.83 | 4.91 | −0.44 | Cytoplasm |
| AetHSP70-9 | AET6Gv20108600 | 665 | 73.2 | 4.85 | −0.47 | ER |
| AetHSP70-10 | AET2Gv21241200 | 679 | 73.09 | 4.92 | −0.32 | ER |
| AetHSP70-11 | AET5Gv20692700 | 614 | 67.32 | 5.65 | −0.33 | ER |
| AetHSP70-12 | AET7Gv21043500 | 664 | 73.11 | 4.87 | −0.42 | ER |
| AetHSP70-13 | AET7Gv21044600 | 667 | 73.53 | 4.87 | −0.39 | ER |
| AetHSP70-14 | AET1Gv20328300 | 575 | 61.89 | 5.17 | −0.21 | Plastid |
| AetHSP70-15 | AET5Gv20312000 | 690 | 73.58 | 4.72 | −0.27 | Plastid |
| AetHSP70-16 | AET5Gv20629700 | 684 | 72.74 | 6.04 | −0.27 | Mitochondrion |
| AetHSP70-17 | AET3Gv20701400 | 683 | 73.34 | 5.57 | −0.34 | Mitochondrion |
| AetHSP70-18 | AET4Gv20882800 | 661 | 70.74 | 5.08 | −0.27 | Mitochondrion |
| AetHSP70-19 | AET2Gv21290700 | 680 | 72.85 | 5.05 | −0.29 | Mitochondrion |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Liu, Y.; Yi, Y.; Liu, J. Genome-Wide Identification and Characterization of HSP70 Gene Family in Tausch’s Goatgrass (Aegilops tauschii). Genes 2025, 16, 19. https://doi.org/10.3390/genes16010019
Xu Y, Liu Y, Yi Y, Liu J. Genome-Wide Identification and Characterization of HSP70 Gene Family in Tausch’s Goatgrass (Aegilops tauschii). Genes. 2025; 16(1):19. https://doi.org/10.3390/genes16010019
Chicago/Turabian StyleXu, Yongmei, Yue Liu, Yanjun Yi, and Jiajia Liu. 2025. "Genome-Wide Identification and Characterization of HSP70 Gene Family in Tausch’s Goatgrass (Aegilops tauschii)" Genes 16, no. 1: 19. https://doi.org/10.3390/genes16010019
APA StyleXu, Y., Liu, Y., Yi, Y., & Liu, J. (2025). Genome-Wide Identification and Characterization of HSP70 Gene Family in Tausch’s Goatgrass (Aegilops tauschii). Genes, 16(1), 19. https://doi.org/10.3390/genes16010019






