From Species to Varieties: How Modern Sequencing Technologies Are Shaping Medicinal Plant Identification
Abstract
1. Introduction
2. Materials and Methods
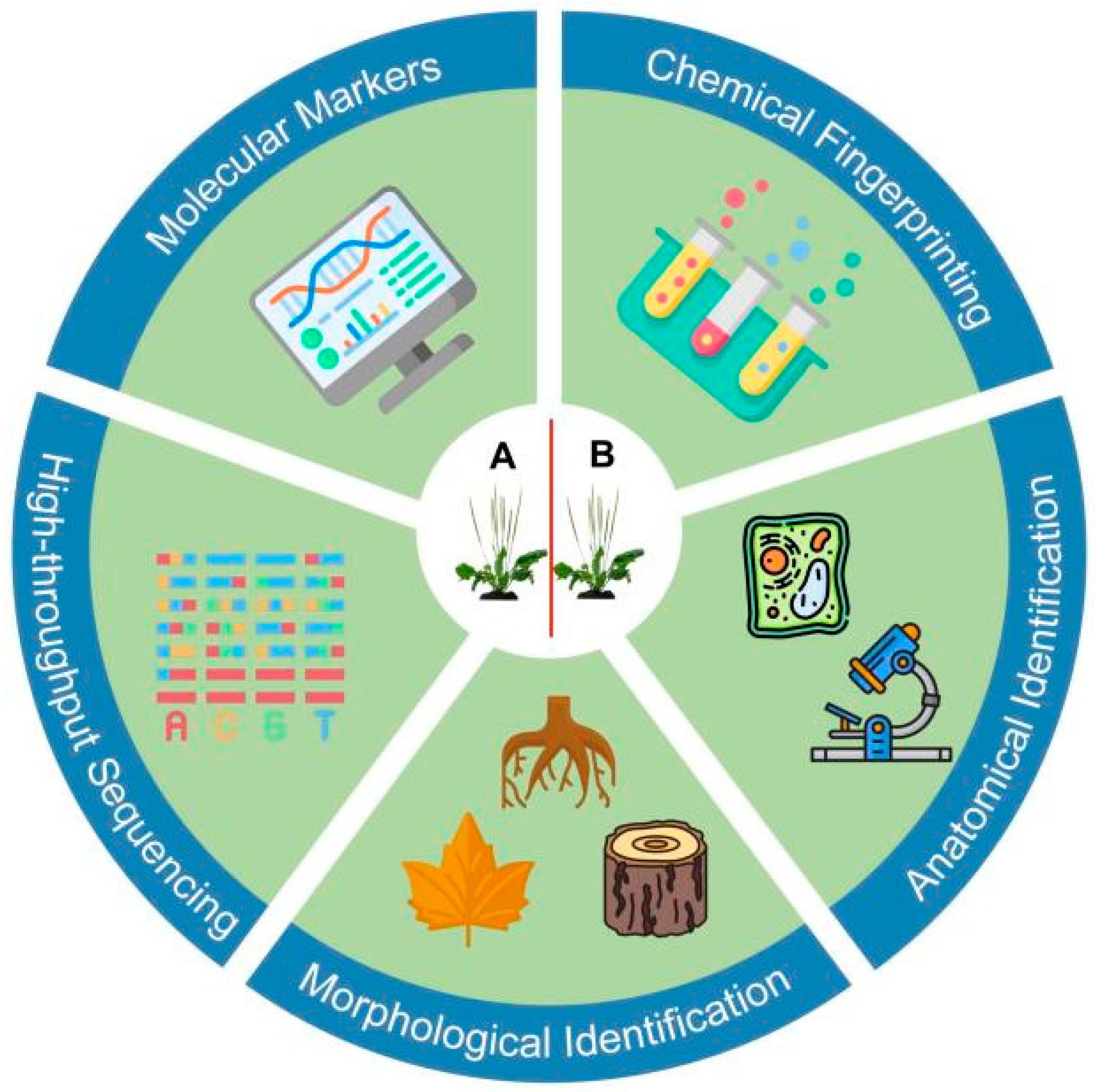
3. Historical Approaches to Medicinal Plant Identification
3.1. Morphological and Anatomical Identification
3.2. Chemical Fingerprinting
3.3. Limitations of Traditional Methods
3.4. Transition to Molecular Methods
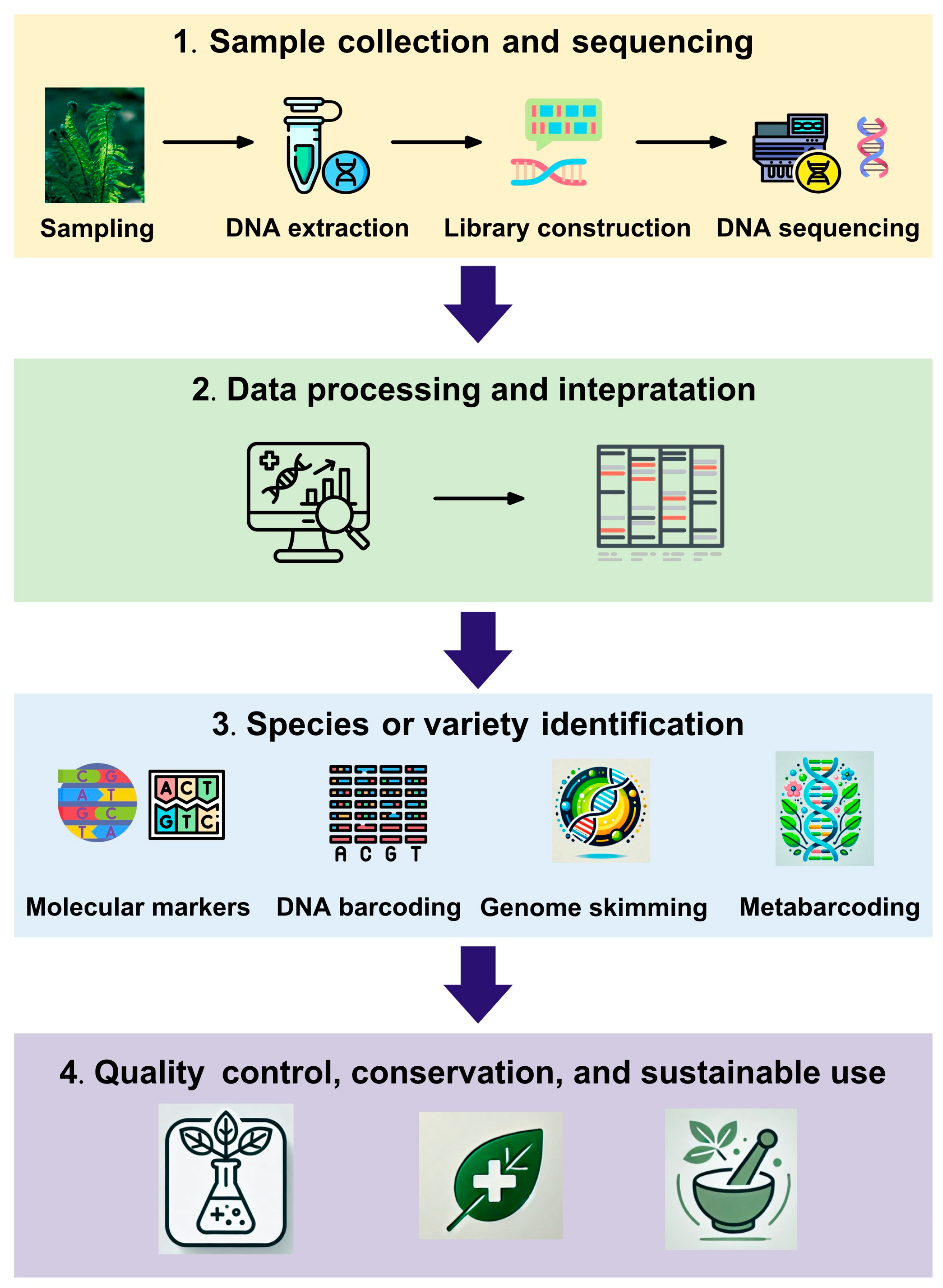
4. Advancements in DNA-Based Identification Methods
4.1. Molecular Markers for Species and Variety Identification
4.2. DNA Barcoding and Super-Barcoding: Principles and Applications
4.3. Advantages of DNA Barcoding over Traditional Methods
4.4. Challenges and Limitations of DNA Barcoding and Super-Barcoding
5. NGS in Plant Identification
5.1. Overview of NGS Technologies
5.2. Genome Skimming for Plant Identification
5.3. Metabarcoding for Complex Herbal Mixtures
5.4. Benefits and Challenges of NGS in Plant Identification
6. TGS in Plant Identification
6.1. Introduction to TGS Platforms
6.2. Advantages of TGS for Plant Genomics
6.3. Applications in Identifying Complex or Hybrid Genomes
6.4. Role of TGS in Enhancing Medicinal Plant Authentication
7. Integration of Multi-Omics Approaches for Comprehensive Identification
7.1. Combining Genomics, Transcriptomics, and Metabolomics
7.2. The Role of Epigenetics and Gene Expression Profiling
7.3. Integrating Sequencing Data with Chemical Profiling
7.4. Integrating Genome Editing-Based Approaches
7.5. Integrating Other Innovative Approaches
7.6. Challenges and the Need for Standardization
8. Challenges, Future Directions, and Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.J.; Khan, I.A. DNA barcoding for the identification of botanicals in herbal medicine and dietary supplements: Strengths and limitations. Planta Med. 2016, 82, 1225–1235. [Google Scholar] [CrossRef]
- Tasneem, S.; Liu, B.; Li, B.; Choudhary, M.I.; Wang, W. Molecular pharmacology of inflammation: Medicinal plants as anti-inflammatory agents. Pharmacol. Res. 2019, 139, 126–140. [Google Scholar] [CrossRef]
- Süntar, I. Importance of ethnopharmacological studies in drug discovery: Role of medicinal plants. Phytochem. Rev. 2020, 19, 1199–1209. [Google Scholar] [CrossRef]
- Nwozo, O.S.; Effiong, E.M.; Aja, P.M.; Awuchi, C.G. Antioxidant, phytochemical, and therapeutic properties of medicinal plants: A review. Int. J. Food Prop. 2023, 26, 359–388. [Google Scholar] [CrossRef]
- Al-Worafi, Y.M. Herbal Medicines Safety Issues. In Drug Safety in Developing Countries; Academic Press: Cambridge, MA, USA, 2020; pp. 163–178. [Google Scholar]
- Duminil, J.; Di Michele, M. Plant species delimitation: A comparison of morphological and molecular markers. Plant Biosyst. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- Crang, R.; Lyons-Sobaski, S.; Wise, R. Plant Anatomy: A Concept-Based Approach to the Structure of Seed Plants; Springer: New York, NY, USA, 2018. [Google Scholar]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Nettle, D.; Bateson, M. Adaptive developmental plasticity: What is it, how can we recognize it and when can it evolve? Proc. R. Soc. B 2015, 282, 20151005. [Google Scholar] [CrossRef] [PubMed]
- Barthélémy, D.; Caraglio, Y. Plant architecture: A dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Ann. Bot. 2007, 99, 375–407. [Google Scholar] [CrossRef]
- Soni, U.; Brar, S.; Gauttam, V.K. Effect of seasonal variation on secondary metabolites of medicinal plants. Int. J. Pharm. Sci. Res. 2015, 6, 3654–3662. [Google Scholar]
- Kessler, A.; Kalske, A. Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Techen, N.; Parveen, I.; Pan, Z.; Khan, I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Nevill, P.G.; Zhong, X.; Tonti-Filippini, J.; Byrne, M.; Hislop, M.; Thiele, K.; Van Leeuwen, S.; Boykin, L.M.; Small, I. Large scale genome skimming from herbarium material for accurate plant identification and phylogenomics. Plant Methods 2020, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Xu, W.; Xin, T.; Song, J. Application of third-generation sequencing to herbal genomics. Front. Plant Sci. 2023, 14, 1124536. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Kelso, J. High-throughput DNA sequencing–concepts and limitations. Bioessays 2010, 32, 524–536. [Google Scholar] [CrossRef]
- Ward, R.M.; Schmieder, R.; Highnam, G.; Mittelman, D. Big data challenges and opportunities in high-throughput sequencing. Syst. Biomed. 2013, 1, 29–34. [Google Scholar] [CrossRef]
- Pieruschka, R.; Schurr, U. Plant phenotyping: Past, present, and future. Plant Phenomics 2019, 2019, 7507131. [Google Scholar] [CrossRef] [PubMed]
- Molina-Montenegro, M.A.; Atala, C.; Gianoli, E. Phenotypic plasticity and performance of Taraxacum officinale (dandelion) in habitats of contrasting environmental heterogeneity. Biol. Invasions 2010, 12, 2277–2284. [Google Scholar] [CrossRef]
- Tátrai, Z.A.; Sanoubar, R.; Pluhár, Z.; Mancarella, S.; Orsini, F.; Gianquinto, G. Morphological and physiological plant responses to drought stress in Thymus citriodorus. Int. J. Agron. 2016, 2016, 4165750. [Google Scholar] [CrossRef]
- Noviana, E.; Indrayanto, G.; Rohman, A. Advances in fingerprint analysis for standardization and quality control of herbal medicines. Front. Pharmacol. 2022, 13, 853023. [Google Scholar] [CrossRef]
- Ali, A.H. High-performance liquid chromatography (HPLC): A review. Ann. Adv. Chem. 2022, 6, 10–020. [Google Scholar]
- Al-Rubaye, A.F.; Hameed, I.H.; Kadhim, M.J. A review: Uses of gas chromatography-mass spectrometry (GC-MS) technique for analysis of bioactive natural compounds of some plants. Int. J. Toxicol. Pharmacol. Res. 2017, 9, 81–85. [Google Scholar] [CrossRef]
- Ciura, K.; Dziomba, S.; Nowakowska, J.; Markuszewski, M.J. Thin layer chromatography in drug discovery process. J. Chromatogr. A 2017, 1520, 9–22. [Google Scholar] [CrossRef]
- Ciechomska, M.; Woźniakiewicz, M.; Machlowska, K.; Klepacki, P.; Kościelniak, P. Differentiation of Solanaceae psychoactive plants based on GC-MS analysis supported by chemometric tools. Microchem. J. 2019, 150, 104098. [Google Scholar] [CrossRef]
- Lv, X.; Li, Y.; Tang, C.; Zhang, Y.; Zhang, J.; Fan, G. Integration of HPLC-based fingerprint and quantitative analyses for differentiating botanical species and geographical growing origins of Rhizoma coptidis. Pharm. Biol. 2016, 54, 3264–3271. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, J.; Li, F.; Liu, X.; Deng, M.; Wu, H. Determination of alkaloid contents in various tissues of Coptis chinensis Franch. by reversed phase-high performance liquid chromatography and ultraviolet spectrophotometry. J. Chromatogr. Sci. 2017, 55, 556–563. [Google Scholar] [CrossRef]
- Zeng, H.; Su, S.; Ang, X.; Sha, X.; Zhu, Z.; Wang, Y.; Guo, S.; Yan, H.; Qian, D.; Duan, J. Comparative analysis of the major chemical constituents in Salvia miltiorrhiza roots, stems, leaves and flowers during different growth periods by UPLC-TQ-MS/MS and HPLC-ELSD methods. Molecules 2017, 22, 771. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Long, H.; Wu, X.; Hou, J.; Gao, L.; Yao, S.; Lei, M.; Zhang, Z.; Guo, D.A.; Wu, W. Quantitative and fingerprint analysis of proanthocyanidins and phenylpropanoids in Cinnamomum verum bark, Cinnamomum cassia bark, and Cassia twig by UPLC combined with chemometrics. Eur. Food Res. Technol. 2021, 247, 2687–2698. [Google Scholar] [CrossRef]
- Kindscher, K.; Wittenberg, R. The naming and classification of Echinacea species. In Echinacea: Herbal Medicine with a Wild History; Kindscher, K., Ed.; Springer: Cham, Switzerland, 2016; pp. 37–45. [Google Scholar]
- Zhou, M.; Gong, X.; Pan, Y. Panax species identification with the assistance of DNA data. Genet. Resour. Crop Evol. 2018, 65, 1839–1856. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Wang, R.; Luo, S.; Nong, S.; Wang, J.; Chen, X. Applications of molecular markers in conserving endangered species. Biodivers. Sci. 2020, 28, 367. [Google Scholar]
- Supple, M.A.; Shapiro, B. Conservation of biodiversity in the genomics era. Genome Biol. 2018, 19, 1–12. [Google Scholar] [CrossRef]
- Klein-Junior, L.C.; de Souza, M.R.; Viaene, J.; Bresolin, T.M.; de Gasper, A.L.; Henriques, A.T.; Vander Heyden, Y. Quality control of herbal medicines: From traditional techniques to state-of-the-art approaches. Planta Med. 2021, 87, 964–988. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Bjørnstad, Å.; Ndjiondjop, M.N. An overview of molecular marker methods for plants. Afr. J. Biotechnol. 2006, 5, 2584–2597. [Google Scholar]
- Shirasawa, K.; Ishii, K.; Kim, C.; Ban, T.; Suzuki, M.; Ito, T.; Muranaka, T.; Kobayashi, M.; Nagata, N.; Isobe, S.; et al. Development of Capsicum EST–SSR markers for species identification and in silico mapping onto the tomato genome sequence. Mol. Breed. 2013, 31, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Abou Zid, S. Authentication of Silybum marianum varieties using RAPD analysis. Plant Tissue Cult. Biotechnol. 2014, 24, 57–63. [Google Scholar]
- Taheri, S.; Lee Abdullah, T.; Yusop, M.R.; Hanafi, M.M.; Sahebi, M.; Azizi, P.; Shamshiri, R.R. Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules 2018, 23, 399. [Google Scholar] [CrossRef] [PubMed]
- Dvornyk, V.; Long, J.R.; Ong, D.H.; Liu, P.Y.; Zhao, L.J.; Shen, H.; Zhang, Y.Y.; Liu, Y.J.; Rocha-Sanchez, S.; Gao, P.; et al. Current limitations of SNP data from the public domain for studies of complex disorders: A test for ten candidate genes for obesity and osteoporosis. BMC Genet. 2004, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.R.; Telles, M.P.C.; Diniz-Filho, J.A.F.; Soares, T.N.; Melo, D.B.; Oliveira, G. Optimizing reproducibility evaluation for random amplified polymorphic DNA markers. Genet. Mol. Res. 2008, 7, 1384–1391. [Google Scholar] [CrossRef]
- Blignaut, M.; Ellis, A.G.; Le Roux, J.J. Towards a transferable and cost-effective plant AFLP protocol. PLoS ONE 2013, 8, e61704. [Google Scholar] [CrossRef]
- Dunning, L.T.; Savolainen, V. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Bot. J. Linn. Soc. 2010, 164, 1–9. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Chen, S.; Yin, X.; Han, J.; Sun, W.; Yao, H.; Song, J.; Li, X. DNA barcoding in herbal medicine: Retrospective and prospective. J. Pharm. Anal. 2023, 13, 431–441. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, J.; Folk, R.A.; Zhao, J.; Liu, J.; He, Z.; Peng, H.; Yang, S.; Wang, C.; Yu, X. Species delimitation of tea plants (Camellia sect. Thea) based on super-barcodes. BMC Plant Biol. 2024, 24, 181. [Google Scholar] [CrossRef]
- Wu, L.; Wu, M.; Cui, N.; Wang, L.; Li, Y.; Li, X.; Chen, S. Plant super-barcode: A case study on genome-based identification for closely related species of Fritillaria. Chin. Med. 2021, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Antil, S.; Abraham, J.S.; Sripoorna, S.; Maurya, S.; Dagar, J.; Makhija, S.; Bhagat, P.; Gupta, R.; Sood, U.; Lal, R.; et al. DNA barcoding, an effective tool for species identification: A review. Mol. Biol. Rep. 2023, 50, 761–775. [Google Scholar] [CrossRef]
- Gesto-Borroto, R.; Medina-Jiménez, K.; Lorence, A.; Villarreal, M.L. Application of DNA barcoding for quality control of herbal drugs and their phytopharmaceuticals. Rev. Bras. Farmacogn. 2021, 31, 127–141. [Google Scholar] [CrossRef]
- Meiklejohn, K.A.; Damaso, N.; Robertson, J.M. Assessment of BOLD and GenBank—Their accuracy and reliability for the identification of biological materials. PLoS ONE 2019, 14, e0217084. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.T.; Shaw, P.C. Application of next-generation sequencing for the identification of herbal products. Biotechnol. Adv. 2019, 37, 107450. [Google Scholar] [CrossRef] [PubMed]
- Dodsworth, S. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 2015, 20, 525–527. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Li, P. Development of genomic resources for the genus Celtis (Cannabaceae) based on genome skimming data. Plant Divers. 2021, 43, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Tang, L.; He, P.; Miao, K.; Yang, C.; Liu, H.; Ji, Y. Genome skimming as an efficient tool for authenticating commercial products of the pharmaceutically important Paris yunnanensis (Melanthiaceae). BMC Plant Biol. 2023, 23, 344. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, M.E. From barcoding single individuals to metabarcoding biological communities: Towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 2014, 29, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Milla, L.; Sniderman, K.; Lines, R.; Mousavi-Derazmahalleh, M.; Encinas-Viso, F. Pollen DNA metabarcoding identifies regional provenance and high plant diversity in Australian honey. Ecol. Evol. 2021, 11, 8683–8698. [Google Scholar] [CrossRef] [PubMed]
- Letsiou, S.; Madesis, P.; Vasdekis, E.; Montemurro, C.; Grigoriou, M.E.; Skavdis, G.; Moussis, V.; Koutelidakis, A.E.; Tzakos, A.G. DNA barcoding as a plant identification method. Appl. Sci. 2024, 14, 1415. [Google Scholar] [CrossRef]
- Urumarudappa, S.K.J.; Tungphatthong, C.; Prombutara, P.; Sukrong, S. DNA metabarcoding to unravel plant species composition in selected herbal medicines on the National List of Essential Medicines (NLEM) of Thailand. Sci. Rep. 2020, 10, 18259. [Google Scholar] [CrossRef] [PubMed]
- Seethapathy, G.S.; Raclariu-Manolica, A.C.; Anmarkrud, J.A.; Wangensteen, H.; de Boer, H.J. DNA metabarcoding authentication of ayurvedic herbal products on the European market raises concerns of quality and fidelity. Front. Plant Sci. 2019, 10, 68. [Google Scholar] [CrossRef]
- Wang, M.; Li, R.; Zhao, Q. Multi-omics techniques in genetic studies and breeding of forest plants. Forests 2023, 14, 1196. [Google Scholar] [CrossRef]
- Xu, S.; Chen, R.; Zhang, X.; Wu, Y.; Yang, L.; Sun, Z.; Zhu, Z.; Song, A.; Wu, Z.; Li, T.; et al. The evolutionary tale of lilies: Giant genomes derived from transposon insertions and polyploidization. The Innovation 2024, 5, 100726. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Lee, G.W.; Ko, S.R.; Go, S.; Kwon, S.Y.; Kim, Y.M.; Shin, A.Y. Two long read-based genome assembly and annotation of polyploidy woody plants, Hibiscus syriacus L. using PacBio and Nanopore platforms. Sci. Data 2023, 10, 713. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, G.; Yang, F.; Liang, Y.; Gao, Q.; Li, X.; Yang, R.; Zhang, G.; Jiang, H.; Yu, L. Multilayered regulation of secondary metabolism in medicinal plants. Mol. Hortic. 2023, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, M.; Dong, S.; Wu, X.; Zhang, G.; He, L.; Jiao, Y.; Chen, S.; Li, L.; Luo, H. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 2023, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Small, E. Evolution and classification of Cannabis sativa (marijuana, hemp) in relation to human utilization. Bot. Rev. 2015, 81, 189–294. [Google Scholar] [CrossRef]
- Radosavljević, I.; Bogdanović, S.; Celep, F.; Filipović, M.; Satovic, Z.; Surina, B.; Liber, Z. Morphological, genetic and epigenetic aspects of homoploid hybridization between Salvia officinalis L. and Salvia fruticosa Mill. Sci. Rep. 2019, 9, 3276. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Leng, L.; Sun, W.; Liu, B.; Feng, X.; Li, X.; Chen, S. Whole-genome sequencing in medicinal plants: Current progress and prospect. Sci. China Life Sci. 2024, 67, 258–273. [Google Scholar] [CrossRef]
- Alami, M.M.; Ouyang, Z.; Zhang, Y.; Shu, S.; Yang, G.; Mei, Z.; Wang, X. The current developments in medicinal plant genomics enabled the diversification of secondary metabolites’ biosynthesis. Int. J. Mol. Sci. 2022, 23, 15932. [Google Scholar] [CrossRef]
- Wang, W.; Xu, J.; Fang, H.; Li, Z.; Li, M. Advances and challenges in medicinal plant breeding. Plant Sci. 2020, 298, 110573. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulou, K.; Boti, M.A.; Adamopoulos, P.G.; Skourou, P.C.; Scorilas, A. Third-generation sequencing: The spearhead towards the radical transformation of modern genomics. Life 2021, 12, 30. [Google Scholar] [CrossRef]
- Shi, J.; Tian, Z.; Lai, J.; Huang, X. Plant pan-genomics and its applications. Mol. Plant 2023, 16, 168–186. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, S.; Li, R.; Zhao, Q. Unraveling the specialized metabolic pathways in medicinal plant genomes: A review. Front. Plant Sci. 2024, 15, 1459533. [Google Scholar] [CrossRef]
- Lei, L.; Goltsman, E.; Goodstein, D.; Wu, G.A.; Rokhsar, D.S.; Vogel, J.P. Plant pan-genomics comes of age. Annu. Rev. Plant Biol. 2021, 72, 411–435. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Y.; Peng, J.; Fan, B.; Xu, D.; Wu, J.; Cao, Z.; Gao, Y.; Wang, X.; Li, S.; et al. High-quality genome assembly and pan-genome studies facilitate genetic discovery in mung bean and its improvement. Plant Commun. 2022, 3, 100352. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Y.; Huang, L.; Cui, X.; Liu, Y. From single-to multi-omics: Future research trends in medicinal plants. Brief. Bioinform. 2023, 24, bbac485. [Google Scholar] [CrossRef]
- Singh, K.S.; van der Hooft, J.J.; van Wees, S.C.; Medema, M.H. Integrative omics approaches for biosynthetic pathway discovery in plants. Nat. Prod. Rep. 2022, 39, 1876–1896. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, W.; Fu, X.; Zhang, Y.; Hassani, D.; Liu, H.; Chen, T.; Yan, X.; Peng, B. Transcriptomic analysis reveals the parallel transcriptional regulation of UV-B-induced artemisinin and flavonoid accumulation in Artemisia annua L. Plant Physiol. Biochem. 2021, 163, 189–200. [Google Scholar] [CrossRef]
- Sharma, B.; Yadav, D.K. Metabolomics and network pharmacology in the exploration of the multi-targeted therapeutic approach of traditional medicinal plants. Plants 2022, 11, 3243. [Google Scholar] [CrossRef] [PubMed]
- Rapp, R.A.; Wendel, J.F. Epigenetics and plant evolution. New Phytol. 2005, 168, 81–91. [Google Scholar] [CrossRef]
- Li, J.; Li, C.; Lu, S. Identification and characterization of the cytosine-5 DNA methyltransferase gene family in Salvia miltiorrhiza. PeerJ 2018, 6, e4461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Liu, X.; Cui, Y.; Wang, Y.H.; Lv, Z.L.; Cheng, L.; Liu, B.; Liu, H.; Liu, X.Y.; Deyholos, M.K.; et al. Genomic, transcriptomic, and metabolomic analyses provide insights into the evolution and development of a medicinal plant Saposhnikovia divaricata (Apiaceae). Hortic. Res. 2024, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Xu, W.; Qi, G.; Xin, T.; Xu, Z.; Lei, H.; Song, J. GAGE is a method for identification of plant species based on whole genome analysis and genome editing. Commun. Biol. 2022, 5, 947. [Google Scholar] [CrossRef] [PubMed]
- Belton, J.M.; McCord, R.P.; Gibcus, J.H.; Naumova, N.; Zhan, Y.; Dekker, J. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pandey, M.K.; Wang, J.; Zhao, K.; Ma, X.; Li, Z.; Zhao, K.; Gong, F.; Guo, B.; Varshney, R.K.; et al. Chromatin spatial organization of wild type and mutant peanuts reveals high-resolution genomic architecture and interaction alterations. Genome Biol. 2021, 22, 315. [Google Scholar] [CrossRef] [PubMed]
- Marand, A.P.; Schmitz, R.J. Single-cell analysis of cis-regulatory elements. Curr. Opin. Plant Biol. 2022, 65, 102094. [Google Scholar] [CrossRef]
- Farmer, A.; Thibivilliers, S.; Ryu, K.H.; Schiefelbein, J.; Libault, M. Single-nucleus RNA and ATAC sequencing reveal the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol. Plant 2021, 14, 372–383. [Google Scholar] [CrossRef]
- Feng, D.; Liang, Z.; Wang, Y.; Yao, J.; Yuan, Z.; Hu, G.; Qu, R.; Xie, S.; Li, D.; Yang, L.; et al. Chromatin accessibility illuminates single-cell regulatory dynamics of rice root tips. BMC Biol. 2022, 20, 274. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Su, Y.; Bian, J.; Han, X.; Guo, H.; Yang, Z.; Chen, Y.; Li, L.; Li, T.; Deng, X.W.; et al. Single-nucleus RNA and ATAC sequencing analyses provide molecular insights into early pod development of peanut fruit. Plant Commun. 2024, 5, 100979. [Google Scholar] [CrossRef]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef]
- Si, X.; Zhang, H.; Wang, Y.; Chen, K.; Gao, C. Manipulating gene translation in plants by CRISPR-Cas9-mediated genome editing of upstream open reading frames. Nat. Protoc. 2020, 15, 338–363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 2018, 36, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Hu, B.; Jiang, W.; Wang, Y.; Yan, J.; Ma, F.; Guan, Q.; Xu, J. Advances in plant epigenome editing research and its application in plants. Int. J. Mol. Sci. 2023, 24, 3442. [Google Scholar] [CrossRef] [PubMed]
- Fal, K.; Le Masson, M.; Berr, A.; Carles, C.C. Manipulating plant development by editing histone methylation with the dCas9 tool: The CUC3 boundary gene as a case study. bioRxiv 2024. [Google Scholar]

| Feature/Method | Molecular Markers | DNA Barcoding | Next-Generation Sequencing | Third-Generation Sequencing |
|---|---|---|---|---|
| Technique | SSR, RAPD, AFLP, SNP | rbcL, matK, ITS | Illumina, Ion Torrent | PacBio, Oxford Nanopore |
| Data scope | Specific loci | Specific gene regions | Large genomic regions, whole-genome | Ultra-long reads, whole-genome |
| Advantages | Cost-effective, simple | Rapid, consistent results | Scalability, high accuracy | Long-read data, structural analysis |
| Limitations | Reproducibility issues, labor-intensive | Limited to species level | High cost, computational demands | Higher cost, complex data handling |
| Scalability | Moderate | High | Very high | Moderate |
| Bioinformatics requirement | Low | Low | High | Very high |
| Suitability for mixed samples | Limited | Limited | Excellent (metabarcoding) | Moderate |
| Discriminatory power | Moderate (depends on marker) | Moderate (species level) | High (genome-wide) | Very high (structural variation) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Lin, H.; Lin, H.; Du, P.; Zhang, S. From Species to Varieties: How Modern Sequencing Technologies Are Shaping Medicinal Plant Identification. Genes 2025, 16, 16. https://doi.org/10.3390/genes16010016
Wang M, Lin H, Lin H, Du P, Zhang S. From Species to Varieties: How Modern Sequencing Technologies Are Shaping Medicinal Plant Identification. Genes. 2025; 16(1):16. https://doi.org/10.3390/genes16010016
Chicago/Turabian StyleWang, Mingcheng, Haifeng Lin, Hongqiang Lin, Panyue Du, and Shuqiao Zhang. 2025. "From Species to Varieties: How Modern Sequencing Technologies Are Shaping Medicinal Plant Identification" Genes 16, no. 1: 16. https://doi.org/10.3390/genes16010016
APA StyleWang, M., Lin, H., Lin, H., Du, P., & Zhang, S. (2025). From Species to Varieties: How Modern Sequencing Technologies Are Shaping Medicinal Plant Identification. Genes, 16(1), 16. https://doi.org/10.3390/genes16010016







