Variability and Number of Circulating Complementary Sex Determiner (Csd) Alleles in a Breeding Population of Italian Honeybees under Controlled Mating
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. DNA Extraction, Library Preparation, Sequence Processing and Alignment
2.3. De Novo Assembly and Analysis of Sample-Specific HVR Allele Consensus Sequences
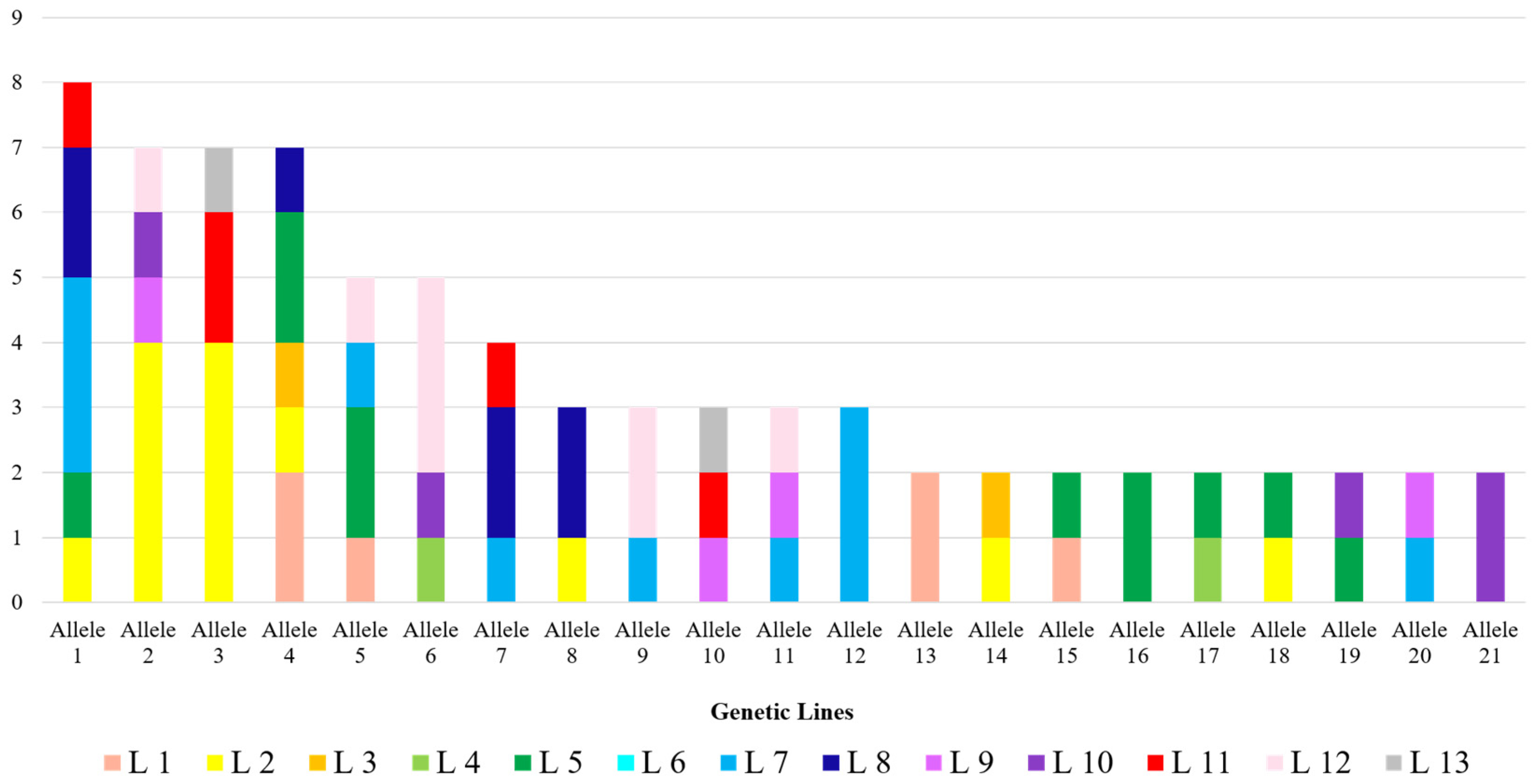
3. Results
Csd HVR Amino Acid Sequence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beye, M.; Hasselmann, M.; Fondrk, M.; Page, R.E.; Omholt, S.W. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 2003, 114, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Dzierzon, J. Neue Verbesserte Bienen-Zucht des Pfarrers Dzierzon zu Carlsmarkt in Schlesien; Ernst’schen: Quedlinburg, Germany, 1861. [Google Scholar]
- Woyke, J. What happens to diploid drone larvae in a honeybee colony. J. Apic. Res. 1963, 2, 73–75. [Google Scholar] [CrossRef]
- Kaskinova, M.D.; Nikolenko, A.G. Csd gene of honeybee: Genetic structure, functioning, and evolution. Russ. J. Genet. 2017, 53, 297–301. [Google Scholar] [CrossRef]
- Beye, M.; Seelmann, C.; Gempe, T.; Hasselmann, M.; Vekemans, X.; Fondrk, M.K.; Page, R.E. Gradual molecular evolution of a sex determination switch through incomplete penetrance of femaleness. Curr. Biol. 2013, 23, 2559–2564. [Google Scholar] [CrossRef]
- Hasselmann, M.; Beye, M. Signatures of selection among sex-determining alleles of the honey bee. Proc. Natl. Acad. Sci. USA 2004, 101, 4888–4893. [Google Scholar] [CrossRef] [PubMed]
- Hasselmann, M.; Beye, M. Pronounced differences of recombination activity at the sex determination locus of the honeybee, a locus under strong balancing selection. Genetics 2006, 174, 1469–1480. [Google Scholar] [CrossRef]
- Charlesworth, D. Sex determination: Balancing selection in the honey bee. Curr. Biol. 2004, 14, 568–569. [Google Scholar] [CrossRef] [PubMed]
- Lechner, S.; Ferretti, L.; Schöning, C.; Kinuthia, W.; Willemsen, D.; Hasselmann, M. Nucleotide variability at its limit? Insights into the number and evolutionary dynamics of the sex-determining specificities of the honeybee Apis mellifera. Mol. Biol. Evol. 2014, 31, 272–287. [Google Scholar] [CrossRef] [PubMed]
- Bovo, S.; Ribani, A.; Utzeri, V.J.; Taurisano, V.; Schiavo, G.; Bolner, M.; Fontanesi, L. Application of Next Generation Semiconductor-Based Sequencing for the Identification of Apis mellifera Complementary Sex Determiner (csd) Alleles from Honey DNA. Insects 2021, 12, 868. [Google Scholar] [CrossRef]
- Adams, J.; Rothman, E.D.; Kerr, W.E.; Paulino, Z.L. Estimation of the number of sex alleles and queen matings from diploid male frequencies in a population of Apis mellifera. Genetics 1977, 86, 583–596. [Google Scholar] [CrossRef]
- Cho, S.; Huang, Z.Y.; Green, D.R.; Smith, D.R.; Zhang, J. Evolution of the complementary sex-determination gene of honeybees: Balancing selection and trans-species polymorphisms. Genome Res. 2006, 2, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Hasselmann, M.; Vekemans, X.; Pflugfelder, J.; Koeniger, N.; Koeniger, G.; Tingek, S.; Beye, M. Evidence for convergent nucleotide evolution and high allelic turnover rates at the complementary sex determiner gene of western and Asian honeybees. Mol. Biol. Evol. 2008, 25, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Hyink, O.; Laas, F.; Dearden, P.K. Genetic tests for alleles of complementary-sex-determiner to support honeybee breeding programmes. Apidologie 2013, 44, 306–313. [Google Scholar] [CrossRef]
- Kaskinova, M.D.; Gataullin, A.R.; Saltykova, E.S.; Gaifullina, L.R.; Poskryakov, A.V.; Nikolenko, A.G. Polymorphism of the Hypervariable region of the csd Gene in the Apis mellifera L. Population in Southern Ural. Russ. J. Genet. 2019, 55, 267–270. [Google Scholar] [CrossRef]
- Kolics, É.; Parrag, T.; Házi, F.; Szepesi, K.; Heltai, B.; Mátyás, K.; Kutasy, B.; Virág, E.; Taller, J.; Orbán, L.; et al. An alternative, high throughput method to identify csd alleles of the honeybee. Insects 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Z.; Wu, X.; Yan, W.; Zeng, Z. Polymorphism analysis of csd gene in six Apis mellifera subspecies. Mol. Biol. Rep. 2012, 39, 3067–3071. [Google Scholar] [CrossRef] [PubMed]
- Zareba, J.; Blazej, P.; Laszkiewicz, A.; Sniezewski, L.; Majkowski, M.; Janik, S.; Cebrat, M. Uneven distribution of complementary sex determiner (csd) alleles in Apis mellifera population. Sci. Rep. 2017, 7, 2317. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, L.; Elsik, C. A scientific note defining allelic nomenclature standards for the highly diverse complementary sex-determiner (csd) locus in honey bees. Apidologie 2021, 52, 749–754. [Google Scholar] [CrossRef]
- Paolillo, G.; De Iorio, M.G.; Filipe, J.F.S.; Riva, F.; Stella, A.; Gandini, G.; Pagnacco, G.; Lazzari, B.; Minozzi, G. Analysis of complementary sex-determiner (csd) allele diversity in different honeybee subspecies from Italy based on NGS Data. Genes 2022, 13, 991. [Google Scholar] [CrossRef]
- Bienefeld, K.; Reinhardt, F.; Pirchner, F. Inbreeding effects of queen and workers on colony traits in the honeybee. Apidologie 1989, 20, 439–450. [Google Scholar] [CrossRef]
- Dickerson, G.E. Inbreeding and heterosis in animals. J. Anim. Sci. 1973, 1973, 54–77. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Meth. Mol. Biol. 2014, 1079, 105–116. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]


| Year | GL | N° |
|---|---|---|
| 2019 | L1Q | 3 |
| L2Q | 9 | |
| L3Q | 2 | |
| L4Q | 2 | |
| L5Q | 7 | |
| L6Q | 1 | |
| 2020 | L7Q | 6 |
| L8Q | 5 | |
| L9Q | 2 | |
| L10Q | 3 | |
| L11Q | 5 | |
| L12Q | 5 | |
| L13Q | 1 |
| ID Sample | Mother | GL | Sequences | ID Allele |
|---|---|---|---|---|
| L1AW | L1AQ | L1Q | KIISSLSNNYNYSNYNNNNNNYNNYKQLCYNINYIEQI | Allele 5 |
| KIISSLSNKTIHNNNNYKYNYNNNNYNNNYNNYKKLYYNINYIEQI | Allele 13 | |||
| L1BW | L1BQ | L1Q | KIISSLSNKTIHNNNNYKYNYNNNNYNNNYNNYKKLYYNINYIEQI | Allele 13 |
| KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 | |||
| L1CW | L1CQ | L1Q | KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 |
| KIISSLSNKTIHNNNNYKYNYNNNNYNNNNYNNNNYKKLQYYNINYIEQI | Allele 15 | |||
| L2AW | L2AQ | L2Q | KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 |
| KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 | |||
| L2BW | L2BQ | L2Q | KIISSLSNNYKYSNYNNYNNNYNNNYNNNYNNNYKKLYKNYIINIEQI | Allele 28 |
| KIISSLSNKTIHNNNNYKYNYNNNNYKNYNNYKKLYYNINYIEQI | Allele 18 | |||
| L2CW | L2CQ | L2Q | KIISSLSNKTIHNNNNYKNYNNYKKLYYNINYIEQI | Allele 33 |
| KIISSLSNNYKYSNYNNYNNYNKKLYYKNYIINIEQI | Allele 8 | |||
| L2DW | L2DQ | L2Q | KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 |
| KIISSLSNNYNYSNYNNYNNNNNYNNYKKLYYNINYIEQI | Allele 26 | |||
| L2EW | L2EQ | L2Q | QIISSLSNNYNYNYNKHNNKLYYNINYIEQI | Allele 31 |
| KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 | |||
| L2FW | L2FQ | L2Q | KIISSLSNKTIHNNNNYNNNNYNNYKKLYYNINYIEQI | Allele 14 |
| KIISSLSNNTIHNNNNYKYNYNNNYNNYNNKKLYYNINYIEQI | Allele 35 | |||
| L2GW | L2GQ | L2Q | KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 |
| KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 | |||
| L2HW | L2HQ | L2Q | KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 |
| KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 | |||
| L2IW | L2IQ | L2Q | KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 |
| KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 | |||
| L3AW | L3AQ | L3Q | KIISSLSNKTIHNNNNYNNNNYNNYKKLYYNINYIEQI | Allele 14 |
| KIISSLSNNTIHNNNYKYNYNNNYNNYKKLYYNINYIEQI | Allele 25 | |||
| L3BW | L3BQ | L3Q | KIISSLSNNTIHNNNYKYNYNNNNYNNNNYNKKLYYNIINIEQI | Allele 32 |
| KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 | |||
| L4AW | L4AQ | L4Q | KIISSLSNNYKYSNYNNYNNNNNYNNNYSKKLYYNIINIEQI | Allele 6 |
| KIISSLSNNYKYSNYNNYNNNYNNYKKLYKNYIINIEQI | Allele 27 | |||
| L4BW | L4BQ | L4Q | KITSSLSNSCNYSNNYNNNYNNTKKLYYNINYIEQI | Allele 30 |
| KIISSLSNKTIHNNNNYKYNYNNNNNNYKNYNNYKKLYYNINYIEQI | Allele 17 | |||
| L5AW | L5AQ | L5Q | KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 |
| KIISSLSNNYISNISNYNNNNNSKKLYYNINYIEQI | Allele 16 | |||
| L5BW | L5BQ | L5Q | KIISSLSNNYNYSNYNNNNNNYNNYKQLCYNINYIEQI | Allele 5 |
| KIISSLSNKTIHNNNNYKYNYNNNNNNYKNYNNYKKLYYNINYIEQI | Allele 17 | |||
| L5CW | L5CQ | L5Q | KIISSLSNKTIHNNNNYKYNYNNNNYNNNNYNNNNYKKLQYYNINYIEQI | Allele 15 |
| KIISSLSNKTIHNNNNYNNNNYNNYKKLYYNIINIEQI | Allele 19 | |||
| L5DW | L5DQ | L5Q | KIISSLSNNYNYSNYNNNNNNYNNYKQLCYNINYIEQI | Allele 5 |
| KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 | |||
| L5EW | L5EQ | L5Q | KIISSLSNNTIHNNNNYNKKLYYNIINIEQI | Allele 29 |
| KIISSLSNKTIHNNNNYKYNYNNNNYKNYNNYKKLYYNINYIEQI | Allele 18 | |||
| L5FW | L5FQ | L5Q | KIISSLSNNYNYSNYNNYNNNNYNNYKKLYYNINYIEQI | Allele 23 |
| KIISSLSNNYKYSNYNNYNNNYNNYNNNYKKLYYNINYIEQI | Allele 34 | |||
| L5GW | L5GQ | L5Q | KIISSLSNNYISNISNYNNNNNSKKLYYNINYIEQI | Allele 16 |
| KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 | |||
| L6AW | L6AQ | L6Q | KIISSLSNNYNYNNCNYKHNKLYYNIINIEQI | Allele 24 |
| KIISSLSNNYNYNNNNYNNYNNNYNNNYNKKLYYNIINIEQI | Allele 22 | |||
| L7AW | L7AQ | L7Q | KIISSLSNKTIHNNNNYKKLYYNINYIEQI | Allele 9 |
| KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 | |||
| L7BW | L7BQ | L7Q | KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNNNNYNKKLYYKNYIINIEQI | Allele 20 |
| KIISSLSKNTIHNNNYKYNYNNNNNYNNNYKKLQYYNINYIEQI | Allele 12 | |||
| L7CW | L7CQ | L7Q | KIISSLSNNYNSNSLLSYNNYNNNNNYNKLYYNINYIEQI | Allele 47 |
| KIISSLSKNTIHNNNYKYNYNNNNNYNNNYKKLQYYNINYIEQI | Allele 12 | |||
| L7DW | L7DQ | L7Q | KIISSLSKNTIHNNNYKYNYNNNNNYNNNYKKLQYYNINYIEQI | Allele 12 |
| KIISSLSNNYNYSNYNNNNNNYNNYKQLCYNINYIEQI | Allele 5 | |||
| L7EW | L7EQ | L7Q | KIISSLSNNTIHNNNYKYNYNNNYNNYNNYKKLYYNINYIEQI | Allele 7 |
| KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 | |||
| L7FW | L7FQ | L7Q | KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 |
| KIISSLSNKTIHNNNNYKNYNNYNNNNYKNYNNYKKLYYNIINIEQI | Allele 11 | |||
| L8AW | L8AQ | L8Q | KIISSLSNNTIHNNNYKYNYNNNYNNYNNYKKLYYNINYIEQI | Allele 7 |
| KIISSLSNNTIHNNNYKYNYNNNNYNKKLYYNIINIEQI | Allele 38 | |||
| L8BW | L8BQ | L8Q | KIISSLSNNYNYNNNYNNYNNNYNKKLYYNINYIEQI | Allele 39 |
| KIISSLSNKTIHNNNKYNYNKYNYNNNNYNNYKKLYYNINYIEQI | Allele 4 | |||
| L8CW | L8CQ | L8Q | KIISSLSNHYNYNNNKYNNYNNDYKKLYYNINYIEQI | Allele 41 |
| KIISSLSNNTIHNNNYKYNYNNNYNNYNNYKKLYYNINYIEQI | Allele 7 | |||
| L8DW | L8DQ | L8Q | KIISSLSNNYKYSNYNNYNNYNKKLYYKNYIINIEQI | Allele 8 |
| KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 | |||
| L8EW | L8EQ | L8Q | KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 |
| KIISSLSNNYKYSNYNNYNNYNKKLYYKNYIINIEQI | Allele 8 | |||
| L9AW | L9AQ | L9Q | KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 |
| KIISSLSNKTIHNNNNYKYNYNNNNYNNNNYNNNYNNNCKKLYYNIINIEQI | Allele 10 | |||
| L9BW | L9BQ | L9Q | KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNNNNYNKKLYYKNYIINIEQI | Allele 20 |
| KIISSLSNKTIHNNNNYKNYNNYNNNNYKNYNNYKKLYYNIINIEQI | Allele 11 | |||
| L10AW | L10AQ | L10Q | KIISSLSNNYSYNNYNNNNYNKKLYYNINYIEQI | Allele 44 |
| KIISSLSNNYNYNNYNNNYKPLYYNINYIEQI | Allele 21 | |||
| L10BW | L10BQ | L10Q | KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 |
| KIISSLSNNYNYNNYNNNYKPLYYNINYIEQI | Allele 21 | |||
| L10CW | L10CQ | L10Q | KIISSLSNKTIHNNNNYNNNNYNNYKKLYYNIINIEQI | Allele 19 |
| KIISSLSNNYKYSNYNNYNNNNNYNNNYSKKLYYNIINIEQI | Allele 6 | |||
| L11AW | L11AQ | L11Q | KIISSLSNNTIHNNNYKYNYNNNNYNNNYNKKLYYKNYIINIEQI | Allele 36 |
| KIISSLSNNYKYSNYNNYNNNYNNYNNNYNNNYKKLYYNINYIEQI | Allele 37 | |||
| L11BW | L11BQ | L11Q | KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 |
| KIISSLSNKTIHNNNNYNNNNNNYNNYNNYKKLYYNVINIEQI | Allele 43 | |||
| L11CW | L11CQ | L11Q | KIISSLSNNRNSNNYNNYNYKKLYYNINYIEQI | Allele 45 |
| KIISSLSNNTIHNNNNYKYNYNNNYNNYNNYNNKKLYYNINYIEQI | Allele 46 | |||
| L11DW | L11DQ | L11Q | KIISSLSNKTIHNNNNYKNYNYKKLYYNIINIEQI | Allele 1 |
| KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 | |||
| L11EW | L11EQ | L11Q | KIISSLSNNTIHNNNYKYNYNNNYNNYNNYKKLYYNINYIEQI | Allele 7 |
| KIISSLSNKTIHNNNNYKYNYNNNNYNNNNYNNNYNNNCKKLYYNIINIEQI | Allele 10 | |||
| L12AW | L12AQ | L12Q | KIISSLSNKTIHNNNNYKYNYNNNYNNNHYNNNYKKLQYYNIINIEQI | Allele 40 |
| KIISSLSNNYKYSNYNNYNNNNNYNNNYSKKLYYNIINIEQI | Allele 6 | |||
| L12BW | L12BQ | L12Q | KIISSLSNNYKYSNYNNYNNNNNYNNNYSKKLYYNIINIEQI | Allele 6 |
| KIISSLSNKTIHNNNNYKYNYNNNNYNNNNYNNYNNNCKKLYYNIINIEQI | Allele 42 | |||
| L12CW | L12CQ | L12Q | KIISSLSNKTIHNNNNYKNYNNYNNNNYKNYNNYKKLYYNIINIEQI | Allele 11 |
| KIISSLSNNYNYSNYNNNNNNYNNYKQLCYNINYIEQI | Allele 5 | |||
| L12DW | L12DQ | L12Q | KITSSLSNNYNSNNYNKYNYNNSKKLYYNINYIEQI | Allele 2 |
| KIISSLSNKTIHNNNNYKKLYYNINYIEQI | Allele 9 | |||
| L12EW | L12EQ | L12Q | KIISSLSNKTIHNNNNYKKLYYNINYIEQI | Allele 9 |
| KIISSLSNNYKYSNYNNYNNNNNYNNNYSKKLYYNIINIEQI | Allele 6 | |||
| L13AW | L13AQ | L13Q | KIISSLSNKTIHNNNNYKYNYNNKYNYNNNNYNKKLYYKNYIINIEQI | Allele 3 |
| KIISSLSNKTIHNNNNYKYNYNNNNYNNNNYNNNYNNNCKKLYYNIINIEQI | Allele 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Iorio, M.G.; Lazzari, B.; Colli, L.; Pagnacco, G.; Minozzi, G. Variability and Number of Circulating Complementary Sex Determiner (Csd) Alleles in a Breeding Population of Italian Honeybees under Controlled Mating. Genes 2024, 15, 652. https://doi.org/10.3390/genes15060652
De Iorio MG, Lazzari B, Colli L, Pagnacco G, Minozzi G. Variability and Number of Circulating Complementary Sex Determiner (Csd) Alleles in a Breeding Population of Italian Honeybees under Controlled Mating. Genes. 2024; 15(6):652. https://doi.org/10.3390/genes15060652
Chicago/Turabian StyleDe Iorio, Maria Grazia, Barbara Lazzari, Licia Colli, Giulio Pagnacco, and Giulietta Minozzi. 2024. "Variability and Number of Circulating Complementary Sex Determiner (Csd) Alleles in a Breeding Population of Italian Honeybees under Controlled Mating" Genes 15, no. 6: 652. https://doi.org/10.3390/genes15060652
APA StyleDe Iorio, M. G., Lazzari, B., Colli, L., Pagnacco, G., & Minozzi, G. (2024). Variability and Number of Circulating Complementary Sex Determiner (Csd) Alleles in a Breeding Population of Italian Honeybees under Controlled Mating. Genes, 15(6), 652. https://doi.org/10.3390/genes15060652






