The Epigenetic Machinery and Energy Expenditure: A Network to Be Revealed
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Description of the Cohort
3.2. Physical, Metabolic and Plasma Nutritional Data
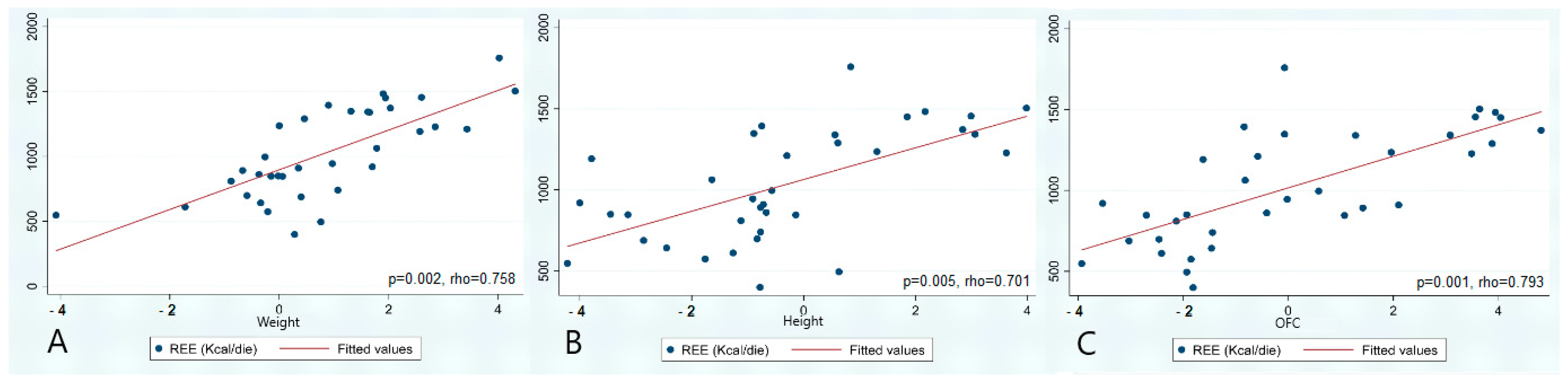
3.3. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larizza, L.; Finelli, P. Developmental disorders with intellectual disability driven by chromatin dysregulation: Clinical overlaps and molecular mechanisms. Clin. Genet. 2019, 95, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, J.A.; Bjornsson, H.T. Mendelian disorders of the epigenetic machinery: Postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 2019, 28, R254–R264. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, H.T. The Mendelian disorders of the Epigenetic Machinery. Genome Res. 2015, 25, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Nava, A.A.; Arboleda, V.A. The omics era: A nexus of untapped potential for Mendelian chromatinopathies. Hum. Genet. 2024, 143, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Psota, T.; Chen, K.Y. Measuring energy expenditure in clinical populations: Rewards and challenges. Eur. J. Clin. Nutr. 2013, 67, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Alsaif, M.; Elliot, S.A.; MacKenzie, M.L.; Prado, C.M.; Field, C.J.; Haqq, A.M. Energy Metabolism Profile in Individuals with Prader-Willi Syndrome and Implications for Clinical Management: A Systematic Review. Adv. Nutr. 2017, 8, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Stevens, C.A.; Hennekam, R.C.M.; Blackburn, B.L. Growth in the Rubinstein-Taybi syndrome. Am. J. Med. Genet. 1990, 6, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Rodriguez-Buritica, D.F.; Northrup, H. Wiedemann-Steiner syndrome: Novel pathogenic variant and review of literature. Eur. J. Med. Genet. 2017, 60, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Schott, D.A.; Blok, M.J.; Gerver, W.J.; Devriendt, K.; Zimmermann, L.J.; Stumpel, C.T. Growth pattern in Kabuki syndrome with a KMT2D mutation. Am. J. Med. Genet. A 2016, 170, 3172–3179. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, M.H.; Vulto-van Silfhout, A.T.; Nillesen, W.M.; Wissink-Lindhout, W.M.; van Bokhoven, H.; Philip, N.; Berry-Kravis, E.M.; Kini, U.; van Ravenswaaij-Arts, C.M.A.; Delle Chiaie, B.; et al. Update on Kleefstra Syndrome. Mol. Syndromol. 2012, 2, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Koolen, D.A.; Pfundt, R.; Linda, K.; Beunders, G.; Veenstra-Knol, H.E.; Conta, J.H.; Fortuna, A.M.; Gillessen-Kaesbach, G.; Dugan, S.; Halbach, S.; et al. The Koolen-de Vries syndrome: A phenotypic comparison of patients with a 17q21.31 microdeletion versus a KANSL1 sequence variant. Eur. J. Hum. Genet. 2016, 24, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mullegama, S.V.; Alaimo, J.T.; Elsea, S.H. Smith-Magenis syndrome and its circadian influence on development, behavior, and obesity—Own experience. Dev. Period. Med. 2015, 19, 149–156. [Google Scholar] [PubMed]
- Bögershausen, N.; Wollnik, B. Mutational landscapes and phenotypic spectrum of SWI/SNF-related intellectual disability disorders. Front. Mol. Neurosci. 2018, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Nikkel, S.M.; Dauber, A.; De Munnik, S.; Connolly, M.; Hood, R.L.; Caluseriu, O.; Hurst, J.; Kini, U.; Nowaczyk, M.J.; Afenjar, A.; et al. The phenotype of Floating-Harbor syndrome: Clinical characterization of 52 individuals with mutations in exon 34 of, SRCAP. Orphanet J. Rare Dis. 2013, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Agwu, J.C.; Shaw, N.J.; Kirk, J.; Chapman, S.; Ravine, D.; Cole, T.R. Growth in Sotos syndrome. Arch. Dis. Child. 1999, 80, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Tatton-Brown, K.; Murray, A.; Hanks, S.; Douglas, J.; Armstrong, R.; Banka, S.; Bird, L.M.; Clericuzio, C.L.; Cormier-Daire, V.; Cushing, T.; et al. Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am. J. Med. Genet. A 2013, 161A, 2972–2980. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.C.; Caballero, B.; Cousins, R.J.; Tucker, K.I.; Ziegler, T.R. Modern Nutrition in Health and Disease, 11th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2014. [Google Scholar]
- Yoon, M.S. The Emerging Role of branched-chain amino acids in insulin resistance and metabolism. Nutrients 2016, 8, 405. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, J.A.; Bjornsson, H.T. Mendelian disorders of the epigenetic machinery: Tipping the balance of chromatin states. Annu. Rev. Genom. Hum. Genet. 2014, 15, 269–293. [Google Scholar] [CrossRef] [PubMed]
- Ramasubbu, K.; Devi Rajeswari, V. Impairment of insulin signaling pathway PI3K/Akt/mTOR and insulin resistance induced AGEs on diabetes mellitus and neurodegenerative diseases: A perspective review. Mol. Cell. Biochem. 2023, 478, 1307–1324. [Google Scholar] [CrossRef] [PubMed]
- Keppler-Noreuil, K.M.; Parker, V.E.; Darling, T.N.; Martinez-Agosto, J.A. Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. American journal of medical genetics. Part C. Semin. Med. Genet. 2016, 172, 402–421. [Google Scholar] [CrossRef]

| Range and Unit of Measurement | RSTS1 (n = 11) | SS1 (n = 10) | WVS (n = 1) | WDSTS (n = 3) | KMS1 (n = 1) | KLEFS1 (n = 5) | KDV (n = 1) | SMS (n = 2) | CSS1 (n = 3) | FLHS (n = 1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Q1; Q3 | Mean | Q1; Q3 | Mean | Q1; Q3 | Mean | Q1; Q3 | Mean | Q1; Q3 | Mean | Q1; Q3 | ||||||
| Age | y | 8.0 | 5.5; 12.5 | 9.2 | 7.0; 9.8 | 7.8 | 8.2 | 7.1–8.7 | 8.9 | 3.8 | 0.9–6.8 | 6.8 | 14.9 | 12.8–17.0 | 7.3 | 3.8–7.8 | 6.4 |
| Sex | 5 M. 6 F | 5 M. 5 F | M | 2 M. 1 F | F | 3 M. 2 F | F | 2 F | 1 M. 2 F | F | |||||||
| W | SD | −0.02 | −0.34; +1.70 | +1.77 | +0.62; +2.03 | +4.31 | −0.59 | −1.72; +1.31 | −0.88 | +0.90 | +0.76; +1.07 | −0.26 | +2.20 | +0.97; +3.43 | +0.06 | −0.67; +0.35 | −4.08 |
| H | SD | −2.86 | −3.79; −1.64 | +2.01 | +0.96; +2.99 | +3.98 | −0.89 | −1.26; −0.83 | −1.12 | −0.75 | −0.77; +0.63 | −0.57 | −0.60 | −0.91; −0.30 | −0.72 | −0.77; −0.14 | −4.22 |
| OFC | SD | −1.85 | −3.03; −1.46 | +3.52 | +1.97; +3.94 | +3.64 | −2.41 | −2.46; −0.07 | −2.13 | −1.44 | −1.81; −0.84 | +0.58 | −0.30 | −0.58; −0.02 | +1.42 | +1.07; +2.10 | −3.93 |
| BMI | SD | +1.46 | +0.14; +2.22 | +0.53 | +0.07; +0.98 | +2.91 | −0.13 | −1.37; +2.14 | −0.33 | +1.47 | +1.18; +2.07 | +0.07 | +0.19 | −0.66; +1.05 | +0.95 | −0.28; +0.99 | −1.59 |
| REE | Kcal/die | 850 | 688; 920 | 1343 | 1290; 1451 | 1504 | 698 | 611; 1348 | 810 | 740 | 495; 1394 | 996 | 1078 | 945; 1211 | 892 | 846; 910 | 547 |
| Prealb. | 20–40 μg/dL | 19 | 17; 20 | 17 | 16; 19 | 16 | 15 | 11; 18 | 19 | 18 | 16; 18 | 20 | 21 | 21; 21 | 16 | 13; 16 | 18 |
| Leucine | 75–107 μmol/L | 109 | 97; 112 | 110 | 96; 121 | 137 | 118 | 117; 159 | 81 | 123 | 108.5; 126 | 131 | 109 | 109;109 | 90 | 82; 128 | 87 |
| Insulin | 2.6–25.0 mIU/L | 9 | 4; 10.1 | 3.3 | 1.5; 5.2 | 7.7 | 3.3 | 3.3; 3.3 | 6.8 | 2.95 | 1.25; 4.15 | 1.3 | 7.65 | 7.2; 8.1 | 4.5 | 1.9; 4.5 | 7.7 |
| Glyc. | 70–110 mg/dL | 76 | 70; 81 | 76 | 68; 81 | 85 | 85 | 75; 93 | 82 | 79 | 71; 82 | 62 | 77.5 | 63; 92 | 77 | 67; 77 | 80 |
| HDL | >65 mg/dL | 49 | 45; 54 | 60 | 53; 68 | 54 | 55 | 35; 58 | 62 | 46 | 46; 53 | 54 | 50 | 40; 60 | 60 | 47; 63 | 65 |
| ApoA1 | 120–176 mg/dL | 123 | 117; 128 | 127.5 | 122.0; 133.5 | 121 | 112 | 103; 131 | 121 | 116 | 104; 117 | 139 | 132.5 | 129;136 | 118 | 114; 135 | 144 |
| ApoA2 | 25.1–34.5 mg/dL | 25.9 | 23.2; 28.4 | 30.2 | 29.1; 32 | 31.3 | 23 | 18.4; 32 | 27.1 | 30 | 24.8; 30.5 | 20.6 | 21.1 | 21.1; 21.1 | 26.4 | 18.8; 26.5 | 31.4 |
| ApoA | 108–225 mg/dL | 132 | 123; 140 | 155 | 133; 164 | 133 | 128 | 100; 150 | 148 | 128 | 124; 131 | 136 | 142 | 142; 142 | 134 | 118; 139 | 161 |
| LDL | <130 mg/dL | 89 | 74; 98 | 93.5 | 78.0; 110.0 | 104 | 77 | 47; 122 | 91 | 68 | 67; 81 | 66 | 104 | 75; 133 | 86 | 82; 90 | 64 |
| ApoB | 60–133 mg/dL | 79 | 68; 84 | 69.5 | 63; 77 | 82 | 57 | 56; 109 | 86 | 69 | 67; 77 | 56 | 88.5 | 66.0; 111 | 74 | 60; 77 | 65 |
| N | Patients | ContA Median (Q1; Q3) | Patients vs. ContA | ContB Median (Q1; Q3) | Patients vs. ContB | ContC Median (Q1; Q3) | Patients vs. ContC | |
|---|---|---|---|---|---|---|---|---|
| Group A | 25 | 850 (688–996) | 940 (733–1343) | 0.112 | 864 (623–960) | 0.54 | - | - |
| RSTS1 | 9 | 850 (688–920) | 1027 (864–1453) | 0.053 | 733 (631–960) | 0.67 | - | - |
| Group B | 10 | 1358 (1290–1455) | 931 (817–1258) | 0.023 * | 822 (663–1158) | 0.001 *** | 1091 (719–1255) | 0.0039 ** |
| RSTS (n = 11) | SS (n = 10) | RSTS vs. SS. p-Value | |||
|---|---|---|---|---|---|
| Median | Q1; Q3 | Median | Q1; Q3 | ||
| Weight (SD) | −0.02 | −0.34; +1.70 | +1.77 | +0.62; +2.03 | 0.016 * |
| Height (SD) | −2.86 | −3.79; −1.64 | +2.01 | +0.96; +2.99 | <0.0001 *** |
| OFC (SD) | −1.85 | −3.03; −1.46 | +3.52 | +1.97; +3.94 | <0.0001 *** |
| BMI (SD) | +1.46 | +0.14; +2.22 | +0.53 | +0.07; +0.98 | 0.118 |
| REE (Kcal/die) | 850 | 688; 920 | 1343 | 1290; 1451 | <0.0001 *** |
| Prealbumin (μg/dL) | 19 | 17; 20 | 17 | 16; 19 | 0.093 |
| Leucine (μmol/L) | 109 | 97; 112 | 110 | 96; 121 | 0.706 |
| Insulin (mIU/L) | 9.0 | 4.0; 10.1 | 3.3 | 1.5; 5.2 | 0.037 |
| Glycemia (mg/dL) | 76 | 70; 81 | 76 | 68; 81 | 0.932 |
| HDL (mg/dL) | 49 | 45; 54 | 60 | 53; 68 | 0.033 |
| ApoA1 (mg/dL) | 123 | 117; 128 | 127.5 | 122.0; 133.5 | 0.340 |
| ApoA2 (mg/dL) | 25.9 | 23.2; 28.4 | 30.2 | 29.1; 32.0 | 0.004 ** |
| ApoA (mg/dL) | 132 | 123; 140 | 155 | 133; 164 | 0.137 |
| LDL (mg/dL) | 89 | 74; 98 | 93.5 | 78.0; 110.0 | 0.325 |
| ApoB (mg/dL) | 79 | 68; 84 | 69.5 | 63; 77 | 0.244 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prada, E.; Marchetti, G.B.; Pires Marafon, D.; Mazzocchi, A.; Scuvera, G.; Pezzani, L.; Agostoni, C.; Milani, D. The Epigenetic Machinery and Energy Expenditure: A Network to Be Revealed. Genes 2025, 16, 104. https://doi.org/10.3390/genes16010104
Prada E, Marchetti GB, Pires Marafon D, Mazzocchi A, Scuvera G, Pezzani L, Agostoni C, Milani D. The Epigenetic Machinery and Energy Expenditure: A Network to Be Revealed. Genes. 2025; 16(1):104. https://doi.org/10.3390/genes16010104
Chicago/Turabian StylePrada, Elisabetta, Giulia Bruna Marchetti, Denise Pires Marafon, Alessandra Mazzocchi, Giulietta Scuvera, Lidia Pezzani, Carlo Agostoni, and Donatella Milani. 2025. "The Epigenetic Machinery and Energy Expenditure: A Network to Be Revealed" Genes 16, no. 1: 104. https://doi.org/10.3390/genes16010104
APA StylePrada, E., Marchetti, G. B., Pires Marafon, D., Mazzocchi, A., Scuvera, G., Pezzani, L., Agostoni, C., & Milani, D. (2025). The Epigenetic Machinery and Energy Expenditure: A Network to Be Revealed. Genes, 16(1), 104. https://doi.org/10.3390/genes16010104







