Abstract
Eggshell color plays important biological roles and attracts the attention of both egg retailers and researchers. However, whether non-coding RNAs are involved in pigment deposition among different eggshell colors remains unknown. In this study, RNA sequencing was used to analyse the uterine gland transcriptome (CircRNA and miRNA) of Changshun chicken blue-shell hens producing four different eggshell color eggs including dark blue PK(DB) and light blue (LB), dark brown and greenish (between blue and pink, DP) and pink (p). We found that miR-192-x, targeting SLC16a7, was expressed in DB, DP, and LB groups compared with the PK group, which indicates that miR-192-x may play a role in the blue eggshell color. KEGG and GO analyses showed that the “metabolic pathways” with targeted genes such BLVRA and HMOX1 were detected in dark and light blue color eggshell chickens, which confirms the different ratios of biliverdin and HO-1 involved in the deposition of blue color. As annotated by connectivity analysis, RASGRF1 and RASGRF2, belonging to the RASGRF family, are involved in the Ras signaling pathway, which plays an important role in cell growth, differentiation, metastasis and apoptosis. Our findings enrich the database of circRNA, miRNAs and genes for chicken uterine tissue, which will be useful in accelerating molecular selection for blue eggshell color layers.
1. Introduction
Internationally, consumer preferences for eggshell color, including unique options such as green eggs, exhibit variance based on regional and cultural factors. For instance, in certain regions of Asia and Europe, eggs with colored shells are seen as distinct and potentially more attractive due to their perceived health advantages and natural appeal [1,2]. In China, preferences for eggshell color also differ, with white and brown eggs being more prevalent. Green eggs, typically laid by specific breeds such as Dongxiang, Lushi chickens and Changshun blue eggshell chickens, are less widely available on the market [2]. Nevertheless, some consumers favor these eggs for their distinctive color and perceived health benefits, while the actual nutritional differences are minimal [3]. Although still a niche market compared to the predominant white and brown eggshell sectors, the demand for green eggs is on the rise [4].
The eggshell pigment is generally formed from the protoporphyrin, biliverdin and zinc chelate of biliverdin, which results in various eggshell colors. The degree of color in an eggshell is mainly determined by the total amount and proportion of pigment [5]. For birds, brown, white, pink and blue are the most common eggshell colors. Protoporphyrin is an antioxidant [6] and a direct precursor of heme, which is involved in the color deposition in pink, brown or yellow eggshells [7]. Biliverdin has antioxidant activity and is a by-product of hemoglobin decomposition, while biliverdin IX, and biliverdin IX zinc chelate are associated with blue and green-blue eggshell colors [8]. As long as these pigments are not found in the uterine gland, the eggshell color turns out to be white [9,10].
Recently, a few studies regarding protein-coding genes have been carried out to illustrate the genetic mechanisms of pigment deposition in poultry. It is generally accepted that the solute carrier organic anion transporter family member 1B3 (SLCO1B3) gene displays functions relating to the regulation of pigment deposition for the blue eggshell phenotype in both domestic and foreign chickens [11]. So far, with the development of omics technology, long non-coding RNA (lncRNA) is deemed to be critical in numerous biological processes including transcriptional regulation [12,13], the progression of cancer [14], and human genetic disorders [15,16]. A previous study has shown that ALAS1, TAL and SLC13A5 may be key genes regulating biliverdin synthesis by integrating microRNA (miRNA) and messenger RNA (mRNA) sequencing in duck oviduct shell glands [17]. In our previous study, we also reported that lncRNAs exert similar functions with mRNA in color formation by modulating pigment disposition in Changshun blue eggshell chickens [18]. In addition, circular RNAs (circRNAs), as a novel category of endogenous non-coding RNAs (ncRNAs), were identified during intron self-splicing from mitochondrial RNAs, ribosomal RNAs, and transfer RNAs. Increasing evidence indicates that circRNAs play an essential role in controlling transcriptional regulation by maintaining homeostasis [19,20,21]. CircRNAs are known to bind with miRNA to prevent mRNA translation, and interact with RNA-associated proteins or influence gene expression by regulating gene splicing or mRNA levels [22]. Nevertheless, little is known as to whether circRNAs or miRNAs contribute to pigment deposition, and the underlying mechanism needs to be further investigated.
In this study, circRNA and miRNA sequencing was used to determine the uterine transcriptome, and a network of these two ncRNAs was developed to enable better understanding of color formation in Changshun chicken eggs. Our objective was to identify and profile ncRNAs in chicken shell glands and obtain candidate circRNAs, miRNAs, and pathways related to blue eggshell deposition in poultry.
2. Materials and Methods
2.1. Animals and Sample Collection
Changshun blue eggshell layers were obtained from a breeding farm, owned by Tiannong Corporation (Guizhou, China). Thirty days were spent observing 331 layers from the 12th generation preserved population that were housed at 210 days old. The approximate laying time of each group of hens was observed and recorded hourly from 8:00 am to 2:00 pm daily for three consecutive days. Three layers consistently producing dark blue (DB) and light blue (LB) eggs, and four layers producing dark brown and greenish (between blue and pink, DP) eggs were chosen during the peak laying period (about 210 days of age). In addition, four layers producing pink (PK) shell eggs were used as the study control group. The uterine shell gland tissues were taken from birds slaughtered at 7 h after egg laying, which was monitored by cameras hanging on opposite cages. The shell gland from each bird was collected and immediately stored in dry ice and then at −80 °C until being further processed. All experiments were performed using protocols approved by the Animal Care Committee of Foshan University (approval ID: FOSU#080).
2.2. RNA Extraction, Strand-Specific Library Construction and Sequencing
Following the extraction of total RNA using the Trizol reagent kit (Invitrogen, Carlsbad, CA, USA), a nucleic acid protein analyzer was used to analyze the RNA total concentration and optical density (OD) of the samples at wavelengths of 260 nm and 280 nm. The OD260/280 ratio needed to be within the range of 1.8–2.0 for the samples to meet the required specifications. The data of quality analysis of total RNA can be found in Supplementary Table S1. Next, polyacrylamide gel electrophoresis (PAGE) was used to separate the RNA molecules within a size range of 18–30 nt. The 36–48 nt RNAs were then enriched after the addition of 3′ adapters. The 5′ adapters were then ligated to the RNAs. The ligation products were reverse transcribed by Polymerase Chain Reaction (PCR) amplification and the 140–160 bp size PCR products were used to generate DNA (cDNA) libraries, which were sequenced using an Illumina HiSeq X10 platform at Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China). Then, each sample was diluted to a concentration of 300 ng/µL using DEPC. The Vazyme III RT SuperMix for qPCR (+gDNA wiper) kit was utilized for the reverse transcription synthesis of cDNA, with careful pipetting and mixing involved. The reaction was carried out at 42 °C for 2 min, followed by the addition of 4 µL of 5× HiScript III qRT SuperMix at 37 °C for 15 min and a final step at 85 °C for 5 s.
2.3. Differential Expression Analysis of mRNA, miRNA and circRNA
After the total RNA was extracted, rRNAs were removed to retain mRNAs and ncRNAs. The enriched mRNAs and ncRNAs were fragmented into short fragments by using a fragmentation buffer and reverse transcripted into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase l, RNase H, dNTP (dUTP instead of dTTP) and a buffer. Next, the cDNA fragments were purified with the QiaQuick PCR extraction kit, end repaired, mixed with poly(A), and ligated to Illumina sequencing adapters. Then, UNG (Uracil-N-Glycosylase) was used to digest the second-strand cDNA. The digested products were size selected by agarosegel electrophoresis, PCR amplified, and sequenced using illumina HiSeqTM 4000 by Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China). The expression levels of mRNAs were quantified using FPKM (fragments per kilobase of transcript per million mapped reads). Using the DESeq2 [23] (https://bioconductor.org/, accessed on 24 July 2020) software package, differentially expressed circRNAs were first screened using the FDR-adjusted p-value. However, there were few differentially expressed circRNAs. Thus, the genes that exhibited an FDR < 0.05 and log2(fc) > 1 were identified as significantly differentially expressed [24,25,26]. TPM (transcripts per million reads) was used to calculate the expression of miRNAs, and edgeR (http://www.rproject.org/, accessed on 24 July 2020) software was used. RPM (back-spliced Reads Per Million mapped reads) was used to calculate the expression of circRNA and edgeR [27] (https://bioconductor.org/, accessed on 24 July 2020) software was also used for analysis. A threshold of p ≤ 0.05 and log2(fc) > 1 determined significant differential expression. In order to identify distinct anchor positions within a splice site, 20mers from both ends of the unmapped reads were extracted and aligned to the reference genome (Ensembl_release92, Galllus_gallus-5.0). Anchor reads that aligned in the reversed orientation (head-to-tail) indicated circRNA splicing and then were subjected to find_circ [28] to identify circRNAs. The anchor alignments were then extended to ensure that the complete reads aligned and the breakpoints were flanked by GU/AG splice sites. A candidate circRNA was used if it was supported by at least two unique back-spliced reads in at least one sample. Clean tags were then searched against the miRBase database (Release 22) to find known miRNAs (existing miRNAs). Not all chicken miRNA sequences are represented in the miRBase database, so alignment with miRNAs from other species was also a dependable way to identify known miRNAs. All of the unannotated tags were aligned with the reference genome (Ensembl_release92, Galllus_gallus-5.0). According to their genome positions and hairpin structures predicted by software miRDeep2 [29] (https://github.com/rajewsky-lab/mirdeep2, accessed on 24 July 2020), novel miRNA candidates were identified.
2.4. Construction of DEcircRNA-DEmiRNA-DEmRNA Regulatory Network
Target gene prediction was performed using miRanda (http://www.microrna.org/, accessed on 26 July 2020) and TargetScan (https://www.targetscan.org/, accessed on 26 July 2020) software with DEmiRNAs. Spearman’s rank correlation coefficients (SCCs) of miRNA and candidate competing endogenouse RNAs were calculated for the above target gene pairs, and target gene pairs with correlation coefficients ≤ −0.5 were screened. Following the calculation of the SCC for the expression between the competing endogenous RNA (ceRNA) pairs acquired in the preceding step, the ceRNA pairs with a correlation coefficient of more than 0.5 were selected as potential ceRNA pairs. ceRNAs with a P value less than 0.05 were screened using the hypergeometric distribution test. The Cytoscape v3.6.0 (http://www.cytoscape.org/, accessed on 27 July 2020) software was used to build a network map for the interaction between circRNA-miRNA-mRNA. The ceRNA connectivity analysis was performed, and in the ceRNA regulatory network, the higher the number of miRNA molecules with miRNA- or cicrRNA-targeting regulatory relationships, the higher the connectivity. Nodes with high connectivity often have important biological significance.
2.5. Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
In light of potential functional importance, RT-qPCR was performed on DEmiRNAs selected from each comparison. Based on the DEmiRNA sequences obtained by sequencing, primers were designed at Sangon Biotech (www.sangon.com/primerDesign, accessed on 29 July 2020). The primer sequence was designed using Primer sequence 5.0 and synthesized by the Shanghai Bioengineering company (Supplementary Table S2). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was selected as a reference gene. The OD 260/OD 280 ratio was measured for each RNA sample and RNA integrity was analyzed by agarose gel electrophoresis. RNA was confirmed to be of high quality and was then reverse transcribed to cDNA using TRUEscript RT MasterMIX in a 20 μL volume containing l000 ng RNA and RNase free to 16 μL, 4 μL 5 × TRUE RT MasterMix under the following conditions: 42 °C for 10 min (Aidlab, PC5801, Hangzhou, China) for RT-qPCR analyses. RT-qPCR was conducted on qTOWER 2.2 touch (Analytik, Jena, Germany) in a 20 μL volume containing 10 μL SYBR × Premix Ex Taq (Aidlab, PC3302, China), 0.5 μL of each forward and reverse primer (10 μM), 1 μL of cDNA, and 8 μL ddH2O under the following conditions: 95 °C for 15 min; 95 °C for 10 s, annealing for 20 s and 72 °C for 20 s for 40 cycles. Each amplification was performed for three control replicates and three case replicates. The amplification efficiencies were close to 100%, using the 2−ΔΔCt method for calculating the relative gene expression levels of a sample. ΔΔCt = (target gene Ct in the experimental group—reference gene Ct in the experimental group)—(target gene in the control group—reference gene in the control group). U6 can be used for miRNA or cirRNA normalization as shown in previous research [30], and was used as an internal reference gene. MiRNA First Strand cDNA Synthesis (Stem-loop Method) (Sangon Biotech, Shanghai, China) was used for cDNA synthesis. The qPCR machine was programmed to be 16 °C for 30 min, then 37 °C for 30 min, and finally 85 °C for 5 min. RT-qPCR was performed on each sample in triplicate using the ChamQ Universal SYBR qPCR Master Mix (Sangon Biotech, China) on QuantStudio5 (Applied Biosystems, Waltham, MA, USA) in a 20 μL reaction volume. The program settings were as follows: pre-denaturation at 95 °C for 30 s, 40 cycles of reaction at 95 °C for 10 s, melting curve at 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. Melting curve analysis was used to check the amplification specificity. The relative expression of target gene transcripts was calculated using the comparative Ct method (2−ΔΔCt) and subjected to statistical analysis with SPSS software (version 19) (https://www.ibm.com/, accessed on 20 May 2020).
3. Results
3.1. Overview of Sequencing and Identification of miRNA in Chicken Uterus
Following quality control, an average of 90.4% of the total high-quality clean reads were mapped to Galgal 5.0, GenBank and Rfam, Exon and Repeat (Supplementary Table S3). Accordingly, 524 miRNAs, known by blasting against the known chicken miRNAs in the ALDB database, were captured, with 276 of these miRNAs expressed in all four groups. In addition, 16, 12, 26 and 18 miRNAs were specifically expressed in DB, DP, LB, and PK groups, respectively (Table 1). In addition, 53 of 142 new miRNAs were seen to be expressed in all four groups (Table 1).

Table 1.
Known and new miRNAs.
Between the DB vs. PK group, there were 31 significantly differentially expressed miRNAs (DEmiRNAs) identified, among which 10 were upregulated, and 21 were downregulated in the DB group compared to the PK group. Between the LB vs. PK group, 53 DEmiRNAs were identified with 36 upregulated and 17 downregulated in the LB group as compared to the PK group. Between the DP vs. PK group, 18 DEmiRNAs were identified, with 10 upregulated and 8 downregulated DEmiRNAs in the former group and latter group, respectively. Between the DP vs. DB group, 41 DEmiRNAs were obtained, of which 23 were upregulated and 18 were downregulated in the DB group compared to the DP group. Additionally, only miR-2995-x and novel-m0026-5p were significantly expressed in the four comparison groups, of which the highest expression was the DB vs. PK group. miR-192-x was seen to be expressed in the DB vs. PK, LB vs. PK, and the DP vs. PK group. The top three up and downregulated miRNAs of significantly differentially expressed miRNA in all the four-comparison groups are shown in Table 2.

Table 2.
Significantly differentially expressed miRNAs.
3.2. Functional Enrichment Analysis of Differently Expressed miRNA and Targeted Genes
Furthermore, based on these miRNAs, Miranda and TargetScan were used to predict target genes (Table 2). Between the DB vs. PK group, there were 2995 target mRNAs identified, of which 723, 1473 and 297 target mRNAs were found by the upregulated miR-2995-x, novel-m0026-5p and gga-miR-3528, while 162, 220, and 220 target mRNAs were obtained by the downregulated miR-423-y, novel-m0066-5p and novel-m0067-5p. Between the LB vs. PK groups, 4555 genes were identified, of which 629, 723 and 1493 target genes were identified by the upregulated miR-224-x, miR-2995-x and novel-m0026-5p, while 556, 556 and 618 target genes were identified by the downregulated gga-miR-2984-3p, gga-miR-6552-3p and gga-miR-6552-5p. Between DP vs. PK, 4245 target genes were found, of which 549, 594 and 723 genes were predicted by the upregulated novel-m0093-3p, novel-m0142-3p and miR-2995-x, while 471, 1420 and 488 target genes were predicted by the downregulated miR-484-x, gga-miR-6544-5p and novel-m0096-3p. Between the comparison of the DB vs. DP group, 3614 genes were identified, with 220, 220 and 300 target genes predicted by the upregulated novel-m0066-5p, novel-m0067-5p and gga-miR-6516-3p and 875, 579 and 1420 genes predicted by gga-miR-217-5p, novel-m0136-5p and gga-miR-6544-5p.
We then performed gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on these target genes. In each comparison group, GO enrichment analysis revealed that these target genes were significantly enriched in the cellular process, metabolic process, cell, cell part, binding and catalytic activity (Supplementary Figure S1A–D). Bubble charts were used to represent the top 20 pathways in the DB vs. PK group (Figure 1A), LB vs. PK group (Figure 1B), DP vs. PK group (Figure 1C), and DB vs. DP group (Figure 1D), respectively. In the DB vs. PK group, KEGG pathways were mainly involved in platinum drug resistance, viral protein interaction with cytokine and cytokine receptors, biosynthesis of unsaturated fatty acids, mineral absorption, fatty acid metabolism; arginine and proline metabolism (Figure 1A). In the LB vs. PK group, KEGG pathways were mainly enriched in SNARE interactions in vesicular transport, drug metabolism-other enzymes, mannose type O-glycan biosyntheis, platinum drug resistance, steroid biosynthesis. (Figure 1B). In the LB vs. PK group, KEGG pathways were involved in metabolic pathways, pyrimidine metabolism, fatty acid metabolism, protein processing in the endoplasmic reticulum; platinum drug resistance (Figure 1C). In the DB vs. DP group, KEGG pathways were involved in protein processing in the endoplasmic reticulum, cell cycle, retinol metabolism and metabolic pathways (Figure 1D).
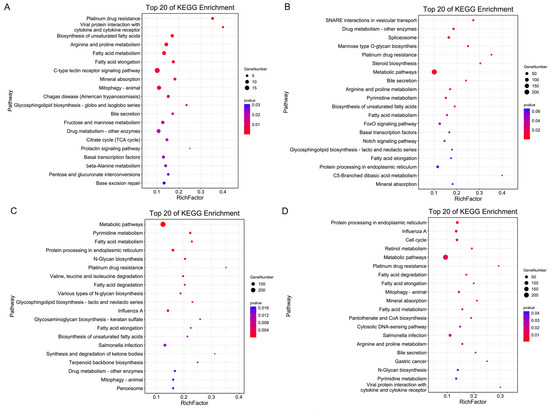
Figure 1.
KEGG pathway enrichment analysis of highly expressed differential miRNA target genes with the top twenty enrichment scores. (A) DB vs. PK. (B) DP vs. PK. (C) LB vs. PK. (D) DB vs. LB. The Y-axis label represents the pathway and the X-axis label represents the rich factor. The colour and size of the bubble represent enrichment significance and the amount of differentially expressed genes enriched in the pathway, respectively. The analysis includes DB, DP, LB, and PK mean chickens producing dark blue, dark brown and greenish, light blue, and pink eggshell eggs, respectively.
3.3. Overview of Sequencing and Identification of circRNA in Chicken Uterus
In total, 861,368,284 reads were obtained. After quality control, the averages of effective reads of Q20 and Q30 were 98.23% and 94.56%, respectively (Supplementary Table S4). More than 0.71% of rRNA was removed from these clean reads of each sample. Afterwards, these reads were mapped to the chicken genome, and an 83.89% mapping rate was obtained (Supplementary Table S5).
In total, 13235 circRNAs were identified, of which there were 7931, 7816, 8791 and 8674 circRNAs in the DB, DP, LB and PK groups, respectively, and 6659 circRNAs were found in all four groups. The length of these circRNAs averaged around 300–400 bp, 400–500 bp and 3000 bp, and the circRNAs were located on chromosomes 1, 2, 3, 4 and Z. Accordingly, these circRNAs were annotated to 5940 annot_exons, 1914 antisense, 1870 exon_intron, 1723 intergenic, 1138 intronic and 650 one_exon (Supplementary Figure S2).
Between the DB vs. PK group, there were 129 differentially expressed circRNAs identified, of which 70 were upregulated and 59 were downregulated. Between the LB vs. PK group, there were 122 differentially expressed circRNAs with 50 upregulated and 70 downregulated circRNAs obtained. Between DP vs. PK, 121 differentially expressed circRNAs were identified, of which 50 were upregulated and 71 downregulated. Novel_circ_001519, novel_circ_012603, novel_circ_001808, novel_circ_001461, novel_circ_003137, novel_circ_011869, novel_circ_006235, novel_circ_006931, and novel_circ_005940 were found to be expressed in all three comparisons.
3.4. Functional Enrichment Analysis of Differentially Expressed circRNA and Target Genes
The target mRNAs sourced from circRNA were further used for the GO and KEGG analyses (Figure 2). Between the DB vs. PK group, 129 target DEGs were assigned to 201 GO terms (p < 0.05), 122 target DEGs were assigned to 261 GO terms in the DB vs. PK group, 121 target DEGs were assigned to 262 GO terms in the DB vs. PK group, and 122 target DEGs were assigned to 249 GO terms in the DB vs. PK group (Supplementary Figure S2).
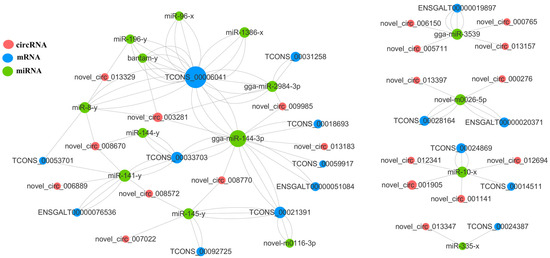
Figure 2.
CeRNA networks. CeRNA networks were constructed based on identified circRNA–miRNA and miRNA–mRNA interactions.
3.5. Construction of the ceRNA Network of Blue-Shell Egg
The ceRNA regulatory network contained 63 circRNA-miRNA-mRNA pairs and included 45 circRNAs, 38 miRNAs, and 37 mRNAs (Figure 3). For the top 10 mRNAs and circRNAs in connectivity, a Sankey diagram of their relationship with miRNA-targeting regulations was drawn (Figure 3). The mRNA in the figure corresponds to the symbol number to find six genes, TCONS_00006041(BBX), TCONS_00021391-GPC6, TCONS_00077632-ZBTB44, TCONS_00062441-ICA1, TCONS_00018693-GRIK1 and TCONS_00028164-RASGRF1.
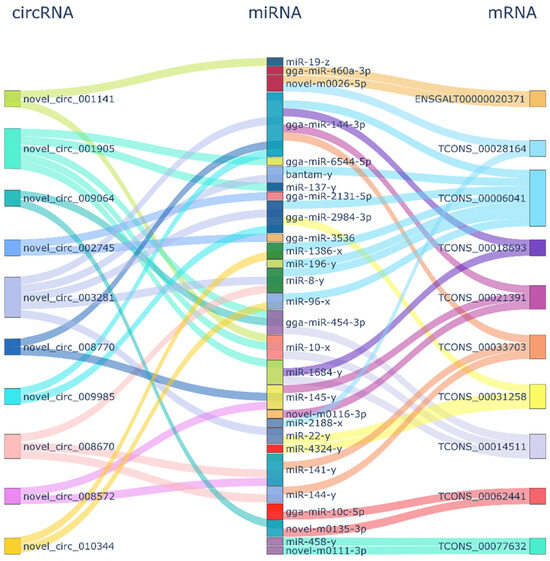
Figure 3.
CircRNA-miRNA-mRNA connectivity.
3.6. Validation of DEmiRNAs by RT-qPCR
The DB vs. PK group, LB vs. PK group, DP vs. PK group, and DB vs. PK group were used to measure the relative expression of DEmiRNAs using RT-qPCR. The relative expression (log2FC) of these DEmiRNAs was similar between the two approaches, although some quantitative differences were found between the RT-qPCR and RNA-seq analytical platforms (Figure 4). Therefore, the RNA-seq results were reliable and could be used for bioinformatics analysis.
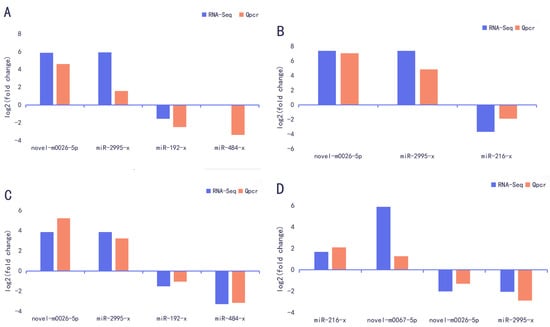
Figure 4.
RT-qPCR validation of differentially expressed miRNAs expression. (A) DB vs. PK. (B) DP vs. PK. (C) LB vs. PK. (D) DB vs. LB.
4. Discussion
There is a growing trend in the market towards the consumption of organic and free-range eggs, particularly those with brown or green shells. These eggs are often perceived by consumers as being associated with higher animal welfare and fewer chemicals, reflecting a broader shift towards sustainable and ethically produced food. In China, there is a vast array of native poultry, including 108 indigenous chicken breeds. Some of these local hens lay eggs with blue shells, which are highly favored by the majority of consumers. These blue eggs are considered precious due to their rarity, and are believed to be more flavorful than white or brown eggs as they come from native birds. Blue eggs have a higher egg white height, Hastelloy units, and calcium and phosphorus content compared to white eggs [31]. The higher Hastelloy unit indicates better protein quality in blue eggs, leading to the conclusion that they are of superior quality compared to white eggs. Additionally, green eggs have been found to have significantly lower cholesterol content than brown eggs [32]. As a result of consumer demand, there has been a deliberate selection process in place for breeders to produce layers that lay blue eggs over the past few decades.
In this study, an miRNA and circRNA network was constructed to illustrate the role of non-coding RNA in the color formation of blue eggshell chickens. To date, miRNAs have been demonstrated to exert essential roles in pigment deposition in several human and animal studies. For example, the significantly upregulated miRNA-21, and downregulated miRNA -320 and miRNA-494 were found to be associated with skin in human melanoma [33]. Furthermore, the hair color of alpacas is determined by the production of melanin in melanocytes, and the target genes are known to be regulated by miRNA [34]. Also, in chickens, it is known that some miRNAs and their target genes play an important role in muscle melanin production [35]. In the current study, small RNA sequencing was performed on the uterine shell gland of four different eggshell color birds. In total, 666 miRNAs were obtained, of which there were 524 known miRNAs and 142 new miRNAs. Subsequently, miR-192-x was found to be expressed in DP vs. PK, DB vs. PK and LB vs. PK, which indicates that it may play a role in blue eggshell color deposition. Interestingly, miR-192 has demonstrated to be associated with the mechanism of tilapia response to carbonate alkalinity stress, and SLC solute vector family member SLC16a7 is the target gene of miR-192 [36]. In addition, miR-192-5p is highly expressed in endometrial epithelial cells in the non-receptive state and is involved in maintaining epithelial cell polarity and anti-adhesion properties on the cell surface [37,38]. Notably, SLC genes are well known to be responsible for blue eggshell deposition in chickens [18]. Thus, we speculate that miR-192-x is a candidate miRNA regulating blue eggshell color regardless, meaning that the underpinning mechanism should be further investigated.
We came to the following conclusions based on the target genes predicted by three of the most differentially up and downregulated miRNAs. The GO terms were similar between four comparisons, which are generally consistent with enrichments in duck shell glands [1], indicating the reliability of these data. The pathway “platinum drug resistance” and target gene SLC31A1 were significant across the DP vs. PK, LP vs. PK and BP vs. PK comparisons, but not between the DP vs. LP group. In addition, the “mineral absorption” with the target SLC family genes was also found to be enriched in dark blue and pink eggshell chicken groups. These results again provide evidence for the possible role of miRNA in regulating blue eggshell color. Interestingly, the “metabolic pathways” with target genes such BLVRA, and HMOX1 were reported in dark and light blue color eggshell chickens. As is known, protoporphyrin reponds to ferrous ions that are haem via iron chelatase, and then degrades into biliverdin using haem oxygenase 1 (HO-1) activity. Further, biliverdin reductase A (BLVRA) converts bilirubin into bilirubin [13]. Our results also confirm that different ratios of biliverdin and HO-1 are involved in the deposition of blue color.
Generally, the vast majorities of circRNAs are often identified in the cytoplasm, and are stable with relatively long half-lives [39]. circRNAs are demonstrated to exert functions in regulating the growth and progression of human cancer, controling the intracellular localization, and tumor autophagy. To date, circRNA studies regarding chicken have been mainly focused on bone growth [40], adipocyte differentiation [41], and follicular development, while the color deposition of chicken eggshell has not yet been reported. In our study, 13235 circRNAs were identified, which is less than the number of circRNAs in a previous study related to follicular development [42]. Accordingly, the ratio of exon, antisense chain, exon–intron, intergenic region, intron and single exon groups is very close to that of a study reporting preadipocyte differentiation in chicken [17].
In terms of biological processes of GO terms, the four comparison groups are mainly focused on metabolic and cellular processes. In terms of cellular components, the four comparator groups were mainly cells, cellular components and organelles. The GO enrichment secondary entries of circRNA-derived genes are similar to those of mRNA and miRNA, indicating the associated genes of mRNA, miRNA and circRNA, which were mainly linked to metabolic and cellular activities by participating and binding activity in cells and cellular components. Furthermore, with regard to the KEGG pathway, “ferroptosis” and the “PPAR signaling pathway” were enriched in both the DB vs. PK and DP vs. PK groups. Heme oxygenase metabolizes heme to produce carbon monoxide, biliverdin and ferric ion. Catalyzing the high expression of unsaturated fatty acids in the cell membrane with the existence of ferrous iron or ester oxygenase may be the possible mechanism of the ferroptosis pathway, which in turn promotes liposome peroxidation and cell apoptosis [43,44]. The concentration of ferric ion affects the efficiency of heme synthesis, which in turn affects the accumulation of protoporphyrin in the epithelial cells of the eggshell gland [17]. The PPAR signaling pathway is associated with lipid metabolism in goldfish [45]. Most of the RNAs in our study were enriched in substance metabolism and transport. Due to their stability, specificity and sensitivity, circRNAs have potential value for eggshell pigment deposition and deserve further investigation.
In the network of circRNA-miRNA-mRNA, various repeated miRNAs were found within comparison groups, i.e., novel-m0026-5p repeated four times, and gga-miR-2984-3p repeated once. All of these are the key targets in miRNA research. Among them, novel_circ_013347-miR-335-x-TCONS_00024387/DUOXA1 and DUOXA1 belong to the NOX protein family, which are membrane-integrated proteins that share a similar core module responsible for catalyzing redox reactions, consisting of a transmembrane region bound by a double heme molecule and an intracellular dehydrogenase structural domain [46,47]. In contrast, biliverdin is the main final metabolite of heme. This network chain, from its composition to its function, is related to eggshell color and needs further investigation. Six genes, including BBX, GPC6, ZBTB44, ICA1, GRIK1 and RASGRF1, were annotated by connectivity analysis. RASGRF1 and RASGRF2 belong to the RASGRF family and are involved in the Ras signaling pathway, which exerts significant effects on development, metabolic and apoptosis [48]. Other genes have scarcely been reported in chickens, but in humans, GPC6 was identified as a biomarker for melanoma metastatic progression [49]. In addition, ICA-1 decreased migration and melanoma cell invasion [50]. We speculate that although the connectivity in the ceRNA network is high, how the circRNAs exert functions on the greenshell trait still needs further investigation.
5. Conclusions
In summary, with respect to transcriptome sequencing of miRNA and circRNA in Changshun chicken shell glands, miR-192-x, miR-2995-x, and novel-m0026-5p are presumed to be related to the regulation of the eggshell’s green color. Novel-m0026-5p, miR-192-x and gga-miR-2984-3p were found as the differential miRNAs in the circRNA-miRNA-mRNA network. Our results provide a catalog of chicken uterine circRNAs and genes worthy of further studies to understand their roles in the selection of blue eggshell color layers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15060812/s1, Figure S1: Differential expression of miRNAs.; Figure S2: GO analysis of highly expressed differential miRNAs target genes.; Table S1: Quality analysis of total RNA.; Table S2: Primers for miRNA RT-qPCR.; Table S3: Small RNA sequencing data quality filtering and annotation.; Table S4: The quality and filtration statistics of sequencing data.; Table S5: Contrast ribosomes and chicken genomes.
Author Contributions
Conceptualization, H.L.; methodology, S.C. and M.Z.; investigation, M.Z.; resources, J.X.; data curation, M.Z.; writing—original draft preparation, S.C., M.Z. and K.C.; writing—review and editing, S.C. and H.L.; visualization, K.C.; supervision, H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by major projects in agricultural biology breeding (2023ZD0406401), Ji Hua Laboratory (X220981UZ230), Qingyuan Science and Technology project of Qingcheng District, Qingyuan City (2020A01), and Poultry Breeding Precision Innovation Team (2019KCXTD006).
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of the Animal Care Committee of Foshan University (approval ID: FOSU#080).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available at the National Center for Biotechnology Information (NCBI—https://www.ncbi.nlm.nih.gov, accessed on 12 July 2021), under the following accession numbers: BioProject PRJNA1029363; submission SUB13907768.
Acknowledgments
We are grateful for the support of Tiannong Food Co., Ltd., Qingyuan city, Guangdong province.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wang, Z.; Meng, G.; Bai, Y.; Liu, R.; Du, Y.; Su, L. Comparative transcriptome analysis provides clues to molecular mechanisms underlying blue-green eggshell color in the Jinding duck (Anas platyrhynchos). BMC Genom. 2017, 18, 725. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.-P.; Lin, X.-Y.; Wu, Y.; Zhou, S.-Y.; Huang, Z.-b.; Huang, Y.-F.; Li, A.; Huang, X.-H. Isolation of blue-green eggshell pigmentation-related genes from Putian duck through RNA-seq. BMC Genom. 2019, 20, 66. [Google Scholar] [CrossRef] [PubMed]
- Ahnen, R.T.; Slavin, J.L. Eggs as Part of a Healthy Eating Pattern. In Eggs as Functional Foods and Nutraceuticals for Human Health; Wu, J., Ed.; The Royal Society of Chemistry: London, UK, 2019. [Google Scholar]
- Yuan, Q.Y.; Lu, L.Z. Progresses in inheritance of genes for avian eggshell color. Yi Chuan 2007, 29, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.R.; Wells, J.W. A Review of eggshell pigmentation. World’s Poult. Sci. J. 1987, 43, 238–246. [Google Scholar] [CrossRef]
- Thompson, C.F.; Hodges, K.E.; Mortimer, N.T.; Vrailas-Mortimer, A.D.; Sakaluk, S.K.; Hauber, M.E. Avian eggshell coloration predicts shell-matrix protoporphyrin content. Can. J. Zool. 2021, 100, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Li, Z.; Yang, N.; Ning, Z. Quantitative expression of candidate genes affecting eggshell color. Anim. Sci. J. 2014, 85, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, G.Y.; Vevers, H.G. A survey of avian eggshell pigments. Comp. Biochem. Physiol. B Comp. Biochem. 1976, 55, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Hamchand, R.; Hanley, D.; Prum, R.O.; Bruckner, C. Expanding the eggshell colour gamut: Uroerythrin and bilirubin from tinamou (Tinamidae) eggshells. Sci. Rep. 2020, 10, 11264. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Yang, Q.L.; Tang, Q.; Liu, R.X.; Hu, J.W.; Li, L.; Bai, L.L.; Liu, H.H. Metabonomics reveals the main small molecules differences between green and white egg shells in ducks. Ital. J. Anim. Sci. 2022, 21, 208–216. [Google Scholar] [CrossRef]
- Wang, Z.P.; Qu, L.J.; Yao, J.F.; Yang, X.L.; Li, G.Q.; Zhang, Y.Y.; Li, J.Y.; Wang, X.T.; Bai, J.R.; Xu, G.Y.; et al. An EAV-HP Insertion in 5⌢ Flanking Region of SLCO1B3 Causes Blue Eggshell in the Chicken. PLoS Genet. 2013, 9, e1003183. [Google Scholar] [CrossRef]
- Afonso, S.; Vanore, G.; Batlle, A. Protoporphyrin IX and oxidative stress. Free Radic. Res. 1999, 31, 161–170. [Google Scholar] [CrossRef]
- Ito, S.; Tsudzuki, M.; Komori, M.; Mizutani, M. Celadon: An eggshell color mutation in Japanese quail. J. Hered. 1993, 84, 145–147. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; Johnson, R.; Abascal, F.; Amin, S.B.; Bader, G.D.; et al. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun. Biol. 2020, 3, 56. [Google Scholar] [CrossRef]
- Sparber, P.; Filatova, A.; Khantemirova, M.; Skoblov, M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med. Genom. 2019, 12, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-coding RNA Gene Loci. Front. Genet. 2020, 11, 527484. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, H.H.; Wang, J.W.; Li, L.; Han, C.C.; Mustafa, A.; Xiong, X. Evidences in duck (Anas platyrhynchos) by transcriptome data for supporting the biliverdin was mainly synthesized by shell gland. Poult. Sci. 2019, 98, 2260–2271. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Chen, K.C.; Xu, J.M.; Li, F.W.; Ding, J.L.; Ma, Z.; Li, G.; Li, H. Insights Into mRNA and Long Non-coding RNA Profiling RNA Sequencing in Uterus of Chickens with Pink and Blue Eggshell Colors. Front. Vet. Sci. 2021, 8, 736387. [Google Scholar] [CrossRef]
- Sun, Y.H.; Qiu, L.M.; Chen, J.J.; Wang, Y.; Qian, J.; Huang, L.R.; Ma, H.T. Construction of circRNA-Associated ceRNA Network Reveals Novel Biomarkers for Esophageal Cancer. Comput. Math. Methods Med. 2020, 2020, 7958362. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Wan, X.C.; Lu, Y.L.; Zhang, Y.Y.; Huang, Y.; Xu, Y.; Liu, Y.J.; Zhao, P.Q.; Xiang, X.X.; Li, L.; et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell. Mol. Med. 2020, 24, 799–813. [Google Scholar] [CrossRef]
- He, Q.Q.; Huang, L.F.; Yan, D.; Bi, J.M.; Yang, M.H.; Huang, J.; Lin, T.X. CircPTPRA acts as a tumor suppressor in bladder cancer by sponging miR-636 and upregulating KLF9. Aging 2019, 11, 11314–11328. [Google Scholar] [CrossRef]
- Xu, Z.H.; Li, P.Y.; Fan, L.; Wu, M.H. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front. Immunol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.Y.; Wang, H.B.; Peng, W.; Yue, B.L.; Fu, C.Q.; Shu, S.; Zhong, J.C.; Wang, H. Circular RNA mapping reveals CircCWC22 as a MiR-3059-x sponge in yak fat deposition by regulating HMGCL. Int. J. Biol. Macromol. 2024, 257, 128531. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Fang, D.L.; Zhang, C.Y.; Zhao, Z.R.; Liu, Y.A.; Zhao, S.J.; Zhang, N.; Xu, J.B. Circular MTHFD2L RNA-encoded CM-248aa inhibits gastric cancer progression by targeting the SET-PP2A interaction. Mol. Ther. 2023, 31, 1739–1755. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Jiang, Y.R.; Lu, Y.X.; Hu, Z.; Du, R.R.; Zhou, Y.X.; Liu, Y.P.; Zhao, X.L.; Tian, Y.F.; Yang, C.W.; et al. Thiram exposure induces tibial dyschondroplasia in broilers via the regulation effect of circ_003084/miR-130c-5p/BMPR1A crosstalk on chondrocyte proliferation and differentiation. J. Hazard. Mater. 2024, 465, 133071. [Google Scholar] [CrossRef]
- Hafner, M.; Landgraf, P.; Ludwig, J.; Rice, A.; Ojo, T.; Lin, C.; Holoch, D.; Lim, C.; Tuschl, T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 2008, 44, 3–12. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Mackowiak, S.D. Identification of Novel and Known miRNAs in Deep-Sequencing Data with miRDeep2; Current Protocols in Bioinformatics; Max Delbrück Center for Molecular Medicine: Berlin, Germany, 2011; Chapter 12; pp. 12.10.11–12.10.15. [Google Scholar] [CrossRef]
- Li, B.X.; Zhang, K.Z.; Ye, Y.Q.; Xing, J.J.; Wu, Y.Y.; Ma, Y.J.; Li, Y.G. Effects of Castration on miRNA, lncRNA, and mRNA Profiles in Mice Thymus. Genes 2020, 11, 147. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Sanz, J.J.; Moreno, J. Egg colour reflects the amount of yolk maternal antibodies and fledging success in a songbird. Biol. Lett. 2006, 2, 334–336. [Google Scholar] [CrossRef]
- Hargitai, R.; Csaba, M.; Miklós, B.; Gil, D.; López-Rull, I.; Solymos, E. Eggshell characteristics and yolk composition in the common cuckoo Cuculus canorus: Are they adapted to brood parasitism? J. Avian Biol. 2010, 41, 177–185. [Google Scholar] [CrossRef]
- Bagheri, M.S.; Polivka, J.; Treskova, I.; Houfkova, K.; Knizkova, T.; Woznica, V.; Fikrle, T.; Pivovarcikova, K.; Svaton, M.; Shetti, D.; et al. Preoperative Plasma miRNA Levels Predict Prognosis in Early-stage Malignant Melanoma. Anticancer Res. 2023, 43, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; He, J.; Jia, X.; Jiang, J.; Bai, R.; Yu, X.; Lv, L.; Fan, R.; He, X.; Geng, J.; et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in alpaca (Lama pacos) skin melanocytes. Domest. Anim. Endocrinol. 2010, 38, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, G.; Liao, J.; Tang, M.; Chen, J. Identification of key microRNAs affecting melanogenesis of breast muscle in Muchuan black-boned chickens by RNA sequencing. Br. Poult. Sci. 2020, 61, 225–231. [Google Scholar] [CrossRef]
- Sirri, F.; Zampiga, M.; Berardinelli, A. Effects of genotype and age on eggshell cuticle coverage and color profile in modern laying hen strains. Poult. Sci. 2022, 101, 101691. [Google Scholar] [CrossRef]
- de Hierro, M.; De Neve, L. Pigment limitation and female reproductive characteristics influence egg shell spottiness and ground colour variation in the house sparrow (Passer domesticus). J. Ornithol. 2010, 151, 833–840. [Google Scholar] [CrossRef]
- Liang, J.J.; Cao, D.R.; Zhang, X.W.; Liu, L.J.; Tan, Q.; Shi, S.; Chen, K.Y.; Liang, J.Y.; Wang, Z.G. miR-192-5p suppresses uterine receptivity formation through impeding epithelial transformation during embryo implantation. Theriogenology 2020, 157, 360–371. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xu, H.; Jiang, Y.; Hu, Z.; Du, R.; Zhao, X.; Tian, Y.; Zhu, Q.; Zhang, Y.; Liu, Y.; et al. Comprehensive analysis of differently expression mRNA and non-coding RNAs, and their regulatory mechanisms on relationship in thiram-induced tibial dyschondroplasia in chicken. Ecotoxicol. Environ. Saf. 2022, 242, 113924. [Google Scholar] [CrossRef]
- Zhang, M.; Han, Y.; Zhai, Y.H.; Ma, X.F.; An, X.L.; Zhang, S.; Li, Z.Y. Integrative analysis of circRNAs, miRNAs, and mRNAs profiles to reveal ceRNAs networks in chicken intramuscular and abdominal adipogenesis. BMC Genom. 2020, 21, 594. [Google Scholar] [CrossRef]
- Shen, M.M.; Li, T.T.; Chen, F.X.; Wu, P.F.; Wang, Y.; Chen, L.; Xie, K.Z.; Wang, J.Y.; Zhang, G.X. Transcriptomic Analysis of circRNAs and mRNAs Reveals a Complex Regulatory Network That Participate in Follicular Development in Chickens. Front. Genet. 2020, 11, 503. [Google Scholar] [CrossRef]
- Jiang, X.J.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, W.K.; Bae, K.H.; Lee, S.C.; Lee, E.W. Lipid Metabolism and Ferroptosis. Biology 2021, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Wang, Y.; Yang, W.T.; Li, Z.; Zhang, X.J.; Zhou, L.; Gui, J.F. Upregulation of the PPAR signaling pathway and accumulation of lipids are related to the morphological and structural transformation of the dragon-eye goldfish eye. Sci. China-Life Sci. 2021, 64, 1031–1049. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008, 275, 3249–3277. [Google Scholar] [CrossRef] [PubMed]
- Magnani, F.; Mattevi, A. Structure and mechanisms of ROS generation by NADPH oxidases. Curr. Opin. Struct. Biol. 2019, 59, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Medarde, A.; Santos, E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim. Biophys. Acta-Rev. Cancer 2011, 1815, 170–188. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, M.; Shats, I.; Krahn, J.M.; Flake, G.P.; Umbach, D.M.; Li, X.; Li, L. Glypican 6 is a putative biomarker for metastatic progression of cutaneous melanoma. PLoS ONE 2019, 14, e0218067. [Google Scholar] [CrossRef]
- Ratnayake, W.S.; Apostolatos, C.A.; Apostolatos, A.H.; Schutte, R.J.; Huynh, M.A.; Ostrov, D.A.; Acevedo-Duncan, M. Oncogenic PKC-ι activates Vimentin during epithelial-mesenchymal transition in melanoma; a study based on PKC-ι and PKC-ζ specific inhibitors. Cell Adh. Migr. 2018, 12, 447–463. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).