Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Gene Family Members
2.2. Physicochemical Characteristics and Subcellular Localization
2.3. Analysis of Conserved Domains and Gene Structure
2.4. Multiple Sequence Alignment and Phylogeny
2.5. Chromosome Localization and Gene Duplication Analysis
2.6. Analysis of Cis-Acting Elements in the Promoter Region
2.7. Gene Expression Pattern Analysis
2.8. Plant Materials, Stress Treatments, and Tissue Sampling
2.9. RNA Isolation and Quantitative Real-Time PCR Analysis
2.10. Protein 3D Structure Interaction Network Prediction
3. Results
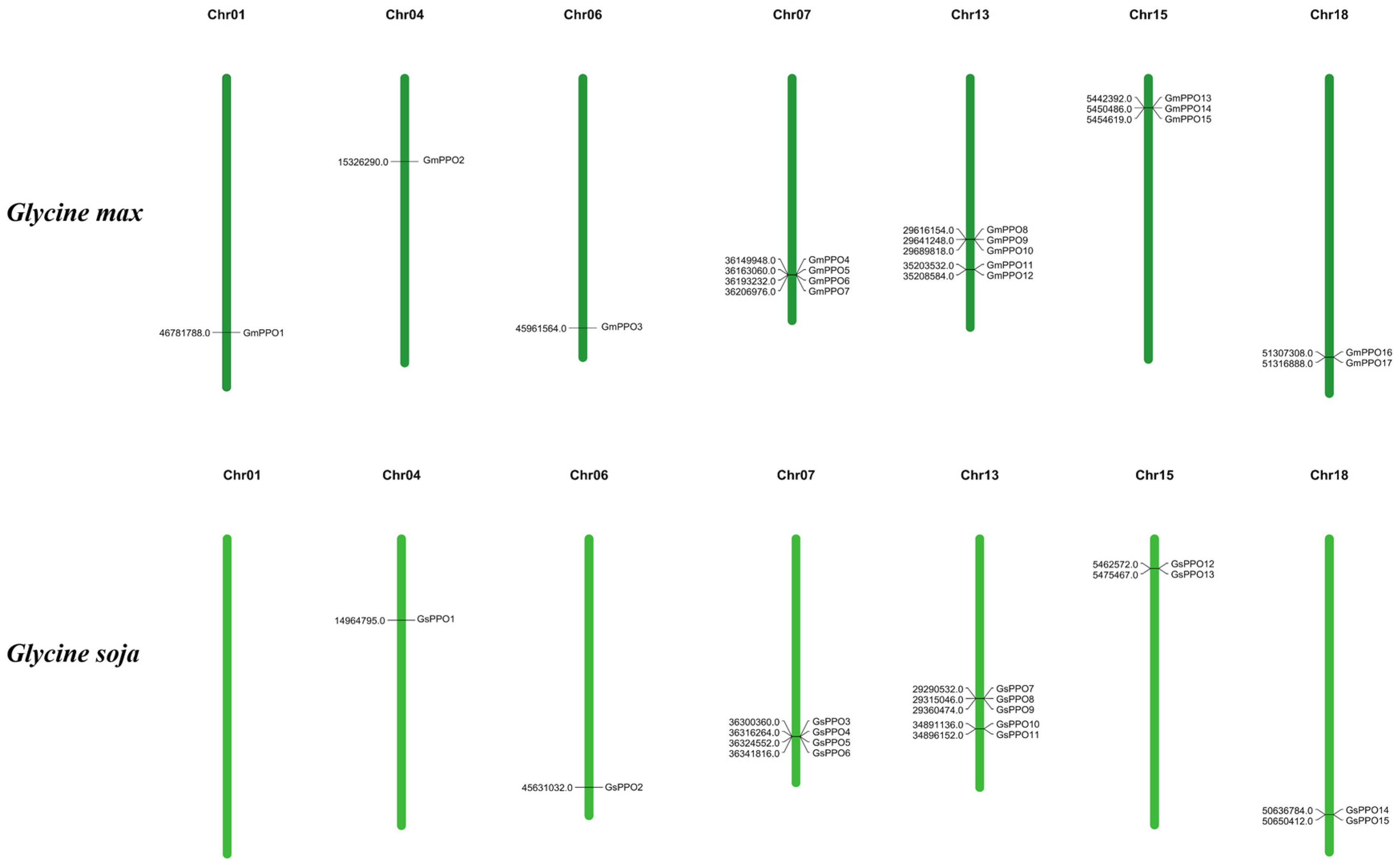
3.1. Identification and Distribution
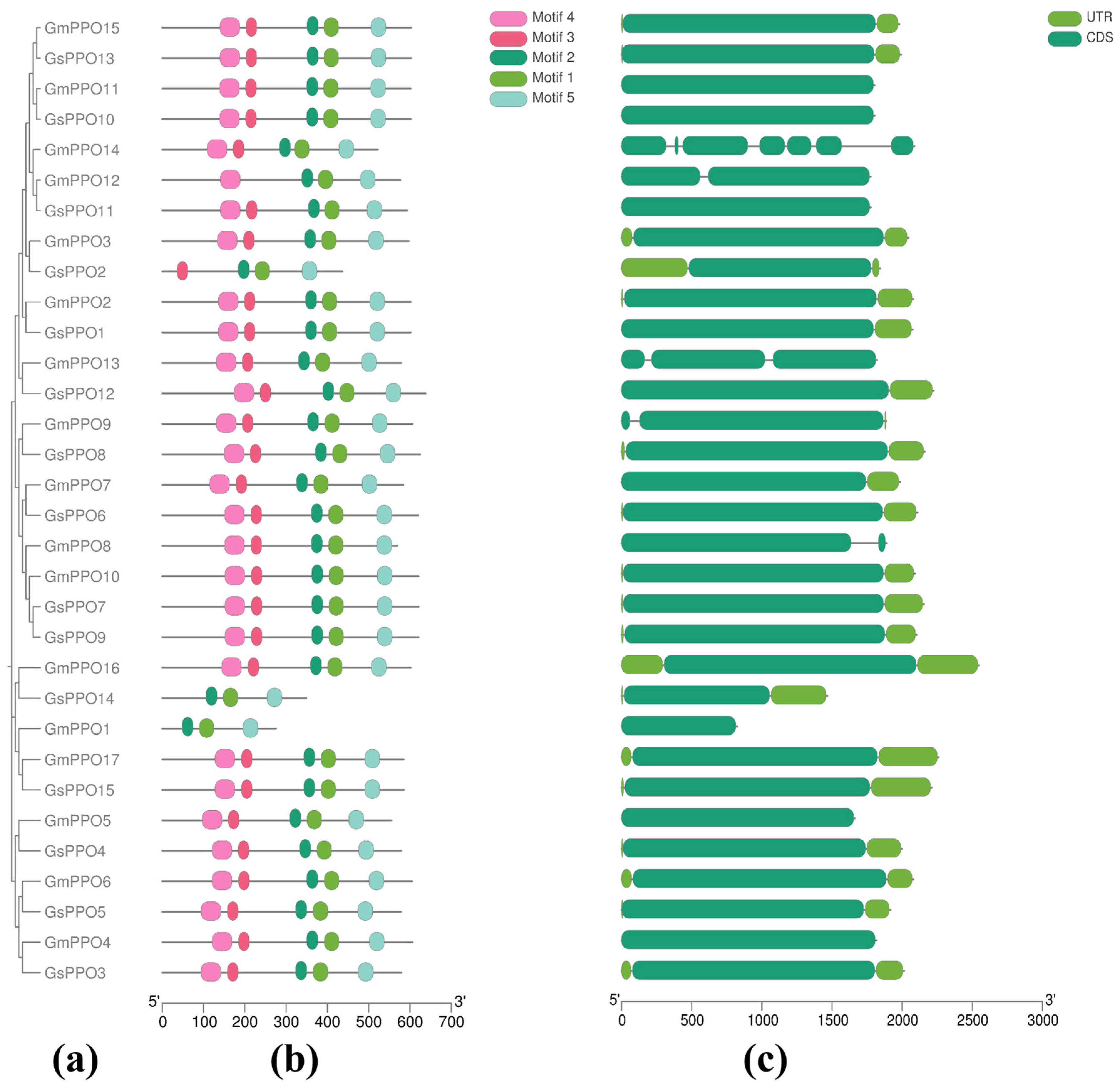
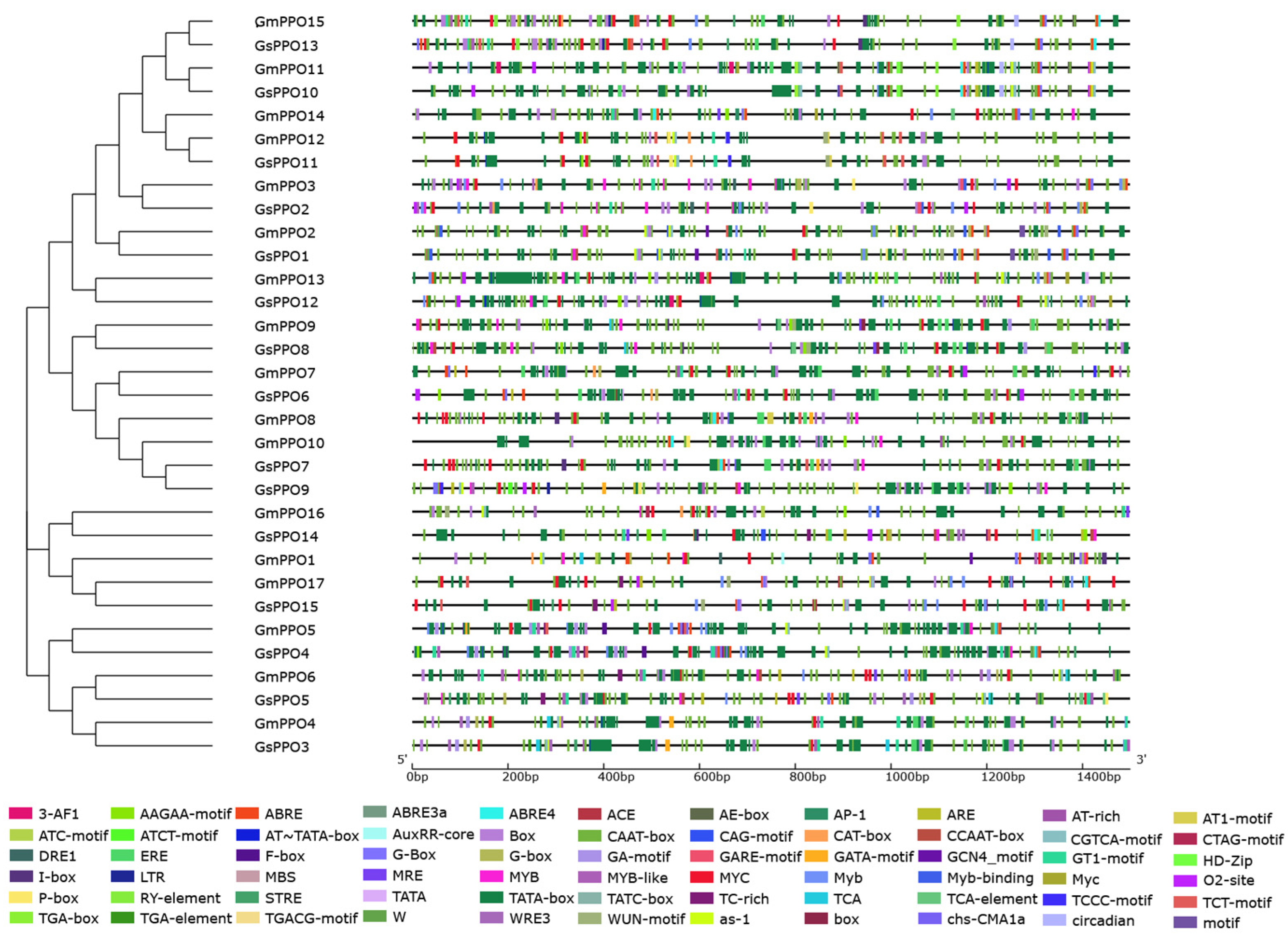
3.2. Analysis of Conserved Domains and Gene Structure
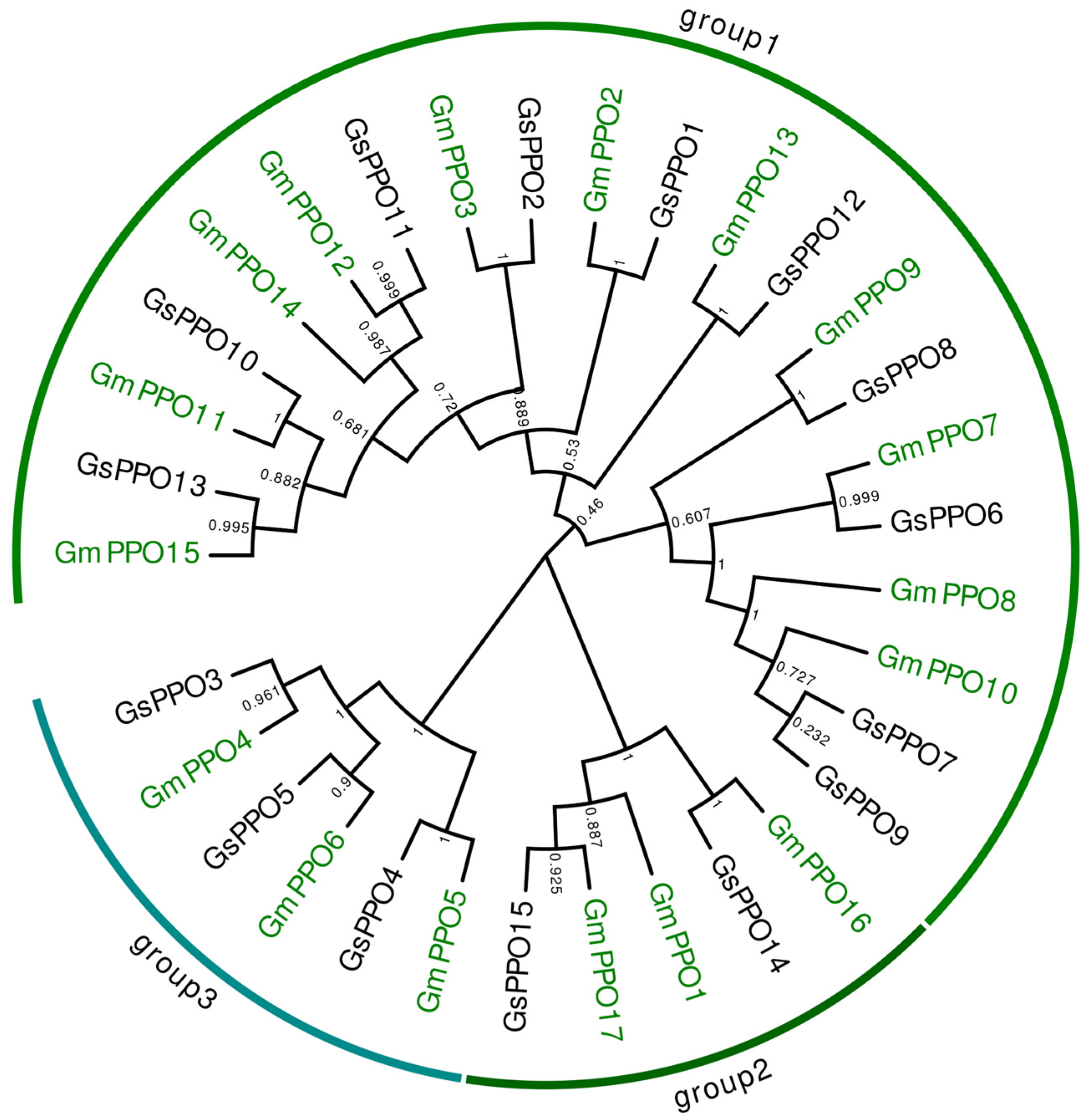
3.3. Phylogenetic Analysis of PPO Gene Families
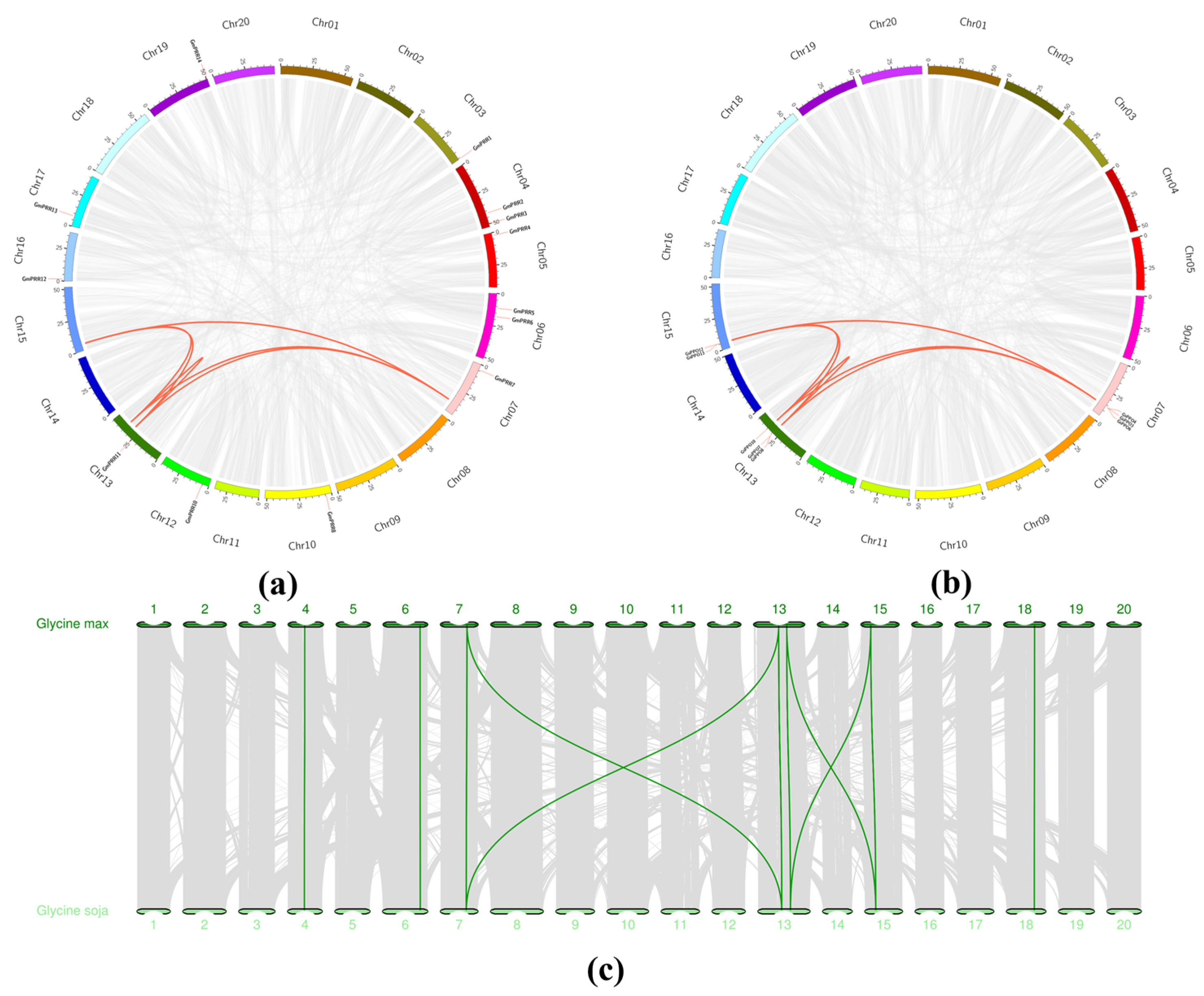
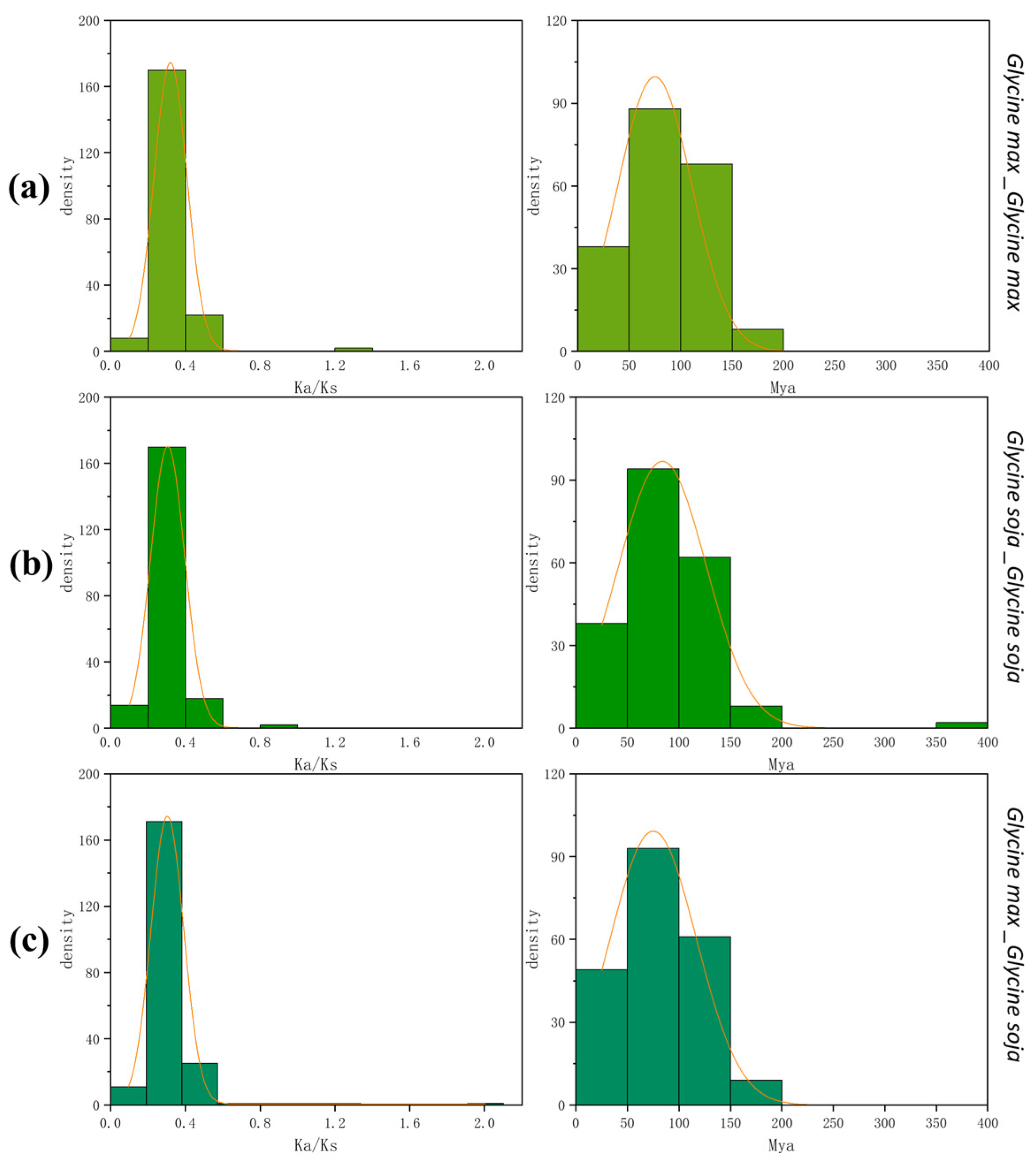
3.4. Chromosomal Location and Gene Duplication Analysis of PPO Genes
3.5. Analysis of the Cis-Acting Elements of the PPO Gene Family
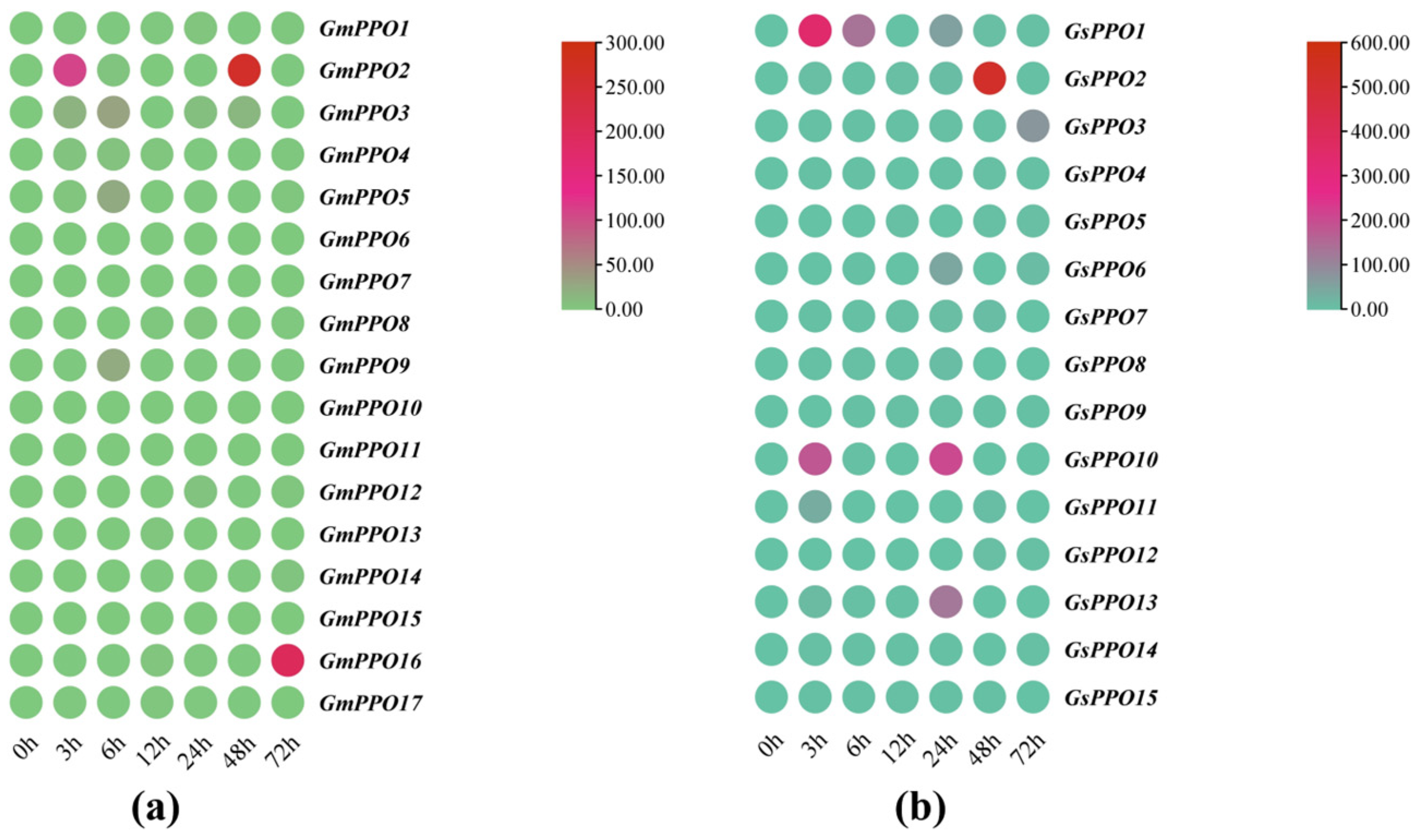
3.6. Analysis of the Gene Expression of PPO Genes Throughout the Growth Period
3.7. Gene Expression Analysis of PPO Genes Under Salt Stress
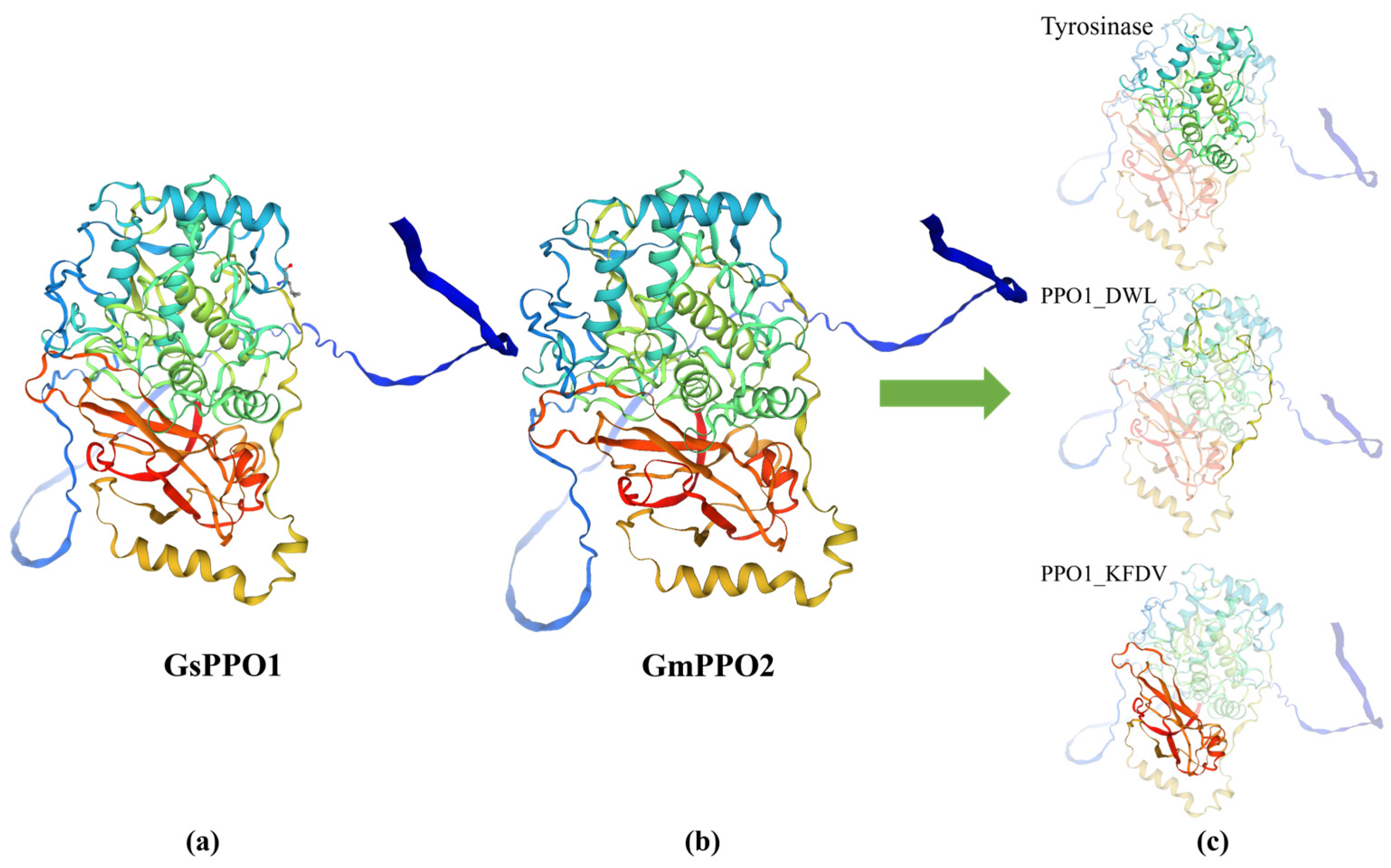
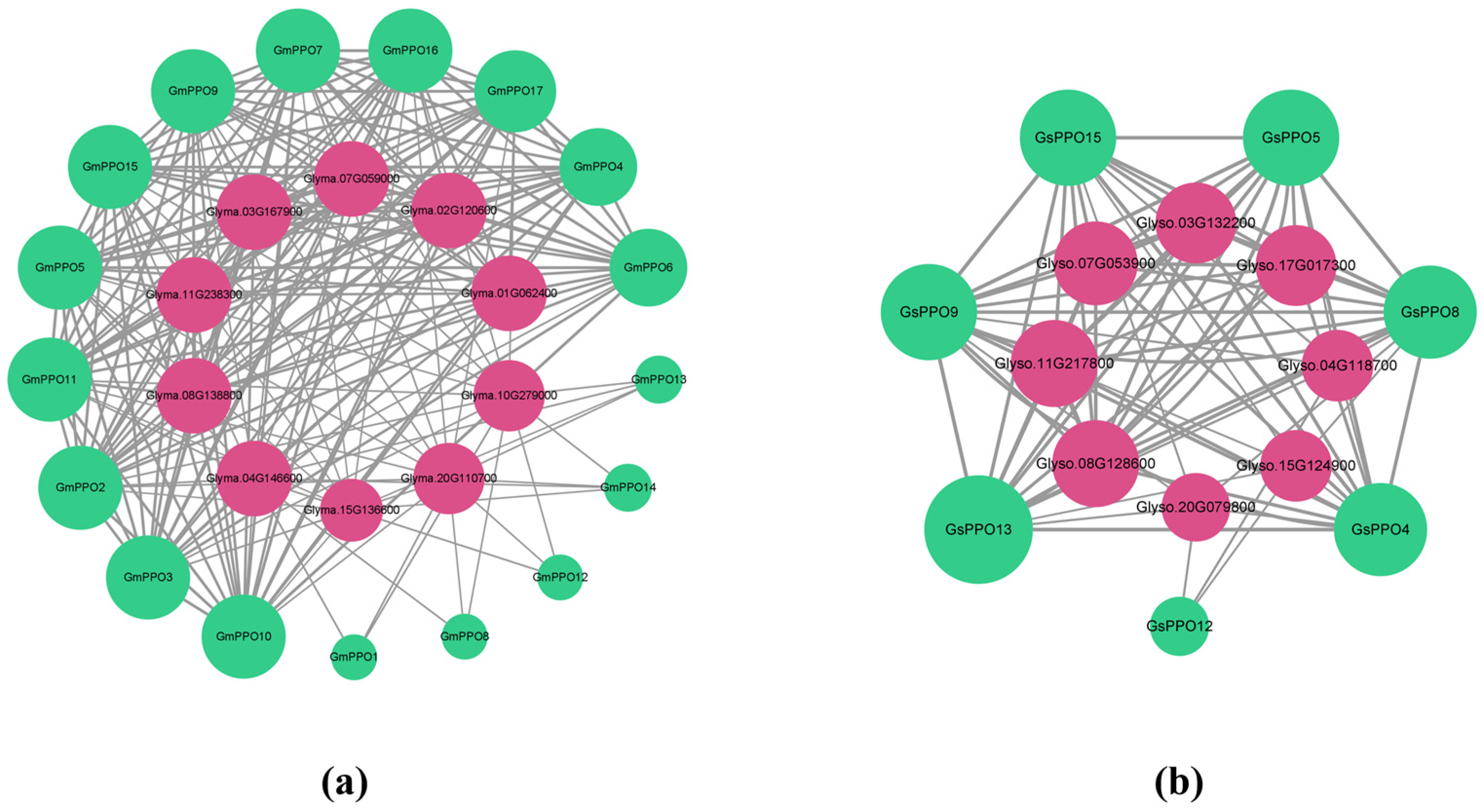
3.8. Prediction of 3D Structure and Protein Interactions of PPO Gene Family Members in Soybean and Wild Soybean
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beecher, B.S.; Carter, A.H.; See, D.R. Genetic mapping of new seed-expressed polyphenol oxidase genes in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2012, 124, 1463–1473. [Google Scholar] [CrossRef]
- Thipyapong, P.; Melkonian, J.; Wolfe, D.W.; Steffens, J.C. Suppression of polyphenol oxidases increases stress tolerance in tomato. Plant Sci. 2004, 167, 693–703. [Google Scholar] [CrossRef]
- Jia, H.F.; Zhao, P.C.; Wang, B.J.; Tariq, P.; Zhao, F.G. Overexpression of Polyphenol Oxidase Gene in Strawberry Fruit Delays the Fungus Infection Process. Plant Mol. Biol. Rep. 2016, 34, 592–606. [Google Scholar] [CrossRef]
- Llorente, B.; López, M.G.; Carrari, F.; Asís, R.; Naranjo, R.D.D.P.; Flawiá, M.M.; Alonso, G.D. Downregulation of polyphenol oxidase in potato tubers redirects phenylpropanoid metabolism enhancing chlorogenate content and late blight resistance. Mol. Breed. 2014, 34, 2049–2063. [Google Scholar] [CrossRef]
- Chen, X.; Yang, B.; Huang, W.; Wang, T.; Li, Y.; Zhong, Z.; Yang, L.; Tian, J. Molecular sciences comparative proteomic analysis reveals elevated capacity for photosynthesis in polyphenol oxidase expression-silenced Clematis terniflora DC. Leaves. Int. J. Mol. Sci. 2018, 19, 3897. [Google Scholar] [CrossRef]
- Bosch, M.; Berger, S.; Schaller, A.; Stintzi, A. Jasmonate-dependent induction of polyphenol oxidase activity in tomato foliage is important for defense against Spodoptera exigua but not against Manduca sexta. BMC Plant Biol. 2014, 14, 257. [Google Scholar] [CrossRef]
- Shikano, I.; Pan, Q.J.; Hoover, K.; Felton, G.W. Herbivore-Induced Defenses in Tomato Plants Enhance the Lethality of the Entomopathogenic Bacterium, Bacillus thuringiensis var. kurstaki. J. Chem. Ecol. 2018, 44, 947–956. [Google Scholar] [CrossRef]
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014, 164, 1191–1203. [Google Scholar] [CrossRef]
- Mahanil, S.; Attajarusit, J.; Stout, M.J.; Thipyapong, P. Overexpression of tomato polyphenol oxidase increases resistance to common cutworm. Plant Sci. 2008, 174, 456–466. [Google Scholar] [CrossRef]
- Guven, R.G.; Guven, K.; Bekler, F.M.; Acer, O.; Alkan, H.; Dogru, M. Purification and characterization of polyphenol oxidase from purslane. Food Sci. Technol. 2016, 62, 6–11. [Google Scholar] [CrossRef]
- Hisabori, T.; Sunamura, E.I.; Kim, Y.; Konno, H. The chloroplast atp synthase features the characteristic redox regulation machinery. Antioxid. Redox Signal. 2013, 19, 1846–1854. [Google Scholar]
- Thygesen, P.W.; Dry, I.B.; Robinson, S.P.; Belknap, W.R.; Vayda, M.E.; Park, W.D. Polyphenol oxidase in potato. A multigene family that exhibits differential expression patterns. Plant Physiol. 1994, 109, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Shahar, T. The Tomato 66.3-kD Polyphenoloxidase Gene: Molecular Identification and Developmental Expression. Plant Cell 1992, 4, 135–147. [Google Scholar]
- Shi, W.K.; Sun, Y.Y.; Shu, Z.M.; Zhang, Y.J.; Liang, Z.S.; Guo, H.B. Cloning and expression analysis of Danshen polyphenol oxidase gene. Northwest Bot. J. 2016, 36, 8. [Google Scholar]
- Tran, L.T.; Taylor, J.S.; Constabel, C. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef]
- Ferjani, A.; Tsukagoshi, H.; Vassileva, V. Model organisms in plant science: Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1279230. [Google Scholar] [CrossRef]
- Wilson, R.F. Soybean: Market driven research needs. Genet. Genom. Soybean 2008, 2, 3–15. [Google Scholar]
- Chao, F.; Ma, Y.; Yuan, L.; Zheng, W.; Rui, Y.; Zhou, Z.; Liu, T. Chloroplast DNA underwent independent selection from nuclear genes during soybean domestication and improvement. J. Genet. Genom. 2016, 43, 217–221. [Google Scholar]
- Zhang, Y.Y.; Hao, X.Y.; Liang, Z.S.; Lin, W.C.; Guo, H.B. Cloning and expression analysis of polyphenol oxidase gene (PPO) in lotus root. J. Agric. Biotechnol. 2011, 19, 634–641. [Google Scholar]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; AAAI Press: Menlo Park, CA, USA, 1994; pp. 28–36. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Kathleen, M.; Yves, M.; Yves, V.P.; Pierre, R.; Stephane, R. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef]
- Li, M.X. Comparative Study on the Adaptability of Cultivated and Wild Soybeans Under Salt and Low Nitrogen Stress. Ph.D. Dissertation, Northeast Normal University, Changchun, China, 2020. [Google Scholar]
- Xu, L.; Dong, Z.B.; Fang, L.; Luo, Y.J.; Wei, Z.Y.; Guo, H.L.; Zhang, G.Q.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.Y.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, 52–58. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Ai, E. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.X.; Suo, H.C.; Hu, X.X.; Qin, Y.Z.; Li, X.B.; Xiong, X.Y. Research progress on polyphenol oxidase genes in plants. Mol. Plant Breed. 2020, 18, 4629–4636. [Google Scholar]
- Matt, L.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar]
- Pfeil, B.E.; Schlueter, J.A.; Shoemaker, R.C.; Doyle, J.J. Placing paleopolyploidy in relation to taxon divergence: A phylogenetic analysis in legumes using 39 gene families. Syst. Biol. 2005, 54, 441–454. [Google Scholar] [CrossRef]
- Schlueter, J.A.; Lin, J.Y.; Schlueter, S.D.; Vasylenkosanders, I.F.; Deshpande, S.; Yi, J.; O’Bleness, M.; Roe, B.A.; Nelson, R.T.; Scheffler, B.E. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genom. 2007, 8, 330. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Dai, A.H.; Yang, S.X.; Zhou, H.K.; Tang, K.Q.; Li, G.; Leng, J.T.; Yu, H.; Zhang, Y.H.; Gao, J.S.; Yang, X. Evolution and Expression Divergence of the CYP78A Subfamily Genes in Soybean. Genes 2018, 9, 611. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Yang, M.; Tang, Y.; Zhang, L.; Zhao, H.; Chen, Q.; Meng, F.; Jiang, J. Dynamics of cis-regulatory sequences and transcriptional divergence of duplicated genes in soybean. Proc. Natl. Acad. Sci. USA 2023, 120, e2303836120. [Google Scholar] [CrossRef]
- Wang, Q.; Li, G.; Zheng, K.; Zhu, X.; Ma, J.; Wang, D.; Tang, K.; Feng, X.; Leng, J.; Yu, H.; et al. The Soybean Laccase Gene Family: Evolution and Possible Roles in Plant Defense and Stem Strength Selection. Genes 2019, 10, 701. [Google Scholar] [CrossRef]
- Mu, H.N.; Wang, L.G.; Sun, T.Z. Cloning and expression analysis of polyphenol oxidase (PPO) gene in Osmanthus fragrans. Mol. Plant Breed. 2017, 15, 6. [Google Scholar]
- Liu, F.J.; Feng, Z.S.; Bai, Y.J.; Wang, M.; Huang, W.; Huang, W.S. Cloning and sequence analysis of polyphenol oxidase gene in seedless white grape. Preserv. Process. 2017, 17, 7. [Google Scholar]
- Zhang, Y.H.; Zhang, X.; Dong, L.; Dong, Q.H. Cloning and expression vector construction of the full-length polyphenol oxidase gene in strawberries. J. Beijing Agric. Univ. 2018, 33, 8. [Google Scholar]
- Sommer, A.; Ne′eman, E.; Steffens, J.C. Import, targeting, and processing of aplant polyphenol oxidase. Plant Physiol. 1994, 105, 1301–1311. [Google Scholar] [CrossRef]
- Hunt, M.D.; Eanneta, N.T.; Yu, H. cDNA cloning and expression of potato polyphenol oxidase. Plant Mol. Biol. 1993, 21, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhai, X.; Zhang, L.; Yang, Y.; Zhu, H.; Lü, H.; Xiong, E.; Chu, S.; Zhang, X.; Zhang, D. Genome-Wide Identification of the Whirly Gene Family and Its Potential Function in Low Phosphate Stress in Soybean (Glycine max). Genes 2024, 15, 833. [Google Scholar] [CrossRef]
- Newman, S.M.; Eannetta, N.T.; Yu, H.; Prince, J.P.; Steffens, J.C. Organisation of the tomato polyphenol oxidase gene family. Plant Mol. Biol. 1993, 21, 1035–1051. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.R.L.; Mccallion, R.F. The Selective Inhibition of ortho- and para-Diphenol Oxidases. Phytochemistry 1980, 19, 373–377. [Google Scholar] [CrossRef]
- Kim, M.Y.; Van, K.; Kang, Y.J.; Kim, K.H.; Lee, S.H. Tracing soybean domestication history: From nucleotide to genome. Breed. Sci. 2012, 61, 445–452. [Google Scholar] [CrossRef]
- Tran, L.T.; Constabel, C.P. The polyphenol oxidase gene family in poplar: Phylogeny, differential expression and identification of a novel, vacuolar isoform. Planta 2011, 234, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]



| Name | Gene ID | Amino Acid Number | MW (Da) | PI | Subcellular Localization Score | |||
|---|---|---|---|---|---|---|---|---|
| Cytoplasm | Nucleus | Chloroplast | Vacuole | |||||
| GmPPO1 | Glyma.01G139800 | 274 | 31,230.1 | 5.84 | 5 | 4 | 3 | |
| GmPPO2 | Glyma.04G121700 | 601 | 67,204.5 | 7.12 | 14 | |||
| GmPPO3 | Glyma.06G270400 | 596 | 66,907.4 | 7.79 | 1 | 12 | ||
| GmPPO4 | Glyma.07G193300 | 605 | 68,939.9 | 6.18 | 2 | 8 | ||
| GmPPO5 | Glyma.07G193400 | 554 | 63,047 | 6.85 | 4 | 5 | ||
| GmPPO6 | Glyma.07G193500 | 604 | 68,829.9 | 6.27 | 1 | 9 | ||
| GmPPO7 | Glyma.07G193600 | 583 | 66,247.4 | 6.22 | 1 | 6 | 5 | |
| GmPPO8 | Glyma.13G183000 | 568 | 63,936.1 | 7.76 | 2 | 10 | ||
| GmPPO9 | Glyma.13G183200 | 605 | 68,257.8 | 7.86 | 3 | 1 | 8 | |
| GmPPO10 | Glyma.13G183500 | 620 | 69,586.2 | 6.29 | 2 | 9 | ||
| GmPPO11 | Glyma.13G242300 | 601 | 67,723.9 | 7.27 | 1 | 10.5 | ||
| GmPPO12 | Glyma.13G242400 | 576 | 64,359.9 | 7.36 | 1 | 12 | ||
| GmPPO13 | Glyma.15G071000 | 578 | 64,686.8 | 7.44 | 12 | |||
| GmPPO14 | Glyma.15G071100 | 521 | 59,024.3 | 8.68 | 1 | 2 | 7 | |
| GmPPO15 | Glyma.15G071200 | 602 | 67,819.2 | 7.67 | 1 | 12 | ||
| GmPPO16 | Glyma.18G225000 | 601 | 68,922.9 | 7.39 | 1 | 1 | 2 | 7 |
| GmPPO17 | Glyma.18G225100 | 584 | 66,815.1 | 6.55 | 1 | 1 | 6 | |
| Name | Gene ID | Amino Acid Number | MW (Da) | PI | Subcellular Localization Score | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cytoplasm | Nucleus | Chloroplast | Peroxisome | Mitochondrion | |||||
| GsPPO1 | Glyso.04G102600 | 602 | 67,204.5 | 7.12 | 14 | ||||
| GsPPO2 | Glyso.06G232000 | 436 | 49,464.6 | 6.38 | 2 | 3 | 3 | 3 | 3 |
| GsPPO3 | Glyso.07G158900 | 579 | 66,147.8 | 6.12 | 4 | 6 | 2 | 2 | |
| GsPPO4 | Glyso.07G159000 | 579 | 65,503.7 | 6.88 | 1 | 11 | 1 | 1 | |
| GsPPO5 | Glyso.07G159100 | 578 | 66,037.8 | 6.21 | 3 | 6 | 2 | 2 | |
| GsPPO6 | Glyso.07G159200 | 620 | 69,897.5 | 6.34 | 12.5 | ||||
| GsPPO7 | Glyso.13G146200 | 621 | 69,572.1 | 6.29 | 2 | 9 | 1 | 1 | |
| GsPPO8 | Glyso.13G146400 | 625 | 70,134.9 | 8.16 | 12.5 | ||||
| GsPPO9 | Glyso.13G146700 | 621 | 69,600.2 | 6.29 | 2 | 9 | 1 | 1 | |
| GsPPO10 | Glyso.13G200100 | 602 | 67,751.9 | 7.27 | 10.5 | 1 | |||
| GsPPO11 | Glyso.13G200200 | 593 | 66,072.9 | 7.35 | 1 | 12 | 1 | ||
| GsPPO12 | Glyso.15G066400 | 638 | 71,107 | 7.18 | 12.5 | 1.5 | |||
| GsPPO13 | Glyso.15G066500 | 603 | 67,819.2 | 7.67 | 1 | 12 | 1 | ||
| GsPPO14 | Glyso.18G171400 | 349 | 39,845.6 | 5.6 | 8 | 3 | 2 | ||
| GsPPO15 | Glyso.18G171500 | 585 | 66,829.2 | 6.62 | |||||
| ID | Functional Annotation | Chain Pair pae min | Chain ptm | Score |
|---|---|---|---|---|
| Glyma.08G138800 | Reproduction | 0.76, 26.45; 27.5, 0.76 | 0.79, 0.82 | 26.45 |
| Glyma.07G059000 | Organic acid metabolic process | 0.76, 26.95; 26.32, 0.76 | 0.76, 0.88 | 26.32 |
| Glyma.11G238300 | Reproduction | 0.76, 26.28; 26.8, 0.76 | 0.79, 0.81 | 26.28 |
| Glyma.10G279000 | Protein dephosphorylation | 0.76, 26.77; 26.21, 0.76 | 0.78, 0.48 | 26.21 |
| Glyma.03G167900 | Organic acid metabolic process | 0.76, 25.57; 26.38, 0.76 | 0.78, 0.89 | 25.57 |
| GmPPO6 | - | 0.76, 24.31; 23.84, 0.76 | 0.73, 0.7 | 23.84 |
| GmPPO17 | - | 0.76, 22.59; 22.89, 0.76 | 0.72, 0.74 | 22.59 |
| GmPPO7 | - | 0.76, 22.5; 22.44, 0.76 | 0.73, 0.75 | 22.44 |
| GmPPO5 | - | 0.76, 23.27; 22.18, 0.76 | 0.73, 0.73 | 22.18 |
| GmPPO10 | - | 0.76, 22.57; 21.94, 0.76 | 0.74, 0.71 | 21.94 |
| Glyma.20G110700 | Protein dephosphorylation | 0.76, 25.05; 21.54, 0.76 | 0.76, 0.84 | 21.54 |
| GmPPO9 | - | 0.76, 21.38; 22.98, 0.76 | 0.72, 0.74 | 21.38 |
| GmPPO4 | - | 0.76, 24.36; 24.01, 0.76 | 0.71, 0.68 | 21.01 |
| GmPPO16 | - | 0.76, 21.92; 19.99, 0.76 | 0.73, 0.73 | 19.99 |
| Glyma.02G120600 | Nitrogen compound metabolic process | 0.76, 16.53; 16.66, 0.76 | 0.76, 0.89 | 16.53 |
| Glyma.04G146600 | Double-strand break repair via homologous recombination | 0.76, 15.82; 17.74, 0.76 | 0.78, 0.85 | 15.82 |
| GmPPO15 | - | 0.76, 14.39; 14.92, 0.76 | 0.61, 0.61 | 14.39 |
| Glyma.15G136600 | Tyrosinase copper-binding domain, and O-acyltransferase WSD1, C-terminal | 0.76, 14.39; 13.92, 0.76 | 0.61, 0.61 | 13.92 |
| GmPPO3 | - | 0.76, 8.12; 7.72, 0.76 | 0.78, 0.78 | 7.72 |
| Glyma.01G062400 | Nitrogen compound metabolic process | 0.76, 6.73; 8.87, 0.76 | 0.78, 0.9 | 6.73 |
| GmPPO11 | - | 0.76, 4.34; 4.31, 0.76 | 0.77, 0.77 | 4.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Z.; Wang, B.; Liu, J.; Liu, N.; Yi, Z.; Li, Z.; Dong, Z.; Zhang, C.; Dong, Y.; Li, Y. Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja. Genes 2025, 16, 17. https://doi.org/10.3390/genes16010017
Song Z, Wang B, Liu J, Liu N, Yi Z, Li Z, Dong Z, Zhang C, Dong Y, Li Y. Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja. Genes. 2025; 16(1):17. https://doi.org/10.3390/genes16010017
Chicago/Turabian StyleSong, Ziye, Bo Wang, Jia Liu, Nianxi Liu, Zhigang Yi, Zhi Li, Zhimin Dong, Chunbao Zhang, Yingshan Dong, and Yuqiu Li. 2025. "Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja" Genes 16, no. 1: 17. https://doi.org/10.3390/genes16010017
APA StyleSong, Z., Wang, B., Liu, J., Liu, N., Yi, Z., Li, Z., Dong, Z., Zhang, C., Dong, Y., & Li, Y. (2025). Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja. Genes, 16(1), 17. https://doi.org/10.3390/genes16010017







