Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF
Abstract
1. Introduction
2. Epidemiological Considerations
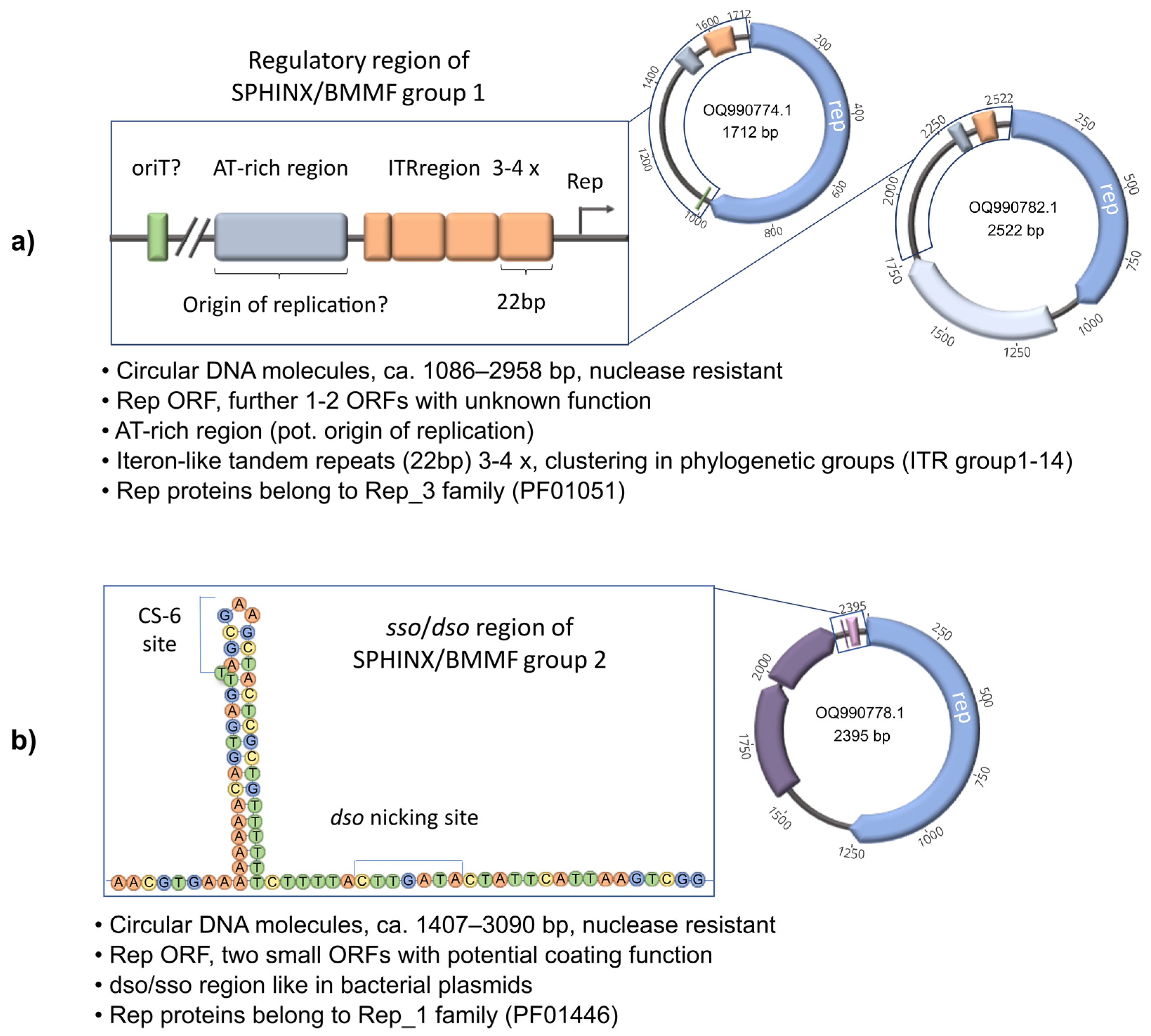
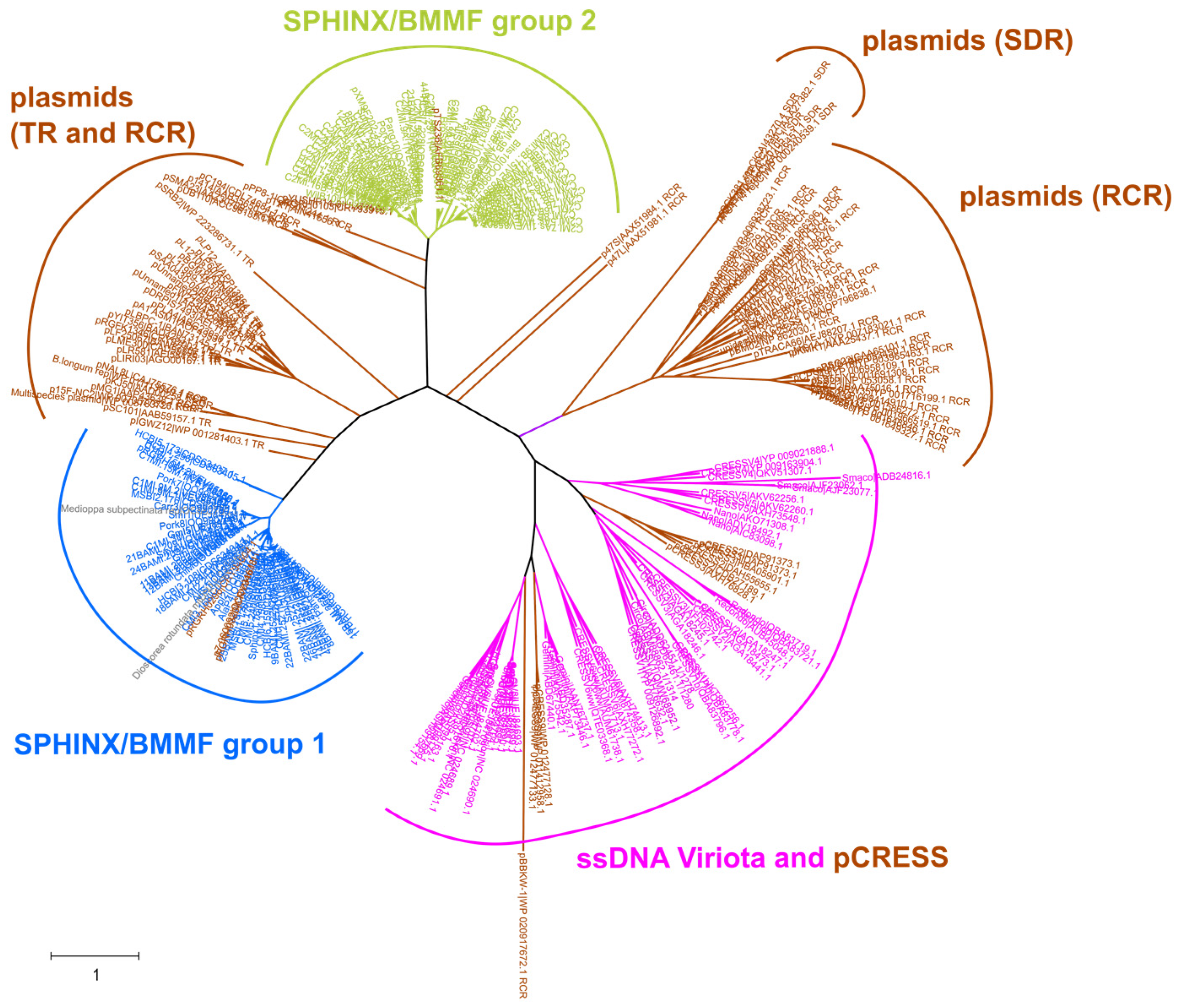
3. Is SPHINX/BMMF DNA Viral or Plasmid DNA?
4. Involvement of SPHINX/BMMF in Neurodegeneration
5. Involvement of SPHINX/BMMF in Cancer Development
6. Immunodetection of SPHINX/BMMF Rep Proteins
7. Infectious Potential of SPHINX/BMMF
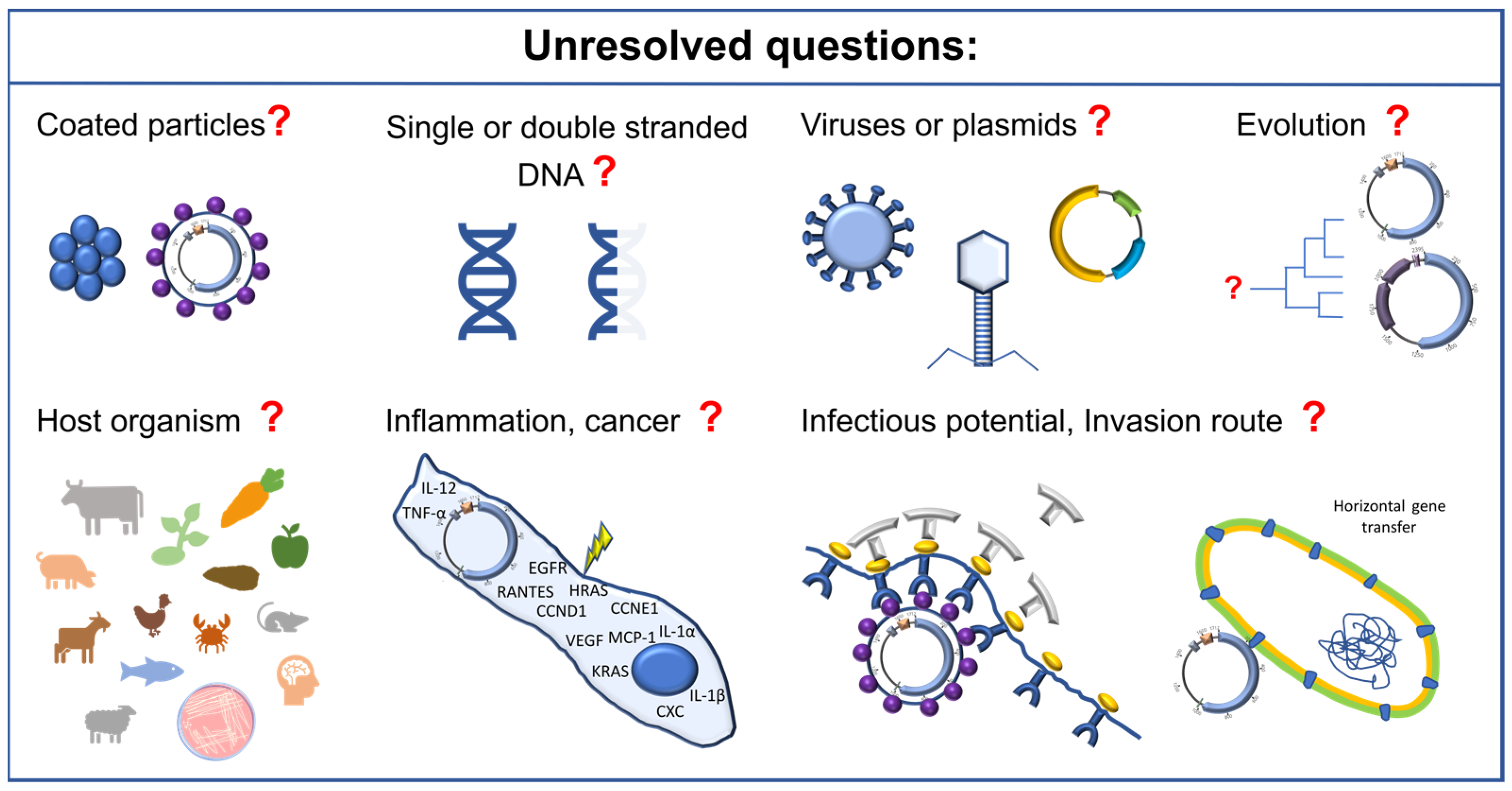
8. Future Perspectives
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lederberg, J. Cell genetics and hereditary symbiosis. Physiol. Rev. 1952, 32, 403–430. [Google Scholar] [CrossRef] [PubMed]
- Kunisada, T.; Yamagishi, H.; Sekiguchi, T. Intracellular location of small circular DNA complexes in mammalian cell lines. Plasmid 1983, 10, 242–250. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H. The search for infectious causes of human cancers: Where and why. Virology 2009, 392, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kuhnle, G.G.C.; Bingham, S.A. Dietary meat, endogenous nitrosation and colorectal cancer. Biochem. Soc. Trans. 2007, 35, 1355–1357. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, R.L.; Pierre, F.; Corpet, D.E. Processed meat and colorectal cancer: A review of epidemiologic and experimental evidence. Nutr. Cancer 2008, 60, 131–144. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H. Red meat consumption and cancer: Reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int. J. Cancer 2012, 130, 2475–2483. [Google Scholar] [CrossRef] [PubMed]
- Whitley, C.; Gunst, K.; Müller, H.; Funk, M.; zur Hausen, H.; de Villiers, E.-M. Novel replication-competent circular DNA molecules from healthy cattle serum and milk and multiple sclerosis-affected human brain tissue. Genome Announc. 2014, 2, e00849-14. [Google Scholar] [CrossRef] [PubMed]
- Gunst, K.; zur Hausen, H.; de Villiers, E.-M. Isolation of bacterial plasmid-related replication-associated circular DNA from a serum sample of a multiple sclerosis patient. Genome Announc. 2014, 2, e00847-14. [Google Scholar] [CrossRef] [PubMed]
- Funk, M.; Gunst, K.; Lucansky, V.; Müller, H.; zur Hausen, H.; de Villiers, E.-M. Isolation of protein-associated circular DNA from healthy cattle serum. Genome Announc. 2014, 2, e00846-14. [Google Scholar] [CrossRef] [PubMed]
- Manuelidis, L. Nuclease resistant circular DNAs copurify with infectivity in scrapie and CJD. J. Neurovirol. 2011, 17, 131–145. [Google Scholar] [CrossRef] [PubMed]
- DKFZ Neuartige Infektionserreger als Krebsrisikofaktoren: Pressekonferenz des Deutschen Krebsforschungszentrums. 2019. Available online: https://www.bing.com/ck/a?!&&p=7c1c3602a7777262JmltdHM9MTcwOTUxMDQwMCZpZ3VpZD0zNDJmMzYzOS1mOWE5LTY1NTUtMzBjYy0yMjAxZjg3YjY0NjAmaW5zaWQ9NTIwOA&ptn=3&ver=2&hsh=3&fclid=342f3639-f9a9-6555-30cc-2201f87b6460&psq=DKFZ+Pressekonferenz+2019&u=a1aHR0cHM6Ly93d3cuZGtmei5kZS9kZS9wcmVzc2UvZG93bmxvYWQvSGludGVyZ3J1bmQtUEstUGxhc21pZG9tZV9maW5hbC5wZGY&ntb=1 (accessed on 20 September 2022).
- Bundesinstitut Für Risikobewertung. Neuartige Erreger in Rind und Kuhmilchprodukten: Weitere Forschung notwendig: Stellungnahme Nr. 014/2019 des BfR vom 18. April 2019; Bundesinstitut Für Risikobewertung: Berlin, Germany, 2019; Available online: https://www.bfr.bund.de/cm/343/neuartige-erreger-in-rind-und-kuhmilchprodukten-weitere-forschung-notwendig.pdf (accessed on 20 March 2020).
- König, M.-T.; Fux, R.; Link, E.; Sutter, G.; Märtlbauer, E.; Didier, A. Circular Rep-Encoding Single-Stranded DNA Sequences in Milk from Water Buffaloes (Bubalus arnee f. bubalis). Viruses 2021, 13, 1088. [Google Scholar] [CrossRef]
- König, M.-T.; Fux, R.; Link, E.; Sutter, G.; Märtlbauer, E.; Didier, A. Identification and Characterization of Circular Single-Stranded DNA Genomes in Sheep and Goat Milk. Viruses 2021, 13, 2176. [Google Scholar] [CrossRef] [PubMed]
- Khvostov, D.V.; Konorov, E.A.; Minaev, M.Y. Detection of bovine meat and milk factors (BMMF) in food samples using highly sensitive real-time PCR. Izv. Vuzov. Food Technol. (Известия вузoв. Пищевая технoлoгия) 2022, 5, 96–99. [Google Scholar] [CrossRef]
- Pohl, S.; Habermann, D.; Link, E.K.; Fux, R.; Boldt, C.L.; Franz, C.M.; Hölzel, C.; Klempt, M. Detection of DNA sequences attributed to bovine meat and milk factors (BMMF/SPHINX) in food-related samples. Food Control 2022, 135, 108779. [Google Scholar] [CrossRef]
- Bundesinstitut für Risikobewertung. Neue Erkenntnisse zu “Bovine Meat and Milk Factors” (BMMF): Gemeinsame Stellungnahme Nr. 036/2022 von BfR und MRI vom 30. November 2022. 2022. Available online: https://www.bfr.bund.de/cm/343/neue-erkenntnisse-zu-bovine-meat-and-milk-factors-bmmf.pdf (accessed on 30 November 2022).
- König, M.-T.; Frölich, K.; Jandowsky, A.; Knauf-Witzens, T.; Langner, C.; Dietrich, R.; Märtlbauer, E.; Didier, A. First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle. Animals 2023, 13, 1492. [Google Scholar] [CrossRef] [PubMed]
- Habermann, D.; Klempt, M.; Franz, C.M.A.P. Identification and Characterization of Novel SPHINX/BMMF-like DNA Sequences Isolated from Non-Bovine Foods. Genes 2023, 14, 1307. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.S.M.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS ONE 2011, 6, e20456. [Google Scholar] [CrossRef] [PubMed]
- Corpet, D.E. Red meat and colon cancer: Should we become vegetarians, or can we make meat safer? Meat Sci. 2011, 89, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Huxley, R.R.; Ansary-Moghaddam, A.; Clifton, P.; Czernichow, S.; Parr, C.L.; Woodward, M. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: A quantitative overview of the epidemiological evidence. Int. J. Cancer 2009, 125, 171–180. [Google Scholar] [CrossRef]
- Veettil, S.K.; Wong, T.Y.; Loo, Y.S.; Playdon, M.C.; Lai, N.M.; Giovannucci, E.L.; Chaiyakunapruk, N. Role of Diet in Colorectal Cancer Incidence: Umbrella Review of Meta-analyses of Prospective Observational Studies. JAMA Netw. Open 2021, 4, e2037341. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Research Fund/American Institute for Cancer Research. Diet, Nutrition, Physical Activity and Colorectal Cancer [Internet]: Continuous Update Project. 2018. Available online: http://dietandcancerreport.org/ (accessed on 1 February 2024).
- World Cancer Research Fund/American Institute for Cancer Research. Continuous Update Project Expert Report 2018: Meat, Fisch and Dairy Products and Their Risk of Cancer. Available online: https://www.wcrf.org/wp-content/uploads/2021/02/Meat-fish-and-dairy-products.pdf (accessed on 1 February 2024).
- Wu, Y.; Huang, R.; Wang, M.; Bernstein, L.; Bethea, T.N.; Chen, C.; Chen, Y.; Eliassen, A.H.; Freedman, N.D.; Gaudet, M.M.; et al. Dairy foods, calcium, and risk of breast cancer overall and for subtypes defined by estrogen receptor status: A pooled analysis of 21 cohort studies. Am. J. Clin. Nutr. 2021, 114, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, S.; Chen, X.; Yang, J.; Zhou, Y.; Du, L.; Li, K. Red/processed meat consumption and non-cancer-related outcomes in humans: Umbrella review. Br. J. Nutr. 2023, 130, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Watling, C.Z.; Kelly, R.K.; Dunneram, Y.; Knuppel, A.; Piernas, C.; Schmidt, J.A.; Travis, R.C.; Key, T.J.; Perez-Cornago, A. Associations of intakes of total protein, protein from dairy sources, and dietary calcium with risks of colorectal, breast, and prostate cancer: A prospective analysis in UK Biobank. Br. J. Cancer 2023, 129, 636–647. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H.; Bund, T.; de Villiers, E.-M. Viruses, Genes, and Cancer; Springer: Cham, Switzerland, 2017; pp. 83–116. [Google Scholar] [CrossRef]
- de Villiers, E.-M.; Gunst, K.; Chakraborty, D.; Ernst, C.; Bund, T.; zur Hausen, H. A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue. Emerg. Microbes Infect. 2019, 8, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Hotz-Wagenblatt, A.; Sarachaga, V.; Burk, A.; Falida, K.; Chakraborty, D.; Nikitina, E.; Tessmer, C.; Whitley, C.; Sauerland, C.; et al. Expression and replication of virus-like circular DNA in human cells. Sci. Rep. 2018, 8, 2851. [Google Scholar] [CrossRef] [PubMed]
- Longkumer, T.; Kamireddy, S.; Muthyala, V.R.; Akbarpasha, S.; Pitchika, G.K.; Kodetham, G.; Ayaluru, M.; Siddavattam, D. Acinetobacter phage genome is similar to Sphinx 2.36, the circular DNA copurified with TSE infected particles. Sci. Rep. 2013, 3, 2240. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, K.J.; Houtti, M.; Litzau, L.A.; Aird, E.J.; Everett, B.A.; Nelson, A.T.; Pornschloegl, L.; Limón-Swanson, L.K.; Evans, R.L.; Evans, K.; et al. Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering. Nucleic Acids Res. 2021, 49, 1046–1064. [Google Scholar] [CrossRef]
- Marogianni, C.; Sokratous, M.; Dardiotis, E.; Hadjigeorgiou, G.M.; Bogdanos, D.; Xiromerisiou, G. Neurodegeneration and Inflammation—An Interesting Interplay in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 8421. [Google Scholar] [CrossRef]
- Piekut, T.; Hurła, M.; Banaszek, N.; Szejn, P.; Dorszewska, J.; Kozubski, W.; Prendecki, M. Infectious agents and Alzheimer’s disease. J. Integr. Neurosci. 2022, 21, 73. [Google Scholar] [CrossRef] [PubMed]
- Bjornevik, K.; Münz, C.; Cohen, J.I.; Ascherio, A. Epstein–Barr virus as a leading cause of multiple sclerosis: Mechanisms and implications. Nat. Rev. Neurol. 2023, 19, 160–171. [Google Scholar] [CrossRef]
- Yeh, Y.-H.; Gunasekharan, V.; Manuelidis, L. A prokaryotic viral sequence is expressed and conserved in mammalian brain. Proc. Natl. Acad. Sci. USA 2017, 114, 7118–7123. [Google Scholar] [CrossRef] [PubMed]
- Botsios, S.; Manuelidis, L. CJD and Scrapie Require Agent-Associated Nucleic Acids for Infection. J. Cell. Biochem. 2016, 117, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Manuelidis, L. Prokaryotic SPHINX 1.8 REP protein is tissue-specific and expressed in human germline cells. J. Cell. Biochem. 2019, 120, 6198–6208. [Google Scholar] [CrossRef] [PubMed]
- Marr, D. A theory of cerebellar cortex. J. Physiol. 1969, 202, 437–470. [Google Scholar] [CrossRef] [PubMed]
- Middleton, S.J.; Racca, C.; Cunningham, M.O.; Traub, R.D.; Monyer, H.; Knöpfel, T.; Schofield, I.S.; Jenkins, A.; Whittington, M.A. High-frequency network oscillations in cerebellar cortex. Neuron 2008, 58, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Lamberto, I.; Gunst, K.; Müller, H.; zur Hausen, H.; de Villiers, E.-M. Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Bund, T.; Nikitina, E.; Chakraborty, D.; Ernst, C.; Gunst, K.; Boneva, B.; Tessmer, C.; Volk, N.; Brobeil, A.; Weber, A.; et al. Analysis of chronic inflammatory lesions of the colon for BMMF Rep antigen expression and CD68 macrophage interactions. Proc. Natl. Acad. Sci. USA 2021, 118, e2025830118. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.; Alikhanyan, K.; Neßling, M.; Richter, K.; Kaden, S.; Ernst, C.; Seitz, S.; Chuprikova, L.; Häfele, L.; Gunst, K.; et al. Structural expression of bovine milk and meat factors in tissues of colorectal, lung and pancreatic cancer patients. Int. J. Cancer 2023, 153, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.; Burk-Körner, A.; Wiesenfarth, M.; Alwers, E.; Heide, D.; Tessmer, C.; Ernst, C.; Krunic, D.; Schrotz-King, P.; Chang-Claude, J.; et al. Bovine meat and milk factor protein expression in tumor-free mucosa of colorectal cancer patients coincides with macrophages and might interfere with patient survival. Mol. Oncol. 2024, 18, 1076–1092. [Google Scholar] [CrossRef] [PubMed]
- Falida, K.; Eilebrecht, S.; Gunst, K.; zur Hausen, H.; de Villiers, E.-M. Isolation of Two Virus-Like Circular DNAs from Commercially Available Milk Samples. Genome Announc. 2017, 5, e00266-17. [Google Scholar] [CrossRef] [PubMed]
- Szigeti-Buck, K.; Manuelidis, L. Prokaryotic SPHINX replication sequences are conserved in mammalian brain and participate in neurodegeneration. J. Cell. Biochem. 2019, 120, 17687–17698. [Google Scholar] [CrossRef] [PubMed]
- Mobaraki, G.; Shi, S.; Smits, K.M.; Severens, K.; Lommen, K.; Rennspiess, D.; Chteinberg, E.; Winnepenninckx, V.; Samarska, I.; Klufah, F.; et al. Bovine Meat and Milk Factor-like Sequences Are Frequently Detected in Renal Cell Carcinoma Tissues. Cancers 2024, 16, 1746. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bordeaux, J.; Welsh, A.; Agarwal, S.; Killiam, E.; Baquero, M.; Hanna, J.; Anagnostou, V.; Rimm, D. Antibody validation. Biotechniques 2010, 48, 197–209. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Definition of Infection. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/infection (accessed on 1 February 2024).
- Fu, X.Y.; Manley, J.L. Factors influencing alternative splice site utilization in vivo. Mol. Cell. Biol. 1987, 7, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Pastrana, D.V.; Lowy, D.R.; Schiller, J.T. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 2005, 119, 445–462. [Google Scholar] [CrossRef] [PubMed]
- de Villiers, E.-M.; Borkosky, S.S.; Kimmel, R.; Gunst, K.; Fei, J.-W. The diversity of torque teno viruses: In vitro replication leads to the formation of additional replication-competent subviral molecules. J. Virol. 2011, 85, 7284–7295. [Google Scholar] [CrossRef] [PubMed]
| Occurrence of Circular SPHINX/BMMF DNA Molecules | References |
|---|---|
| Mouse and hamster cell lines and tissues (TSE-infected) | [10] |
| Human sera, brain (MS patients), peritumoral colon cancer tissue | [7,8,42,43,44,45] |
| Bovine milk and blood | [7,9,46] |
| Water buffalo milk, sheep and goat milk, blood and feces of African and Asian cattle | [13,14,18] |
| PCR detection in white and red meats, seafood, fruits, vegetables, grain and baby food, feces (pig, chicken) and saliva (pig) | [15,16] |
| Pork, wild boar meat, chicken meat, Alaska pollock, pangasius, black tiger shrimp, apple, carrot, sprouts | [19] |
| Detection of Rep protein 1 | References |
| Rodent and human neuronal cells and brain, esp. synapses | [37,47] |
| Rodent and human smooth muscles of uterus, arteries, gastrointestinal tract, ovary, testis, skin, etc. | [39] |
| Human peritumoral tissues of colon, lung and pancreas cancer, co-localization with interstitial macrophages, but not in cancer tissue | [43,44,45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habermann, D.; Franz, C.M.A.P.; Klempt, M. Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF. Genes 2024, 15, 678. https://doi.org/10.3390/genes15060678
Habermann D, Franz CMAP, Klempt M. Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF. Genes. 2024; 15(6):678. https://doi.org/10.3390/genes15060678
Chicago/Turabian StyleHabermann, Diana, Charles M. A. P. Franz, and Martin Klempt. 2024. "Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF" Genes 15, no. 6: 678. https://doi.org/10.3390/genes15060678
APA StyleHabermann, D., Franz, C. M. A. P., & Klempt, M. (2024). Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF. Genes, 15(6), 678. https://doi.org/10.3390/genes15060678






