Prediction of Protein-DNA Interface Hot Spots Based on Empirical Mode Decomposition and Machine Learning
Abstract
1. Introduction
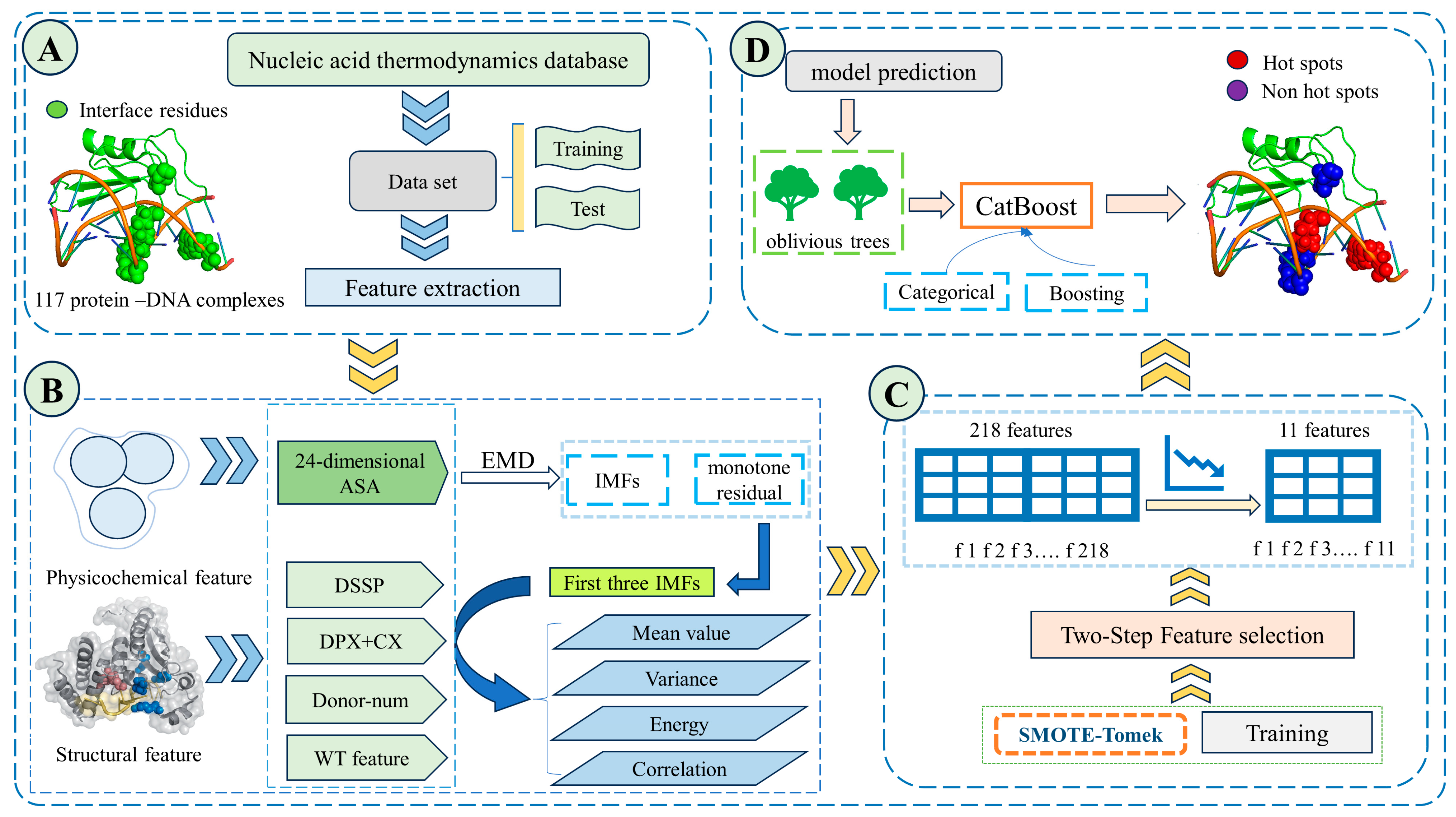
2. Materials and Methods
2.1. Datasets
2.2. Feature Extraction
2.2.1. Solvent-Accessible Surface Area Characteristics
2.2.2. Secondary Structure Features
2.2.3. Depth Index and Protrusion Index
2.2.4. Number of Hydrogen Bonds
2.2.5. Wavelet Transform Features
2.2.6. EMD Feature
2.3. Data Balancing
2.4. Feature Selection
2.5. Model Construction
2.6. Performance Evaluation
3. Results
3.1. Comparison of Various Data-Balancing Approaches
3.2. Comparison of Various Feature Selection Approaches
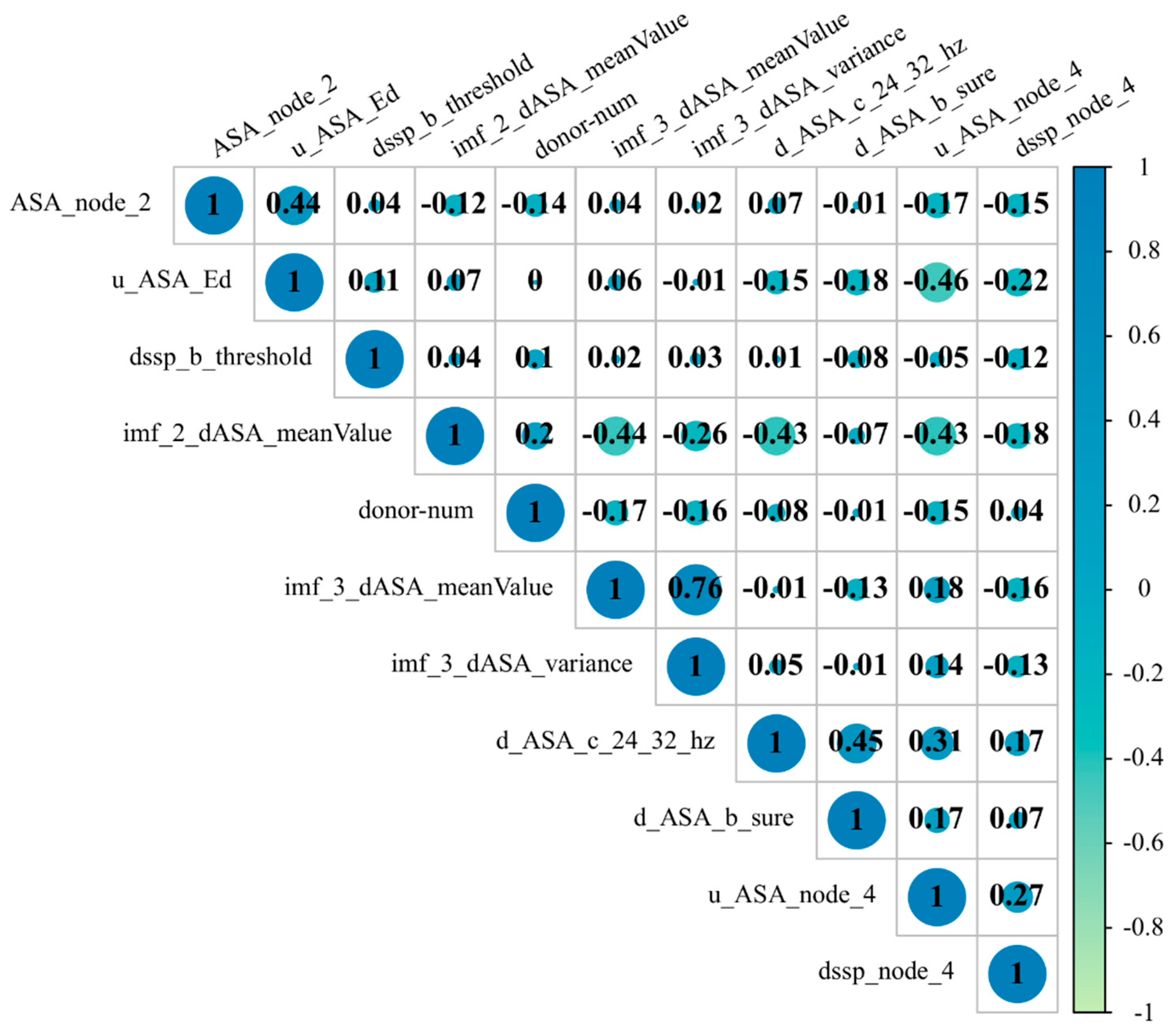
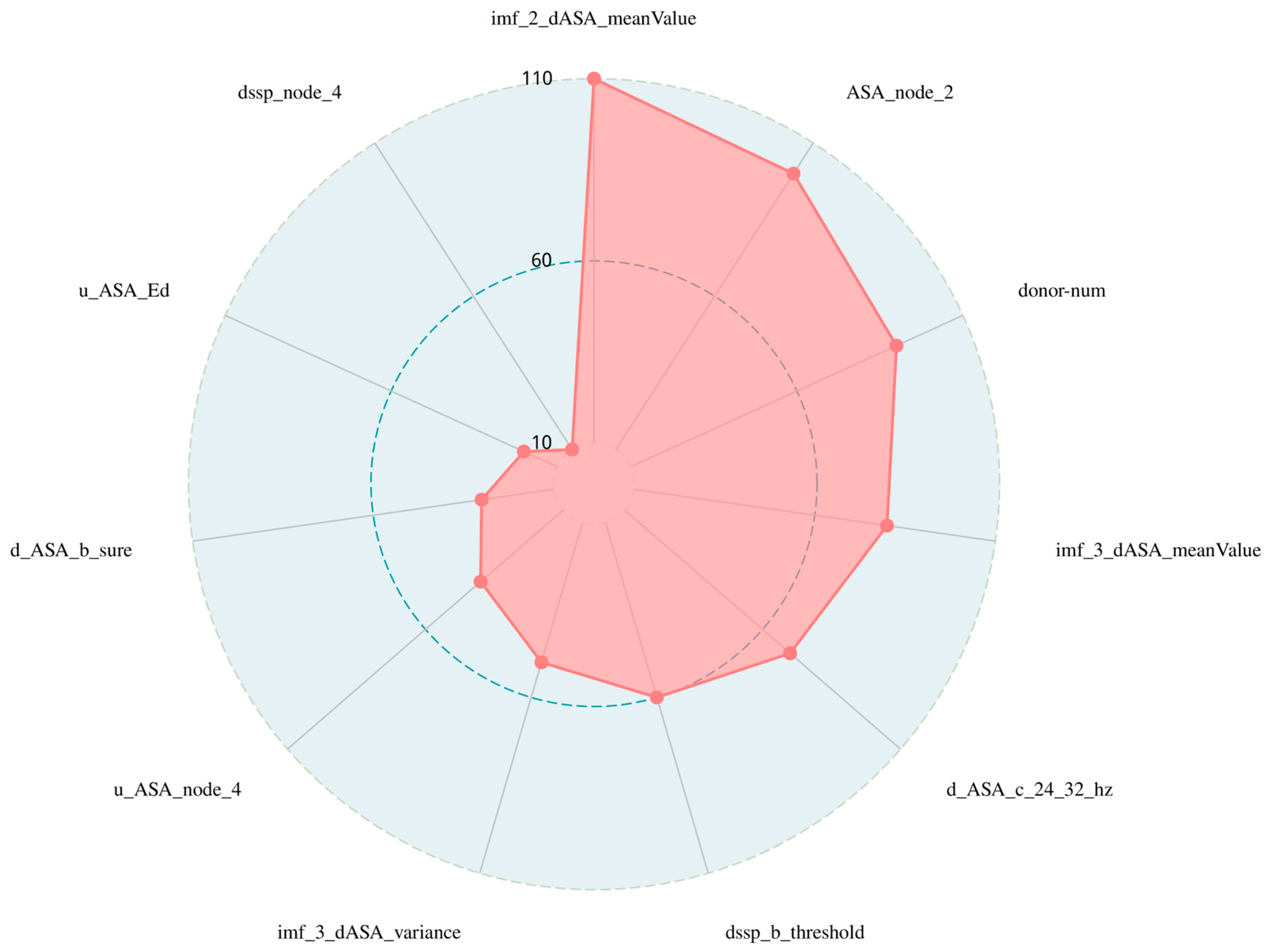
3.3. Importance Ranking of Features and the Best Subset
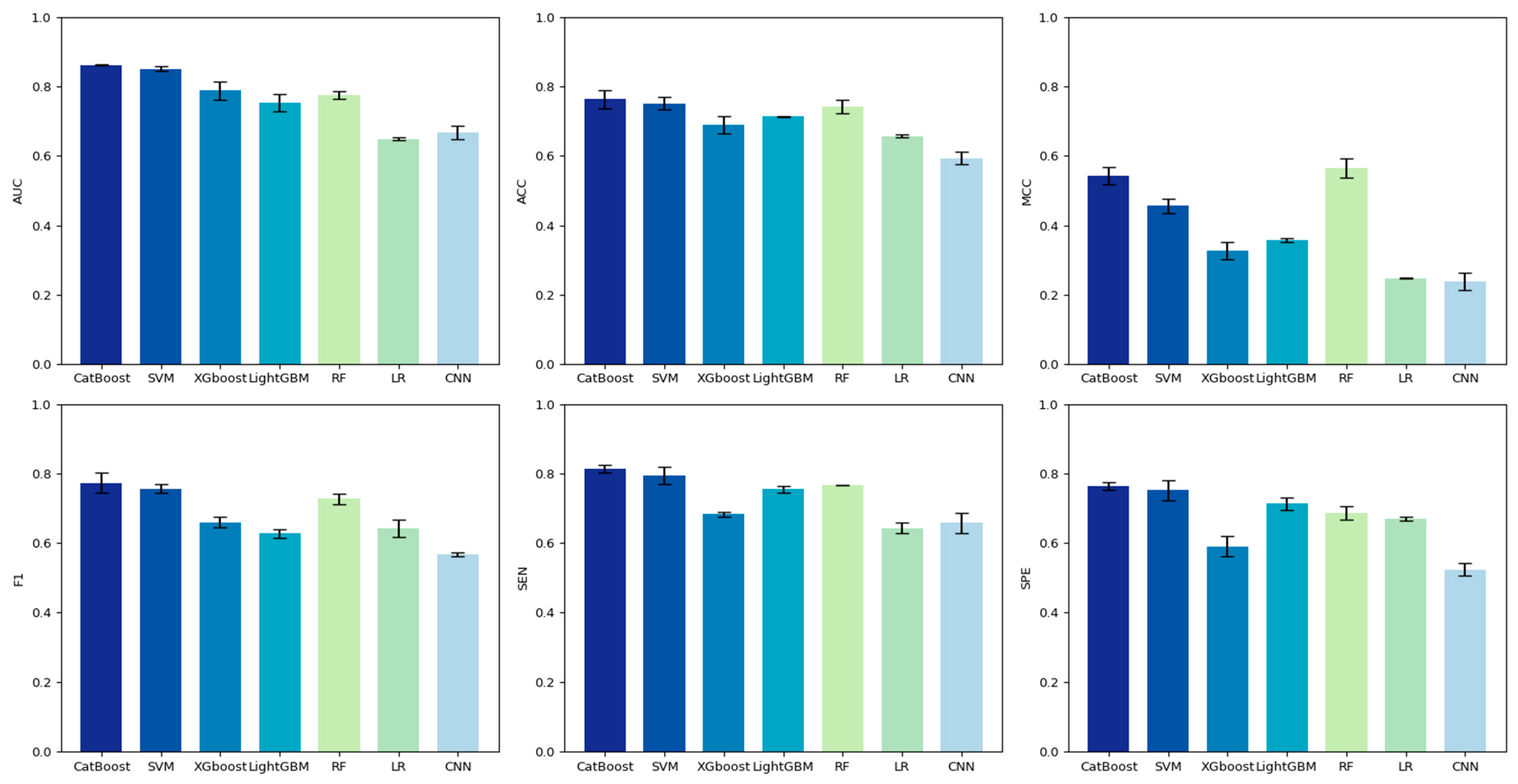
3.4. Comparison of Different Classification Models
3.5. Performance Comparison of Different Methods on the Test Set
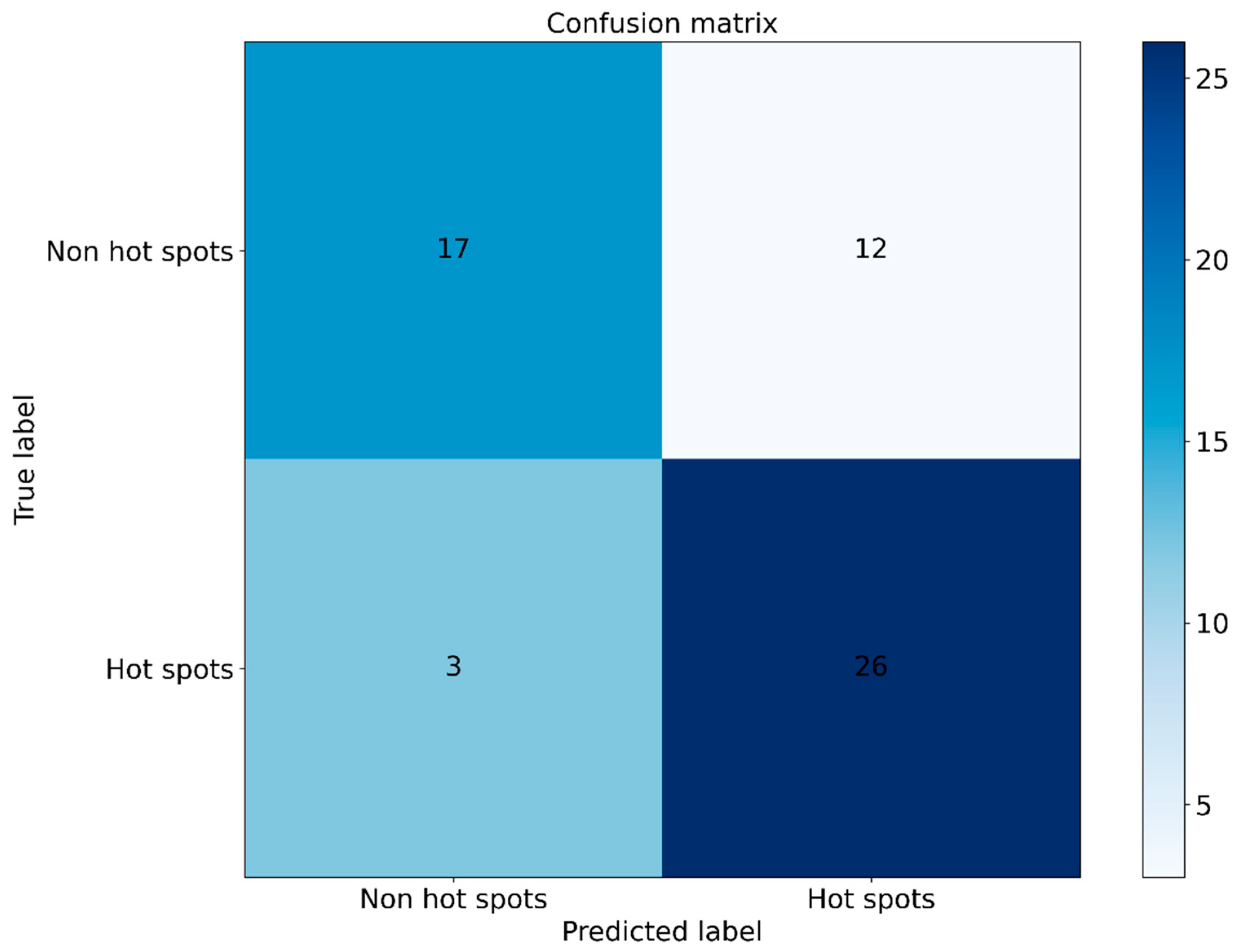
3.6. Case Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santoro, C.; Mermod, N.; Andrews, P.C.; Tjian, R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: Cloning and expression of multiple cDNAs. Nature 1988, 334, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999, 27, 4658–4670. [Google Scholar] [CrossRef] [PubMed]
- Clackson, T.; Wells, J.A. A Hot Spot of Binding Energy in a Hormone-Receptor Interface. Science 1995, 267, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Moreira, I.S.; Fernandes, P.A.; Ramos, M.J. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins Struct. Funct. Bioinform. 2007, 68, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Tjong, H.; Zhou, H.-X. DISPLAR: An accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007, 35, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Sun, L.; Jia, Z.; Li, L.; Alexov, E. Predicting protein-DNA binding free energy change upon missense mutations using modified MM/PBSA approach: SAMPDI webserver. Bioinformatics 2018, 34, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Alexov, E.; Zhang, N.; Chen, Y.; Zhao, F.; Yang, Q.; Simonetti, F.L.; Li, M. PremPDI estimates and interprets the effects of missense mutations on protein-DNA interactions. PLOS Comput. Biol. 2018, 14, e1006615. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Panday, S.K.; Peng, Y.; Alexov, E. SAMPDI-3D: Predicting the effects of protein and DNA mutations on protein-DNA interactions. Bioinformatics 2021, 37, 3760–3765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, L.; Zheng, C.-H.; Xia, J. A feature-based approach to predict hot spots in protein-DNA binding interfaces. Brief. Bioinform. 2020, 21, 1038–1046. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, H.; Xu, Z.; Yue, Z.; Li, K. Prediction of hot spots in protein-DNA binding interfaces based on discrete wavelet transform and wavelet packet transform. BMC Bioinform. 2023, 24, 129. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Zhao, L.; Li, M.; Liu, M.; Li, K.; Bin, Y.; Xia, J. An improved DNA-binding hot spot residues prediction method by exploring interfacial neighbor properties. BMC Bioinform. 2021, 22, 253. [Google Scholar] [CrossRef]
- Li, K.; Zhang, S.; Yan, D.; Bin, Y.; Xia, J. Prediction of hot spots in protein-DNA binding interfaces based on supervised isometric feature mapping and extreme gradient boosting. BMC Bioinform. 2020, 21, 381. [Google Scholar] [CrossRef]
- Yao, L.; Wang, H.; Bin, Y. Predicting Hot Spot Residues at Protein-DNA Binding Interfaces Based on Sequence Information. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 1–11. [Google Scholar] [CrossRef]
- Pan, Y.; Zhou, S.; Guan, J. Computationally identifying hot spots in protein-DNA binding interfaces using an ensemble approach. BMC Bioinform. 2020, 21, 384. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, H.-F.; Liu, R. Systematic comparison and prediction of the effects of missense mutations on protein-DNA and protein-RNA interactions. PLOS Comput. Biol. 2021, 17, e1008951. [Google Scholar] [CrossRef]
- Jonathan, B.; Putra, P.H.; Ruldeviyani, Y. Observation Imbalanced Data Text to Predict Users Selling Products on Female Daily with SMOTE, Tomek, and SMOTE-Tomek. In Proceedings of the 2020 IEEE International Conference on Industry 4.0, Artificial Intelligence, and Communications Technology (IAICT), Bali, Indonesia, 7–8 July 2020; pp. 81–85. [Google Scholar]
- Boudraa, A.O.; Cexus, J.C. EMD-Based Signal Filtering. IEEE Trans. Instrum. Meas. 2007, 56, 2196–2202. [Google Scholar] [CrossRef]
- Nakariyakul, S. Gene selection using interaction information for microarray-based cancer classification. In Proceedings of the 2016 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Chiang Mai, Thailand, 5–7 October 2016; pp. 1–5. [Google Scholar]
- Hancock, J.T.; Khoshgoftaar, T.M. CatBoost for big data: An interdisciplinary review. J. Big Data 2020, 7, 94. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Liu, C.; Zhang, Y.; Pan, Y.; Wang, Z.; Wang, J.; Wen, T.; Deng, L. Nabe: An energetic database of amino acid mutations in protein–nucleic acid binding interfaces. Database 2021, 2021, baab050. [Google Scholar] [CrossRef]
- Harini, K.; Srivastava, A.; Kulandaisamy, A.; Gromiha, M.M. ProNAB: Database for binding affinities of protein–nucleic acid complexes and their mutants. Nucleic Acids Res. 2022, 50, D1528–D1534. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Sj, H. NACCESS-Computer Program. Available online: http://www.bioinf.manchester.ac.uk/naccess/ (accessed on 20 December 2023).
- Jones, S.; Daley, D.T.A.; Luscombe, N.M.; Berman, H.M.; Thornton, J.M. Protein–RNA interactions: A structural analysis. Nucleic Acids Res. 2001, 29, 943–954. [Google Scholar] [CrossRef]
- Dai, W.; Wu, A.; Ma, L.; Li, Y.-X.; Jiang, T.; Li, Y.-Y. A novel index of protein-protein interface propensity improves interface residue recognition. BMC Syst. Biol. 2016, 10, 112. [Google Scholar] [CrossRef]
- Jones, S.; Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Sander, C.; Scharf, M.; Vriend, G. The PDBFINDER database: A summary of PDB, DSSP and HSSP information with added value. Bioinformatics 1996, 12, 525–529. [Google Scholar] [CrossRef][Green Version]
- Xia, J.-F.; Zhao, X.-M.; Song, J.; Huang, D.-S. APIS: Accurate prediction of hot spots in protein interfaces by combining protrusion index with solvent accessibility. BMC Bioinform. 2010, 11, 174. [Google Scholar] [CrossRef]
- Pintar, A.; Carugo, O.; Pongor, S. DPX: For the analysis of the protein core. Bioinformatics 2003, 19, 313–314. [Google Scholar] [CrossRef]
- Mihel, J.; Šikić, M.; Tomić, S.; Jeren, B.; Vlahoviček, K. PSAIA—Protein Structure and Interaction Analyzer. BMC Struct. Biol. 2008, 8, 21. [Google Scholar] [CrossRef]
- Ellis, J.J.; Broom, M.; Jones, S. Protein–RNA interactions: Structural analysis and functional classes. Proteins Struct. Funct. Bioinform. 2007, 66, 903–911. [Google Scholar] [CrossRef]
- Laurent, J.; Touvrey, C.; Gillessen, S.; Joffraud, M.; Vicari, M.; Bertrand, C.; Ongarello, S.; Liedert, B.; Gallerani, E.; Beck, J.; et al. T-cell activation by treatment of cancer patients with EMD 521873 (Selectikine), an IL-2/anti-DNA fusion protein. J. Transl. Med. 2013, 11, 5. [Google Scholar] [CrossRef]
- Hu, J.; Yang, Y.D.; Kihara, D. EMD: An ensemble algorithm for discovering regulatory motifs in DNA sequences. BMC Bioinform. 2006, 7, 342. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, J.; Xu, W. Identification of eukaryotic exons using empirical mode decomposition and modified Gabor-wavelet transform. In Proceedings of the 33rd Chinese Control Conference, Nanjing, China, 28–30 July 2014; pp. 7151–7155. [Google Scholar]
- Weng, B.; Xuan, G.; Kolodzey, J.; Barner, K.E. Empirical mode decomposition as a tool for DNA sequence analysis from terahertz spectroscopy measurements. In Proceedings of the 2006 IEEE International Workshop on Genomic Signal Processing and Statistics, College Station, TX, USA, 28–30 May 2006; pp. 63–64. [Google Scholar]
- Li, B.-Q.; Feng, K.-Y.; Chen, L.; Huang, T.; Cai, Y.-D. Prediction of Protein-Protein Interaction Sites by Random Forest Algorithm with mRMR and IFS. PLoS ONE 2012, 7, e43927. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, Y. A feature selection algorithm of music genre classification based on ReliefF and SFS. In Proceedings of the 2015 IEEE/ACIS 14th International Conference on Computer and Information Science (ICIS), Las Vegas, NV, USA, 28 June–1 July 2015; pp. 539–544. [Google Scholar]
- Nguyen, C.; Wang, Y.; Nguyen, H.N. Random forest classifier combined with feature selection for breast cancer diagnosis and prognostic. J. Biomed. Sci. Eng. 2013, 6, 10. [Google Scholar] [CrossRef]
- Sanz, H.; Valim, C.; Vegas, E.; Oller, J.M.; Reverter, F. SVM-RFE: Selection and visualization of the most relevant features through non-linear kernels. BMC Bioinform. 2018, 19, 432. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; He, X.; Xu, J.; Zhang, R.; Lu, Y. Scattering Feature Set Optimization and Polarimetric SAR Classification Using Object-Oriented RF-SFS Algorithm in Coastal Wetlands. Remote Sens. 2020, 12, 407. [Google Scholar] [CrossRef]
- Ogunleye, A.; Wang, Q.G. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 2131–2140. [Google Scholar] [CrossRef]
- Jhaveri, S.; Khedkar, I.; Kantharia, Y.; Jaswal, S. Success Prediction using Random Forest, CatBoost, XGBoost and AdaBoost for Kickstarter Campaigns. In Proceedings of the 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 27–29 March 2019; pp. 1170–1173. [Google Scholar]
- Wang, D.; Zhang, Y.; Zhao, Y. LightGBM: An Effective miRNA Classification Method in Breast Cancer Patients. In Proceedings of the 2017 International Conference on Computational Biology and Bioinformatics, Newark, NJ, USA, 18–20 October 2017; pp. 7–11. [Google Scholar]
- Ke, G.; Xu, Z.; Zhang, J.; Bian, J.; Liu, T.-Y. DeepGBM: A Deep Learning Framework Distilled by GBDT for Online Prediction Tasks. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 384–394. [Google Scholar]
- Ibrahim, A.A.; Ridwan, R.L.; Muhammed, M.M.; Abdulaziz, R.O.; Saheed, G.A. Comparison of the CatBoost Classifier with other Machine Learning Methods. Int. J. Adv. Comput. Sci. Appl. (IJACSA) 2020, 11. [Google Scholar] [CrossRef]
- Torgo, L.; Ribeiro, R.P.; Pfahringer, B.; Branco, P. SMOTE for Regression. In Portuguese Conference on Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2013; pp. 378–389. [Google Scholar]
- Haibo, H.; Yang, B.; Garcia, E.A.; Shutao, L. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, 1–8 June 2008; pp. 1322–1328. [Google Scholar]
- Pavletich, N.P.; Pabo, C.O. Zinc Finger-DNA Recognition: Crystal Structure of a Zif268-DNA Complex at 2.1 Å. Science 1991, 252, 809–817. [Google Scholar] [CrossRef]
- Zheng, L.; Jia, J.; Finger, L.D.; Guo, Z.; Zer, C.; Shen, B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011, 39, 781–794. [Google Scholar] [CrossRef]


| Dataset | Number of Variants | Amount of PDBs | Number of Hot Spots | Number of Non-Hot Spots | Ratio |
|---|---|---|---|---|---|
| Training | 271 | 92 | 102 | 179 | 0.569 |
| Test | 68 | 25 | 29 | 29 | 0.500 |
| Number | Feature | Feature Description |
|---|---|---|
| 1 | imf_2_dASA_meanValue | The mean of the second IMF digital signaling component after the EMD of dASA |
| 2 | ASA_node_2 | The baud sign of the second node of the third layer of ASA after WPT processing |
| 3 | donor-num | Number of hydrogen bonds |
| 4 | imf_3_dASA_meanValue | The mean of the third IMF digital signaling component after the EMD of dASA |
| 5 | d_ASA_c_24_32_hz | Absolute energy of the eighth node after WPT processing by uASA |
| 6 | dssp_b_threshold | Threshold of secondary structural features after WPT |
| 7 | imf_3_dASA_variance | The variance of the third IMF digital signaling component after the EMD of dASA |
| 8 | u_ASA_node_4 | uASA: the baud sign of the fourth node of the third layer after WPT processing |
| 9 | d_ASA_b_sure | Shannon entropy of dASA features after WPT |
| 10 | u_ASA_Ed | The Ed of the third layer wavelet approximation coefficient is obtained using the uASA wavelet transform |
| 11 | dssp_node_4 | DSSP: the baud sign of the fourth node of the third layer after WPT processing |
| Method | SEN | SPE | PRE | F1 | MCC | ACC | AUC |
|---|---|---|---|---|---|---|---|
| EC-PDH | 0.808 | 0.761 | 0.752 | 0.769 | 0.540 | 0.762 | 0.859 |
| Approach | SEN | SPE | PRE | F1 | MCC | ACC | AUC |
|---|---|---|---|---|---|---|---|
| SMOTE-Tomek | 0.808 | 0.761 | 0.752 | 0.769 | 0.540 | 0.762 | 0.859 |
| SMOTE | 0.782 | 0.779 | 0.748 | 0.762 | 0.531 | 0.758 | 0.848 |
| ADASYN | 0.721 | 0.572 | 0.653 | 0.612 | 0.351 | 0.651 | 0.735 |
| Random Repetitive Oversampling | 0.688 | 0.611 | 0.658 | 0.642 | 0.465 | 0.712 | 0.762 |
| Unprocessed | 0.189 | 0.645 | 0.364 | 0.266 | 0.324 | 0.642 | 0.721 |
| Approach | SEN | SPE | PRE | F1 | MCC | ACC | AUC |
|---|---|---|---|---|---|---|---|
| mRMR-SFS (11) | 0.808 | 0.761 | 0.752 | 0.769 | 0.540 | 0.762 | 0.859 |
| mRMR (12) | 0.705 | 0.724 | 0.737 | 0.708 | 0.465 | 0.713 | 0.789 |
| SFS (15) | 0.678 | 0.701 | 0.721 | 0.692 | 0.429 | 0.695 | 0.776 |
| RF-SFS (16) | 0.756 | 0.762 | 0.756 | 0.745 | 0.536 | 0.728 | 0.841 |
| RF (21) | 0.751 | 0.714 | 0.741 | 0.736 | 0.512 | 0.745 | 0.832 |
| SVM-RFE (22) | 0.681 | 0.649 | 0.624 | 0.526 | 0.215 | 0.621 | 0.694 |
| Feature Combination | SEN | SPE | PRE | F1 | MCC | ACC | AUC |
|---|---|---|---|---|---|---|---|
| EMD and wavelet transform features | 0.813 | 0.764 | 0.756 | 0.772 | 0.543 | 0.764 | 0.863 |
| Wavelet transform features | 0.784 | 0.779 | 0.746 | 0.765 | 0.533 | 0.754 | 0.847 |
| EMD features | 0.801 | 0.765 | 0.744 | 0.769 | 0.539 | 0.766 | 0.855 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Z.; Li, Z.; Li, M.; Yue, Z.; Li, K. Prediction of Protein-DNA Interface Hot Spots Based on Empirical Mode Decomposition and Machine Learning. Genes 2024, 15, 676. https://doi.org/10.3390/genes15060676
Fang Z, Li Z, Li M, Yue Z, Li K. Prediction of Protein-DNA Interface Hot Spots Based on Empirical Mode Decomposition and Machine Learning. Genes. 2024; 15(6):676. https://doi.org/10.3390/genes15060676
Chicago/Turabian StyleFang, Zirui, Zixuan Li, Ming Li, Zhenyu Yue, and Ke Li. 2024. "Prediction of Protein-DNA Interface Hot Spots Based on Empirical Mode Decomposition and Machine Learning" Genes 15, no. 6: 676. https://doi.org/10.3390/genes15060676
APA StyleFang, Z., Li, Z., Li, M., Yue, Z., & Li, K. (2024). Prediction of Protein-DNA Interface Hot Spots Based on Empirical Mode Decomposition and Machine Learning. Genes, 15(6), 676. https://doi.org/10.3390/genes15060676






