Abstract
Alternanthera sessilis is considered the closest relative to the invasive weed Alternanthera philoxeroides in China, making it an important native species for studying the invasive mechanisms and adaptations of A. philoxeroides. Chloroplasts play a crucial role in a plant’s environmental adaptation, with their genomes being pivotal in the evolution and adaptation of both invasive and related species. However, the chloroplast genome of A. sessilis has remained unknown until now. In this study, we sequenced and assembled the complete chloroplast genome of A. sessilis using high-throughput sequencing. The A. sessilis chloroplast genome is 151,935 base pairs long, comprising two inverted repeat regions, a large single copy region, and a small single copy region. This chloroplast genome contains 128 genes, including 8 rRNA-coding genes, 37 tRNA-coding genes, 4 pseudogenes, and 83 protein-coding genes. When compared to the chloroplast genome of the invasive weed A. philoxeroides and other Amaranthaceae species, we observed significant variations in the ccsA, ycf1, and ycf2 regions in the A. sessilis chloroplast genome. Moreover, two genes, ccsA and accD, were found to be undergoing rapid evolution due to positive selection pressure. The phylogenetic trees were constructed for the Amaranthaceae family, estimating the time of independent species formation between A. philoxeroides and A. sessilis to be approximately 3.5186–8.8242 million years ago. These findings provide a foundation for understanding the population variation within invasive species among the Alternanthera genus.
1. Introduction
Alternanthera philoxeroides, commonly known as alligator weed, is a perennial herb within the Alternanthera genus of the Amaranthaceae family. It originates from the Paraguay and Parana River basins in South America. Over time, it has expanded its presence across a vast geographical range, spanning from 32 degrees north to 38 degrees south latitude, making it a globally pervasive invasive weed that poses significant threats to both the environment and the economy [1,2,3]. Alternanthera sessilis, another perennial herb belonging to the same Alternanthera genus as A. philoxeroides, is believed to be native to tropical and subtropical regions of Asia, northeastern Australia, and the wetlands of tropical America [3,4,5]. These two species share similarities in morphology, with upright or prostrate growth habits, and the ability of all stem nodes to develop roots. Their flowers are axillary in nature. While A. sessilis has sessile inflorescences, A. philoxeroides exhibits different characteristics. Their distribution areas overlap within China, with A. sessilis primarily found along the wet edges of East China, South China, Central China, and Southwest China. Conversely, A. philoxeroides is predominantly distributed in the broader regions to the south of the Yangtze River and gradually extends into sporadic areas in North China. It holds a larger territory and exhibits greater competitive strength compared to A. sessilis.
Invasive weeds have a remarkable capacity for rapid adaptation to new environments, making them excellent subjects for studying adaptive changes in plants [6,7,8]. One common approach is to compare the adaptability and invasiveness of alien invasive species with their local relatives. As a native species, A. sessilis has been frequently employed in studies of the adaptation and invasive mechanisms of A. philoxeroides, yielding valuable insights. In contrast to A. sessilis, A. philoxeroides demonstrates superior photosynthetic capacity, a faster stem growth rate, a broader temperature tolerance range, enhanced competitive abilities, and a greater capacity for invasion [9]. A. philoxeroides holds distinct advantages over A. sessilis, whether facing biotic or abiotic stressors [10]. Following exposure to herbivores and nematodes, A. philoxeroides displays increased branching, facilitating its expansion and invasion [11]. Its defense responses surpass those of A. sessilis, making it more resistant to the intrusion of pathogenic microorganisms [12]. Furthermore, A. philoxeroides exhibits a wider environmental adaptability range and more robust phenotypic plasticity. It thrives in various aquatic environments and demonstrates heightened tolerance to waterlogging [13]. Its osmotic adjustment capabilities exceed those of its native congener, A. sessilis [14,15]. Additionally, A. philoxeroides excels in clonal integration, enabling it to outcompete A. sessilis within its ecological niche [16]. These findings shed light on the invasion mechanism employed by A. philoxeroides to a considerable extent.
Chloroplasts are cellular organelles responsible for photosynthesis in plants and the provision of energy for growth. They also serve as vital hubs for plant signal integration, actively participating in adaptation to environmental stress [17]. Chloroplasts possess a circular genome consisting of four main sections: a large single copy region (LSC), a small single copy region (SSC), and two identical inverted regions (IRs) separated by two unique single copy regions. Typically, chloroplast genomes (cp genome) have a size ranging from 107 kb to 218 kb, containing approximately 120 to 130 genes. The number and arrangement of chloroplast genes (cp genes) exhibit a high degree of conservation, albeit with occasional insertions, deletions, and rearrangements. These attributes of high conservation and slow evolution of chloroplast genomes offer an effective means of distinguishing groups that are challenging to classify based on morphology [18,19]. Cp genes also prove effective in the identification of invasive plants, such as the combined utilization of matK and nucleic ITS [20].
The cp genome plays a crucial role in elucidating the relationships and evolutionary dynamics between invasive species and their congeners [21]. By comparing cp genomes across different regions or among related species, researchers can analyze origins, evolutionary pathways, and spread patterns. For example, this approach has been applied to examine invasive and native individuals of Jacobaea vulgaris [22], invasive Mikania micrantha and its native species M. cordata [23], and Sonchus asper and S. oleraceus [24]. This method has increasingly become a potent tool for plant molecular systematics, phytogeography, and the investigation of intraspecific polymorphism and interspecific divergence [21,25,26]. With advancements in next-generation sequencing technologies, there has been a growing interest in studying the cp genomes of invasive species within specific regions. Despite the sequencing and reporting of the cp genome of A. philoxeroides [27], research on the cp genome sequences of its native congener, A. sessilis, and their comparative analysis remains limited.
In this study, the complete cp genome of A. sessilis was sequenced and assembled. The analysis of hotspot regions and repeat sequences was carried out in comparison to the cp genome of the invasive weed, A. philoxeroides. Furthermore, highly divergent regions between A. sessilis and A. philoxeroides were identified. Using protein-coding genes, a phylogenetic tree of the Amaranthaceae family was constructed, and the divergence time between A. sessilis and A. philoxeroides was estimated.
2. Materials and Methods
2.1. Plants Collection
Alternanthera sessilis were gathered from a natural population in Meizhou City, Guangdong, China, and subsequently cultivated within a greenhouse at Shanxi Agriculture University (Taigu, China). Once they had bloomed, their seeds were harvested, dried, and stored in a refrigerator for future cultivation. These seeds were then sown in small black square pots measuring 7 cm × 7 cm and cultured under controlled conditions at a temperature of 25 ± 1 °C with a photoperiod of 16 h of light and 8 h of darkness for six weeks to obtain test plants. Fresh leaves were collected after exposure to more than 6 h of light and were promptly frozen using liquid nitrogen for subsequent DNA extraction.
2.2. DNA Extraction and Sequencing
The frozen leaves of A. sessilis, as described earlier, were pulverized into a fine powder using liquid nitrogen to facilitate total DNA extraction. Total DNA was extracted utilizing the Plant DNA Isolation Kit (Tiangen, Beijing, China). Subsequently, the total DNA was fragmented through ultrasound treatment. The resulting DNA fragments underwent purification, end repair, and adapter ligation. Following PCR enrichment, the fragments were separated through agarose gel electrophoresis and subsequently employed for constructing a DNA library. This library was subjected to sequencing on an Illumina NovaSeq platform (San Diego, CA, USA), generating paired-end reads with a length of 150 bases (PE150).
2.3. Assembly and Annotation of A. sessilis Chloroplast Genome
Raw sequencing data were subjected to filtration using fastp 0.20.0 (https://github.com/OpenGene/fastp, accessed on 4 December 2023), which entailed the removal of adapter sequences and reads with an average quality score falling below Q5 or containing more than five ambiguous bases (N) to obtain high-quality clean reads. To simplify the assembly process, these clean reads were aligned against the chloroplast genome database from Genepioneer Biotechnologies (Nanjing, China) using bowtie2 v2.2.4 (http://bowtiebio.sourceforge.net/bowtie2/index.shtml, accessed on 4 December 2023) to specifically identify sequencing reads corresponding to the chloroplast genome. These selected cp genome sequencing reads were subsequently assembled into contigs using SPAdes v3.10.1 (http://cab.spbu.ru/software/spades/, accessed on 4 December 2023). These contigs were further organized into scaffolds using SSPACE v2.0 (https://www.baseclear.com/services/bioinformatics/basetools/sspace-standard/, accessed on 4 December 2023). To obtain a complete, circular cp genome, any gaps between scaffolds were filled using Gapfiller v2.1.1 (https://sourceforge.net/projects/gapfiller/, accessed on 4 December 2023).
Annotation of the CDS within the A. sessilis cp genome was carried out using Prodigal v2.6.3 (https://www.github.com/hyattpd/Prodigal, accessed on 4 December 2023). Separately, the prediction of transfer RNA (tRNA) and ribosomal RNA (rRNA) was performed using Aragorn [28] v1.2.38 (http://130.235.244.92/ARAGORN/, accessed on 4 December 2023), tRNAscan-SE [29] (http://trna.ucsc.edu/tRN-Ascan-SE/, accessed on 4 December 2023), and Hmmer v3.1b2 (http://www.hmmer.org/, accessed on 4 December 2023). Additionally, sequence alignment and annotation were conducted based on the gene sequences of related species, and the assembled sequences were subjected to blast v2.6 (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 4 December 2023) for further annotation. The final annotation result was obtained after manually removing redundancy. The complete cp genome sequence of A. sessilis was deposited in the NCBI GenBank with the specific accession number PP239384.
2.4. Analysis Data Collection
The cp genome sequences of A. philoxeroides, which were used for comparative analysis, were retrieved from the NCBI GenBank (accession number: NC_042798.1). These samples were collected in Jinan, China.
2.5. Chloroplast Genome Structure
The analysis of inverted repeat regions (IRs) within the A. sessilis cp genome was conducted using GeSeq [30] (https://chlorobox.mpimp-golm.mpg.de/geseq.html, accessed on 18 January 2024). Verification of the large single copy region (LSC), small single copy region (SSC), and IRs was performed using Geneious 10.1 [31]. Visualization of the structure of the A. sessilis cp genome was achieved using OGDraw [32] (https://chlorobox.mpimpgolm.mpg.de/OGDraw.html, accessed on 18 January 2024).
2.6. Codon Usage Bias Analysis of A. sessilis and A. philoxeroides cp Genomes
Codon usage bias is a widespread phenomenon observed across various species and stages of life. This bias is considered a result of long-term evolution and is influenced by multiple factors, with directional mutation and neutral selection being primary contributors [33,34]. Relative synonymous codon usage (RSCU), a commonly employed parameter for studying codon usage bias, represents the ratio between actual and expected codon occurrences. It aids in the analysis of gene function and evolutionary patterns. An RSCU value of 1 signifies no codon bias, while values greater than 1 indicate a higher occurrence of a codon compared to other synonymous codons, and vice versa. The RSCU values for the cp genomes of A. sessilis and A. philoxeroides were calculated using the protein-coding genes from these cp genomes. This analysis was performed using codon usage analysis in MEGA 11.0 [35].
2.7. Repeat Sequence Analysis
Repetitive sequences, distributed widely throughout the genome, are believed to play a crucial role in gene recombination and rearrangement. The cp genome evolves at a relatively slow pace, with repetitive sequences in non-coding regions exhibiting a higher degree of variability. This characteristic facilitates the characterization of genetic variation at lower taxonomic levels and aids in addressing population genetic inquiries [25,36,37]. The identification of repeats in the cp genome holds great significance for the development of novel molecular markers. REPuter [38] (https://bibiserv.cebitec.unibieleeld.de/reputer/, accessed on 18 January 2024) was employed to detect various types of repeat sequences within the cp genomes of both A. sessilis and A. philoxeroides. For the analysis of simple tandem repeats, Tandem Repeats Finder [39] (http://tandem.bu.edu/trf/trf.html, accessed on 18 January 2024) was utilized. Simple sequence repeats (SSRs) were identified using MISA [40] (https://webblast.ipk-gatersleben.de/misa/, accessed on 18 January 2024). The SSRs were searched for mononucleotide to hexanucleotide repeat motifs with a minimum repeat number of 10, 5, 4, 3, 3, and 3 for mo, di, tri, tetra, penta, and hexanucleotide repeats, respectively. The compound SSR was identified when the length of a sequence between two SSRs to register was <100 bp.
2.8. Analysis of Hotspots and ka/ks and Identification of Highly Divergent Regions
The cp genome sequences of A. sessilis and A. philoxeroides were aligned using MAFFT 7.037 [41]. Nucleotide polymorphism (Pi) within the cp genomes of these species was analyzed using DnaSP 6.0 [42] to identify regions with high variability, employing a parameter of a 200 bp step size and 600 bp window length. Seventy-one common protein-coding genes were selected to assess the frequency of synonymous and non-synonymous substitution events using DnaSP 6.0, providing insights into evolutionary selection pressures.
2.9. Contraction and Expansion Analysis of IRs Boundaries
The contraction and expansion of IRs have a substantial impact on the size of the cp genome [26,43]. Irscope [44] (https://irscope.shinyapps.io/irapp/, accessed on 18 January 2024) was employed to analyze the contraction and expansion of IRs within the cp genomes of A. sessilis and A. philoxeroides.
2.10. Genome Analysis and Comparison with Other Amaranthaceae Species cp Genomes
Using the annotation of the A. sessilis cp genome as a reference, a comprehensive comparison was conducted with the cp genomes of other Amaranthaceae species using mVISTA [45] (http://genome.lbl.gov/vista/mvista/submit.shtml, accessed on 18 January 2024). This analysis aimed to assess the distinctions between their respective cp genomes.
2.11. Phylogenetic Analysis and Divergence Time Estimate
To construct the phylogenetic tree, we utilized 59 common protein-coding genes from a total of 28 species. This set included 25 species from the Amaranthaceae family and 3 outgroups consisting of 2 species from the Achatocarpaceae family and Dianthus caryophyllus. Sequence alignment was carried out using MAFFT 7.037 [41]. The maximum likelihood (ML) tree was constructed with MEGA 11.0 [35], employing the best model GTR + G + I and 1000 bootstrap replicates. ModelFinder [46] was utilized to determine the best-fit model for constructing Bayesian inference phylogenies. The Bayesian phylogenetic tree was generated using Mybayes 3.2.6 within Phyosuite 1.1.16 [47], employing the best-fit model GTR + F + I + G4 with 2 parallel runs and 2,000,000 generations. The initial 25% of the sampled data was discarded as burn-in.
Divergence times were estimated using the RelTime-ML method with the local molecular clock in MEGA 11 [35]. Calibration points for divergence times were derived from TimeTree [48] (http://timetree.org/, accessed on 23 January 2024), specifically, the divergence time of the Amaranthus genus and Chenopodium genus (24.5–73.8 MYA) and the Suaeda genus and Salicornia genus (12.1–39.7 MYA), based on data from 10 and 5 studies, respectively. The time tree was constructed using the maximum likelihood method and the GTR + G + I model.
3. Results
3.1. Sequencing, Assembly, and Annotation of A. sessilis cp Genome
High-quality clean reads, totaling 2.54 GB with a Q30 of 92.86%, were obtained and employed for the assembly of the complete cp genome of A. sessilis. The A. sessilis cp genome is 151,935 bp in length and follows the typical quadripartite structure, comprising two inverted repeat regions (IRs), a large single copy region (LSC), and a small single copy region (SSC). The LSC region spans 84,449 bp, while the SSC region is 17,298 bp long, with a pair of Irs, each covering 25,095 bp (Figure 1). The overall GC content of the A. sessilis cp genome is 36.3%, while the GC contents of LSC, SSC, and IRs are 33.3%, 29.8%, and 42.5%, respectively (Table 1). It is noteworthy that IRs exhibit the highest GC content, primarily due to the presence of high-GC-content rRNA genes. A comparison of the A. sessilis cp genome with previously reported cp genomes from the Amaranthaceae species revealed similarities with the A. philoxeroides cp genome (Table 1).
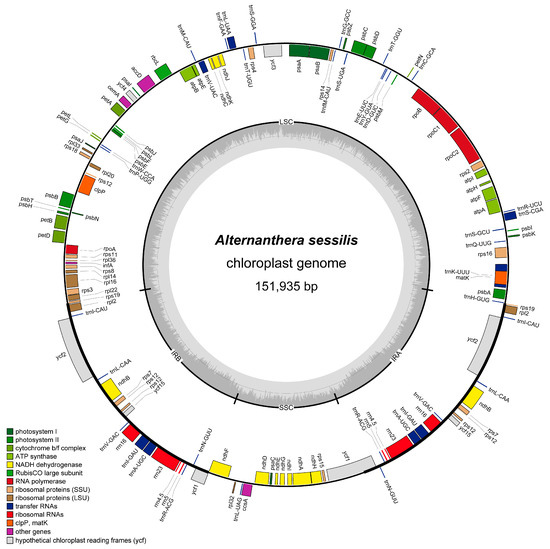
Figure 1.
Chloroplast genome map of Alternanthera sessilis. Genes coding forward are on the outer circle, while genes coding backward are on the inner circle. The gray circle inside represents the GC content.

Table 1.
Comparison of basic characteristics of chloroplast genomes in Amaranthaceae species.
The A. sessilis cp genome encodes a total of 128 genes, including 8 rRNA-coding genes, 37 tRNA-coding genes, 4 pseudogenes, and 83 protein-coding genes. Among the protein-coding genes, 44 are related to photosynthesis, 24 are involved in self-replication, and the remaining 10 have diverse functions (Table 2). In comparison to its close relative, the A. philoxeroides cp genome, A. sessilis possesses three additional tRNA-coding genes: trnG-GCC, trnS-CGA, and trnfM-CAU. Furthermore, the gene trnM-CAU is present in a single copy in the A. sessilis cp genome, whereas there are two copies in the A. philoxeroides cp genomes. Additionally, the A. sessilis cp genome contains two unique protein-coding genes, rpl22 and rps15, absent from the A. philoxeroides cp genome. Notably, the ndhA genes have different structures in the two species, with ndhA in A. sessilis having one intron, whereas that in A. philoxeroides is an all-exon structure. A. sessilis also harbors two specific pseudogenes, ycf15 and ycf1, which contain introns. Within the A. sessilis cp genome, eight tRNA-coding genes possess one intron. Among them, two copies of trnI-GAU and trnA-UGC are located in IRs, while the remaining four tRNA-coding genes are situated in the LSC region. Notably, trnK-UUU contains the largest intron, spanning 2538 bp, and encodes the matK gene. Eleven protein-coding genes in the A. sessilis cp genome contain introns, primarily associated with self-replication and photosynthesis. Nine of these genes possess one intron, predominantly situated in the LSC region, except for ndhB in IRs and ndhA in SSC. Two protein-coding genes, clpP, and ycf3, each exhibit two introns (Table 3).

Table 2.
Genes in the chloroplast genome of Alternanthera sessilis and Alternanthera philoxeroides.

Table 3.
Genes containing introns in the A. sessilis chloroplast genome.
3.2. Codon Usage Bias of A. sessilis and A. philoxeroides cp Genomes
The usage of synonymous codons in the cp genomes of A. sessilis was assessed using relative synonymous codon usage (RSCU) and compared with that of A. philoxeroides. In both genomes, Leu was found to have the highest amino acid frequency, accounting for 10.60% in A. sessilis and 10.37% in A. philoxeroides, while Cys exhibited the lowest frequency at 1.17% in A. sessilis and 1.68% in A. philoxeroides (Figure 2). Regarding start codons, in the A. sessilis cp genome, ACG was used as the start codon for psbL, while GTG was utilized for rps19, ndhD, and ycf1. In the A. philoxeroides cp genome, psbL and ndhD employed ACG as the start codon, while only rps19 used GTG. The RSCU values for stop codons UAA, UAG, and UGA in the A. sessilis cp genome were 1.63, 0.72, and 0.65, respectively. UAA was preferred as the primary stop codon in the A. sessilis cp genome. In contrast, a more balanced preference for stop codon usage was observed in the A. philoxeroides chloroplast genome, with RSCU values of 1.16 for UAA, 0.95 for UAG, and 0.89 for UGA (Table S1).
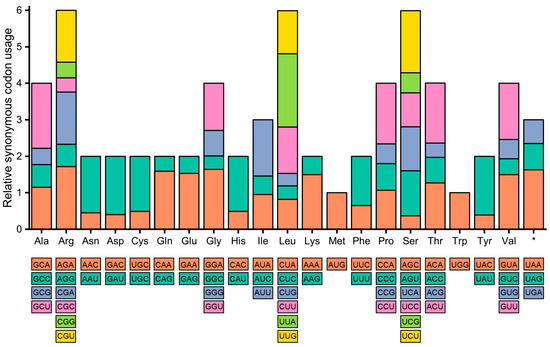
Figure 2.
Relative synonymous codon usage in the A. sessilis chloroplast genomes. *: Terminator.
3.3. SSRs and Long Repeated Sequences
In the cp genomes of both A. sessilis and A. philoxeroides, a total of 96 and 113 SSRs of four types were identified, respectively. Generally, A. philoxeroides exhibits a higher abundance of SSRs compared to A. sessilis. Based on the length of the repeating motifs, the A. sessilis cp genome contains 74 single-nucleotide repeat sequences, 10 dinucleotide repeats, 3 trinucleotide repeats, and 9 tetranucleotide repeats. In contrast, A. philoxeroides has an equal number of dinucleotide repeats but shows a higher occurrence of single-nucleotide, trinucleotide, and tetranucleotide repeats (Figure 3A). Regarding the type of repeating motif, in the A. sessilis cp genome, the most abundant is the T single-nucleotide repeats, followed by A single-nucleotide repeats. These two types of motif repeats account for 60.81% and 37.83%, respectively, out of all single-nucleotide repeats (Figure 3B). A similar distribution pattern was observed in the A. philoxeroides cp genome (Figure 3B).
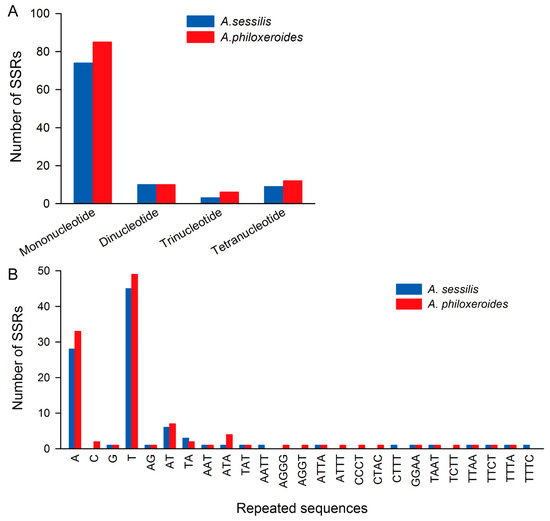
Figure 3.
Number of simple sequence repeats in A. sessilis and A. philoxeroides chloroplast genomes. (A) Number of simple sequence repeats of different types in A. sessilis and A. philoxeroides chloroplast genomes based on the repeating motif length. (B) Number of simple sequence repeats with different motif types in A. sessilis and A. philoxeroides chloroplast genomes.
Forty-nine and fifty repetitive sequences longer than 30 base pairs were identified in the cp genomes of A. sessilis and A. philoxeroides, respectively. These included 21 forward repeats and 28 palindrome repeats in the A. sessilis cp genome and 19 forward repeats, 29 palindrome repeats, and 2 reverse repeats in the A. philoxeroides cp genome (Figure 4A). The majority of these large repetitive sequences are located in the LSC and IR regions (Figure 4B). Repeats with lengths ranging from 30 to 40 base pairs account for 61.2% and 64% of the total repetitive sequences in the cp genomes of A. sessilis and A. philoxeroides, respectively (Figure 4C).
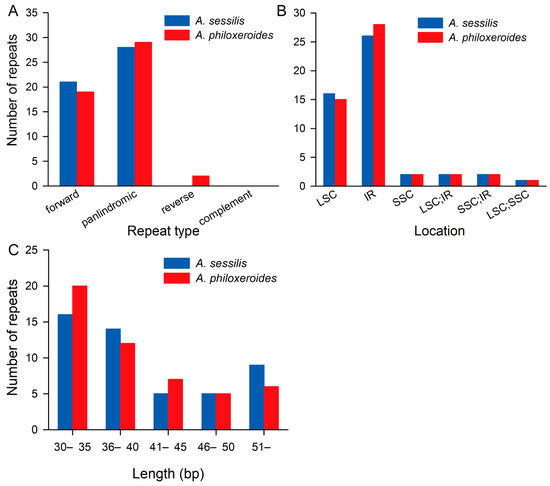
Figure 4.
Number of repetitive sequences in A. sessilis and A. philoxeroides chloroplast genomes. (A) Number of repetitive sequences of different types in A. sessilis and A. philoxeroides chloroplast genomes. (B) Number of repetitive sequences in different locations in A. sessilis and A. philoxeroides chloroplast genomes. (C) Number of repetitive sequences of different lengths in A. sessilis and A. philoxeroides chloroplast genomes.
3.4. Divergence Hotspots and Ka/Ks
Despite the structural similarity between the cp genomes of A. sessilis and A. philoxeroides, notable nucleotide differences exist. Nucleotide polymorphism (Pi) was used as an indicator to measure nucleic acid divergence, ranging from 0 to 0.0433, with an average value of 0.01159. Nine regions with high nucleotide polymorphisms were identified, including trnK-rps16, trnC-petN, petN-trnD, trnT, petL-petG, rps19-rpl2, ndhF-trnL, ccsA, and ycf1 (Pi > 0.033). These highly variable regions are primarily distributed in the LSC and SSC regions, while the IR region remains more conserved. Only two regions exhibit higher Pi values: ycf2-trnL in the IRa region and ycf2 in the IRb region, with Pi values of 0.02833 (Figure 5).
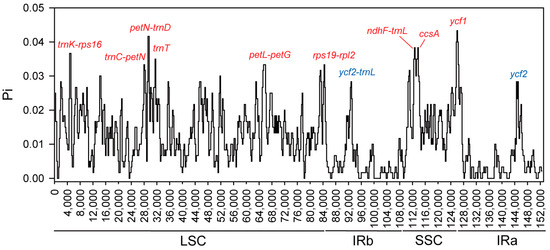
Figure 5.
The nucleotide polymorphism for cp genomes of A. sessilis and A. philoxeroides calculated using DnaSP 6.0 employing parameters of a 200 bp step size and 600 bp window length. Eleven most divergent regions are suggested as mutation hotspots. The name of regions in red indicate these regions are located in LSC region or SSC region, and those in blue indicate the regions are located in IRs.
Synonymous and non-synonymous substitution rates were analyzed for the 73 protein-coding genes shared by the cp genomes of A. sessilis and A. philoxeroides. The synonymous substitution rate ranged from 0 to 0.0789, with rps19 exhibiting the highest synonymous substitution rate. The non-synonymous substitution rate ranged from 0 to 0.0294, and infA displayed the highest non-synonymous substitution rate (Table S2).
The ratio of synonymous substitution rate to non-synonymous substitution rate (Ka/Ks) was further calculated to assess the selection pressure on 57 protein-encoding genes, with Ka/Ks ratios ranging from 0 to 3.475. Out of these, 55 genes had Ka/Ks ratios below 1, indicating a bias toward purification selection. Notably, 16 genes, including atpF and psbA, exhibited a Ka/Ks ratio of 0, suggesting that they are under strong purification selection pressure. However, two genes, ccsA and accD, displayed Ka/Ks ratios above 1, specifically, 1.237 and 3.475, respectively. This suggests that these two genes, especially accD, are rapidly evolving under positive selection influence and may play a crucial role in the evolution of the species. For the remaining 16 genes, the Ka/Ks ratio could not be calculated due to Ks = 0 (Figure 6).
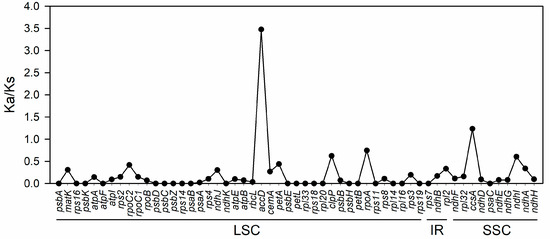
Figure 6.
The Ka/Ks ratio of 55 protein-coding genes in A. sessilis and A. philoxeroides calculated using DnaSP 6.0. Ka/Ks values of AccD and CcsA were more than 1.
3.5. Comparison of Chloroplast Genomes in the Amaranthaceae Family
Among ten species in the Amaranthaceae sensu stricto, their cp genomes exhibit remarkable structural conservation. However, noticeable variations in cp genome size are attributed to the contraction and expansion of the IR boundaries (Figure 7). The IR regions in these cp genomes vary in size, ranging from 24,346 to 25,539 base pairs (bp) (Table 1). Regarding the LSC/IRb boundary, all Amaranthaceae species, except for A. philoxeroides, harbor the rpl22 exclusively within the LSC region, devoid of any cross-boundary coding. With the exception of Amaranthus hupochondriacus and Celosia cristata, the remaining eight species feature the rps19 proximal to rpl22, extending into the IRb regions with segments spanning 72 to 223 bp, overlapping the LSC/IRb boundary. In A. sessilis and A. philoxeroides, the rps19 copy predominantly resides in the LSC region, with minor portions extending into the IRb region (87 bp and 72 bp, respectively). Furthermore, an additional rps19 copy is found solely within the IRa regions of A. sessilis, A. philoxeroides, and A. tricolor. At the SSC/IRb boundary, a pseudogene of ycf1 is present in A. sessilis, Amaranthus tricolor, Amaranthus caudatus, and Amaranthus hybridus, primarily located in IRbs, extending into the SSC region by 10–15 bp. The premature termination of ORF was observed in the aforementioned pseudogene ycf1 as a result of the contraction and expansion of the IR boundaries. Conversely, ndhF genes are primarily situated in the SSC regions, overlapping the SSC/IRb boundary and containing segments approximately 1–34 bp within the IRbs. Moving to the SSC/IRa boundary, ycf1 genes in these Amaranthaceae species primarily inhabit the SSC regions, extending 1387–1778 bp into the IRa regions. Notably, A. hypochondriacus and C. cristata lack ycf1. Concerning the LSC/IRa boundary, the trnH genes in the cp genomes of C. argentea and C. acpitata are predominantly found in the IRb region, while, in the other species, they are mainly located in the LSC region, with a distance ranging from 1 to 25 bp from the boundary (Figure 7).

Figure 7.
Comparison of the border positions of the LSC, IR, and SSC regions among ten Amaranthaceae species chloroplast genomes. Gene names are indicated in the boxes and their lengths in the corresponding regions are displayed above the boxes.
Using the A. sessilis cp genome sequence as a reference, six known cp genome sequences from five related genera were aligned. The results revealed a high degree of similarity among these sequences, with most regions displaying over 50 percent similarity. Considering the chloroplast structure, relatively high similarity was noted in the IR regions, while lower levels of similarity were observed in the LSC and SSC regions. From the perspective of gene structure, extremely high sequence similarity was found in both exon and UTR regions, except for ccsA, ycf1, and ycf2, which exhibited relatively high diversities. The non-coding region displayed low similarity and significant variation, suggesting its potential as a hotspot for the development of new molecular markers. The intergenic regions, trnK-rps16, petN-trnD, petL-petG, and ndhF-trnL, showed large diversity, consistent with the results of single nucleotide polymorphism analysis. Furthermore, significant diversity was observed in the gene introns, such as those within petD, rpl16, and ndhA. In our focused study on the cp genome of A. sessilis and A. philoxeroides, two regions of low similarity were identified between ndhB and ycf2, adjacent to trnL-CAA (Figure 8).
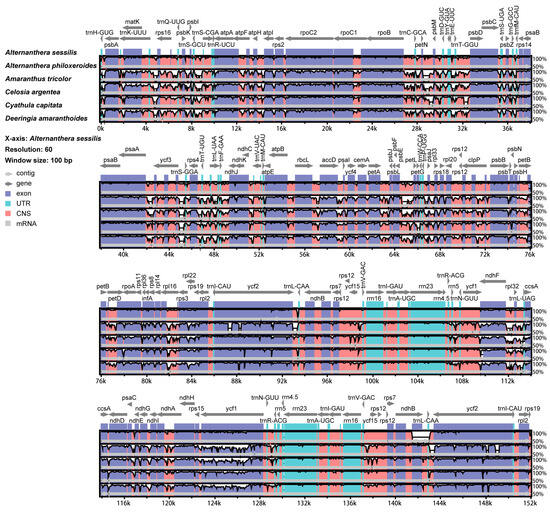
Figure 8.
Similarity of chloroplast genome sequences among six Amaranthaceae species from different genera. Sequence identity is portrayed with a cut-off of 50% identity. The Y-scale axis represents the percent identity within 50–100%. Grey arrows indicate genes with their orientation and position. Genome regions are color-coded as purple blocks for the conserved coding genes (exon), light red blocks for the conserved non-coding sequences in intergenic regions (CNS), and aqua blue blocks for UTR. The lines below the alignment indicate the chloroplast genomes. Black-bordered white peaks that are shown in genome regions indicate the divergent regions with sequence variation among six Amaranthaceae species.
3.6. Phylogenetic Analysis and Estimation of Divergence Time
For the construction of phylogenetic trees, we utilized fifty-nine chloroplast protein-coding genes from 25 species within Amaranthaceae s.l as the inner group, while Achatocarpus nigricans, Achatocarpus pubescens, and Dianthus caryophyllus were employed as the outgroups. The phylogenetic trees were generated using both the maximum likelihood method and the Bayesian inference method independently. Remarkably, both methods yielded similar tree topologies (Figure 9 and Figure 10). In these trees, A. sessilis and A. philoxeroides clustered together with the Cyathula species, forming one branch, while the genera Amaranthus, Celosia, and Deeringias grouped into another branch. These genera are part of Amaranthaceae s.s and formed a single branch collectively. Species belonging to Chenopodium, Beta, Haloxylon, Salicornia, and Suaeda genera constituted a distinct branch. Notably, the Chenopodiaceae branch and the Amaranthaceae s.s branch together formed a monophyletic group.
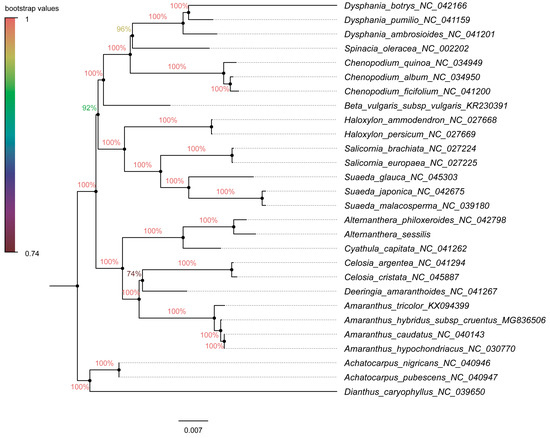
Figure 9.
The phylogenetic tree constructed using the maximum likelihood method in MEGA 11.0 with the GTR + G + I model, employing 59 chloroplast gene sequences from 28 species. The chloroplast genome sequences of Achatocarpus nigricans, Achatocarpus pubescens, and Dianthus caryophyllus were used as the outgroup. The numbers on the branches indicate the bootstrap values.
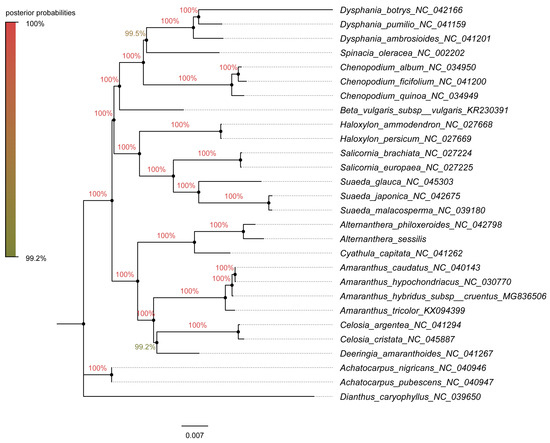
Figure 10.
The phylogenetic tree constructed using the Bayesian inference method in Mybayes 3.2.6 under the GTR + I + G + F model, incorporating 59 chloroplast gene sequences from 28 species. The chloroplast genome sequences of A. nigricans, A. pubescens, and D. caryophyllus were employed as the outgroup. The numbers on the branches indicate the posterior probabilities.
Additionally, a time tree was constructed using the aforementioned phylogenetic trees and analyzed to estimate divergence times. The results indicated that Amaranthaceae s.s. and Chenopodiaceae likely originated during the Paleogene period, approximately 47.24 MYA. The divergence between the Alternanthera genus and the Cyathula genus occurred around 20.04 MYA. The independent speciation events for A. philoxeroides and A. sessilis took place roughly between 3.5186 and 8.8242 MYA (Figure 11).
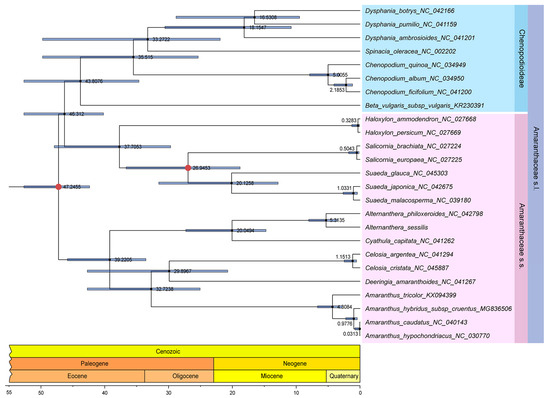
Figure 11.
The time tree estimated under the local molecular clock using the RelTime-ML method in MEGA 11.0. The circular representation with full red indicates the divergence times of the Amaranthus and Chenopodium genus (estimated at 24.5–73.8 MYA) as well as the Suaeda and Salicornia genus (estimated at 12.1–39.7 MYA), derived from 10 studies and 5 studies, respectively, in TimeTree, used for estimating the divergence time between A. sessilis and A. philoxeroides.
4. Discussion
In this study, we conducted sequencing and annotation of the chloroplast genome of A. sessilis. A comparative analysis of cp genomes was performed between relative local species A. sessilis and invasive weed A. philoxeroides. This study aimed to identify differences in the cp genomes of these two Alternanthera species, particularly focusing on hotspot regions and repeat sequences. We also investigated the accD and ccsA genes, which appear to be evolving rapidly due to positive selection. The divergence time between A. sessilis and A. philoxeroides was estimated.
Overall, the cp genomes of both A. sessilis and A. philoxeroides have a size of approximately 150 kb, with GC percentages ranging from 36.3% to 36.8% (Figure 1 and Table 1). These cp genomes exhibit the typical tetrad structure observed in other Amaranthaceae species. Changes in cp genome sizes are typically attributed to variations in the inverted repeat (IR) region, gene loss, and alterations in gene spacer regions [49,50,51]. Our results reveal that various regions within the cp genomes of Amaranthaceae s.s. species have undergone length changes, ranging from a few dozen to thousands of nucleotides. Notably, there is a size variation of approximately 1 kb within the IR regions. To the best of our knowledge, A. sessilis and A. philoxeroides possess medium-sized IR regions. Among Amaranthaceae s.s. species, the smallest IRs are found in A. tricolor, measuring 24,346 bp, while the largest IRs are observed in the cp genome of C. capitata, with a length of 25,539 bp.
When comparing the genes and gene numbers in the cp genomes of A. sessilis and A. philoxeroides with those of other Amaranthaceae species, it is evident that the protein-coding genes in A. sessilis and A. philoxeroides are similar to those found in other Amaranthaceae species. However, A. hypochondriacus and C. cristata exhibit a lower number of protein-coding genes. Interestingly, although both A. sessilis and A. philoxeroides have the same number of protein-coding genes, their genes are different. In the cp genome of A. sessilis, genes such as rpl22 and rps15 are present, while rpl23 is absent. Conversely, the cp genome of A. philoxeroides (NC_042798), collected from Jinan, exhibits the opposite situation (Table 2). Another A. philoxeroides cp genome from Shiyan (MK450441) [27], which contains rpl22 and rps15 but lacks rpl23, is similar to A. sessilis. In general, the loss of plastid genes can be attributed to two main reasons. Firstly, non-essential gene loss results in permanent absence. Secondly, the gene may transfer from the plastid genome to the nuclear genome [52,53]. The loss of rpl23 in the cp genomes of A. sessilis is a common occurrence in Amaranthaceae s.l. Eleven species have demonstrated the loss of rpl23, while other species have shown pseudogenization of rpl23 [54]. It is suggested that both rpl22 and rpl23 are essential for leaf development, and their loss can lead to leaf deformities [55]. In Leguminosae, rpl22 transfers to the nucleus before being lost from the cp genome [56]. This suggests that there may be nuclear transfer of rpl23/rpl22 in the cp genomes of A. sessilis and A. philoxeroides, resulting in the loss of rpl23 or rpl22, which requires further investigation. Furthermore, the absence of rps15, although not essential for plastid translation, can reduce the expression efficiency of chloroplast genes, decrease accumulation levels in photosynthesis complexes, affect ribosomal small subunit specificity, and impact leaf pigments, consequently retarding plant growth and development. Cold stress can exacerbate these negative effects [57]. While the S15 protein encoded by the nucleus can be transported into plastids, it does not fully compensate for the loss of plastid-encoded rps15 [55]. In the cp genomes of A. philoxeroides, the sample from Jinan lacks rps15, whereas the sample from Shiyan possesses it, suggesting that invasive weed A. philoxeroides in northern regions may have other cold adaptation mechanisms to mitigate the negative effects caused by the loss of rps15. All these differences between the two A. philoxeroides cp genomes may be attributed to their different geographical locations, highlighting the potential impact of latitude on population variations in alien species like A. philoxeroides, which needs more evidence.
The boundaries of the IR region in A. sessilis and the other nine Amaranthaceae cp genomes were highly conserved, with minimal variation. This conservation aligns with the typical pattern of IR region boundaries in most angiosperm species [58]. The expansion and contraction of the IR region can lead to the pseudogenization of genes [59]. In our study, we observed that pseudogenes ψrps19 and ψycf1 were located at the IR boundary in the chloroplast genome of A. sessilis (Figure 7), similar to findings for Mikania micrantha and its native congener, Mikania cordata [23]. Specifically, ψrps19 spanned the JLA (LSC/IRb) boundary within Amaranthaceae s.s. Compared to the normal gene, we observed an 83 bp reduction in length at its 3′-end. ψycf1 crossed the JSB (SSC/IRb) boundary and exhibited base deletions, causing a forward shift in its stop codon position and resulting in a size reduction to 1483 bp. Generally, such genes may be transferred to the nuclear genome [60,61]. However, it is worth noting that additional intact copies of rps19 and ycf1 were also present in the chloroplast genome of A. sessilis, raising the possibility of nuclear-transferred genes, which requires further investigation. Additionally, we found two copies of pseudogene ψycf15 in the IR region, both with sizes of 519 bp and incomplete ORFs due to base deletions. In chloroplast genomes, such as those observed in ANA-grade species, monocots, most rosids, etc., ycf15 genes are fragmented or completely lost due to nuclear gene transfers within the same plant lineages [61,62] or horizontal transfers from different plant lineages [63].
The nucleotide polymorphism in the chloroplast genomes of A. sessilis and the related species A. philoxeroides was notably high, and significant nucleic acid divergence regions were observed (Figure 5). These divergence regions were primarily situated in single-copy regions and could serve as hotspots for designing DNA barcodes for species identification. They offer potential molecular markers for investigating genetic variation in invasive plants within the Amaranthaceae family. Notably, regions like trnK-rps16 and trnC-petN, characterized by substantial nucleic acid divergence, have been successfully used for identifying traditionally challenging classified species such as oaks [64] and the Fragaria genus [19]. Furthermore, these regions can contribute to the phylogenetic analysis of related species, as observed in wild grapes [18]. Additionally, large nucleic acid divergences were also identified in the petL-petG, rps19-rpl2, ndhF-trnL, ccsA, and ycf1 regions. Genes like petN, petL, and petG encode subunits of cytochrome b6f, which play a crucial role in photosynthetic electron transport [65], suggesting that these divergences may be related to adaptations to varying light conditions. Furthermore, our study revealed that in the cp genomes of these two Alternanthera species, accD and ccsA are under positive selection pressure (Figure 6). The accD encodes the beta subunit of the enzyme acetyl-CoA carboxylase, which supplies malonyl-CoA primarily for fatty acid synthesis [66], playing an important role in leaf development [67]. This positive selection of accD was also observed in the chloroplast genome of Firmiana [68]. CcsA encodes cytochrome c biogenesis protein, which mediates the attachment of heme to c-type cytochromes [69]. It has been shown that ccsA was under positive selection in the epiphytic orchid [70] and Erigeron [71]. Both of them have been found to contribute to adaption to environmental changes in other species, for example, Lilium Ledebourii and the species in Malvaceae subfamilies [72,73].
Amaranthaceae s.l. is ranked as the second-largest family within the core Caryophyllales, with a sister group relationship to Achatocarpaceae indicated by molecular phylogenetic evidence [74]. Within this family, Amaranthaceae s.s. forms a clearly monophyletic group. In this study, phylogenetic trees were constructed using 59 chloroplast protein-coding genes across 28 species, employing both the maximum likelihood and Bayesian inference methods. Regardless of the methods and models used, the results were highly consistent, displaying similar topological structures (Figure 9 and Figure 10), which aligned with Yao’s plastid phylogenomic report of Caryophyllales [54]. The phylogenetic relationships within the Alternanthera genus were supplemented in our study, a detail not covered in the previous tree. Notably, a strong affinity was observed between the Alternanthera genus, comprising A. sessilis and A. philoxeroides, and the Cyathula genus. The incorporation of a broader range of chloroplast protein-coding genes from various genera within Amaranthaceae s.l. in our phylogenetic analysis provided a clearer understanding of the evolutionary and ancestral relationships within this family compared to previous studies. Although previous research had separately examined the phylogenetic relationships within the Alternanthera genus in the Americas using combined sequences from the cp genome (trnL-F and rpl16) and nuclear sequences (ITS sequences) [75], it did not elucidate the relationship between A. philoxeroides and A. sessilis. The use of complete cp genome sequences to explain their evolution was advantageous for closely related taxa and offered higher resolution [26]. It is important to note, however, that complete cp genome sequences for Amaranthaceae species remain limited. Further research is needed to explore the evolution and ancestral relationships at the subfamily and genus levels within Amaranthaceae.
Subsequently, the divergence time between A. philoxeroides and A. sessilis was estimated to have occurred approximately 3.5186–8.0242 million years ago, during the transition from the Miocene to the Quaternary period (Figure 11). This timeframe coincided with the wide proliferation of C4 plants and a shift in climate toward an ice age. The Alternanthera genus comprises species with various photosynthetic pathways, including C3, C4, and C3-C4 intermediate species [75], with A. sessilis being classified as a C3 species [76]. Our results indicated that the rapidly evolving genes in these two Alternanthera species were associated with photoadaptation and environmental adaptation, potentially contributing to their invasive capabilities.
5. Conclusions
In this study, the complete chloroplast genome of A. sessilis was sequenced, assembled, and compared with its relative species. This cp genome is 151,935 base pairs long with a typical quadripartite structure and contains 128 genes, including 8 rRNA-coding genes, 37 tRNA-coding genes, 4 pseudogenes, and 83 protein-coding genes. This chloroplast genome exhibited high conservation in structure and gene contents with other relative species; however, some regions showed signification variations and rapidly evolving genes involved in photosynthesis and environmental adaption were identified. The phylogenetic trees indicated that the Alternanthera genus is closely related to the Cyathula genus, and A. philoxeroides and A. sessilis were estimated to have diverged approximately 3.5186–8.8242 million years ago. Our findings lay the foundation for understanding the population variation and environmental adaptability within invasive species among the Alternanthera genus.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes15050544/s1, Table S1: Relative synonymous codon usage of cp genomes in A. sessilis and A. philoxeroides; Table S2: Synonymous and non-synonymous substitution rates and Ka/Ks ratio of 55 protein-coding genes in A. sessilis and A. philoxeroides.
Author Contributions
Conceptualization and methodology, R.M. and D.J.; performing the experiments: Y.W. and Q.C.; software: Y.W., X.Z. and J.H.; validation, X.Z., Q.C. and D.J.; formal analysis, Y.W., J.Y. and D.J.; writing—original draft preparation: Y.W.; writing—review and editing, D.J. and R.M.; project administration and funding acquisition, R.M. and D.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the special fund for Science and Technology Innovation Teams of Shanxi Province (no. 202304051001006); Research Program Sponsored by State Key Laboratory of Sustainable Dryland Agriculture (in preparation), Shanxi Agricultural University (no. 202003-4); Excellent Doctor Introduction Award Program of Shanxi Province (no. SXBYKY2022047); Fundamental Research Program of Shanxi Province (no. 20210302123386); Research Program Sponsored by Ministerial and Provincial Co-Innovation Centre for Endemic Crops Production with High-quality and Efficiency in Loess Plateau, Taigu 030801, China (no. SBGJXTZX-26); and Cultivation and Innovation Program for Scientific research, College of Plant Protection, Shanxi Agricultural University (no. ZBXY23A-10).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete chloroplast genome sequence of Alternanthera sessilis is publicly available online in the NCBI GenBank with the specific accession number PP239384.
Acknowledgments
We thank the assistance of everyone in the Biosafety and Biocontrol Laboratory, Shanxi Agricultural University, Shanxi, China, for their generous help.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Julien, M.H.; Skarratt, B.; Maywald, G. Potential geographical distribution of alligator weed and its biological control by Agasicles hygrophila. J. Aquat. Plant Mang. 1995, 33, 55–60. [Google Scholar]
- Coulson, J.R. Biological Control of Alligatorweed, 1959–1972: A Review and Evaluation; US Department of Agriculture, Agricultural Research Service: Washington, DC, USA, 1977. [Google Scholar]
- Pan, X.-Y. Invasive Alternanthera philoxeroides: Biology, ecology and management. Acta Phytotaxon. Sin. 2007, 45, 884–900. [Google Scholar] [CrossRef]
- Global Invasive Species Database (2024) Species Profile: Alternanthera sessilis. Available online: http://www.iucngisd.org/gisd/speciesname/Alternanthera+sessilis (accessed on 22 April 2024).
- Fan, S.; Yu, D.; Liu, C. The invasive plant Alternanthera philoxeroides was suppressed more intensively than its native congener by a native generalist: Implications for the biotic resistance hypothesis. PLoS ONE 2013, 8, e83619. [Google Scholar] [CrossRef]
- Baucom, R.S.; Holt, J.S. Weeds of agricultural importance: Bridging the gap between evolutionary ecology and crop and weed science. New Phytol. 2009, 184, 741–743. [Google Scholar] [CrossRef]
- Neve, P.; Barney, J.N.; Buckley, Y.; Cousens, R.D.; Graham, S.; Jordan, N.R.; Lawton-Rauh, A.; Liebman, M.; Mesgaran, M.B.; Schut, M.; et al. Reviewing research priorities in weed ecology, evolution and management: A horizon scan. Weed Res. 2018, 58, 250–258. [Google Scholar] [CrossRef]
- Vigueira, C.C.; Olsen, K.M.; Caicedo, A.L. The red queen in the corn: Agricultural weeds as models of rapid adaptive evolution. Heredity 2013, 110, 303–311. [Google Scholar] [CrossRef]
- Chu, S.; Cong, S.; Li, R.; Hou, Y. Host range of Herpetogramma basalis (Lepidoptera: Crambidae), a biological control agent for the invasive weed Alternanthera philoxeroides (Centrospermae: Amaranthaceae) in China. J. Insect Sci. 2019, 19, 1–7. [Google Scholar] [CrossRef]
- Sun, Y.; Ding, J.; Frye, M.J. Effects of resource availability on tolerance of herbivory in the invasive Alternanthera philoxeroides and the native Alternanthera sessilis. Weed Res. 2010, 50, 527–536. [Google Scholar] [CrossRef]
- Qin, H.; Guo, W.; Li, X. Density-dependent interactions between the nematode Meloidogyne incognita and the biological control agent Agasicles hygrophila on invasive Alternanthera philoxeroides and its native congener Alternantera sessilis. BioControl 2021, 66, 837–848. [Google Scholar] [CrossRef]
- Manoharan, B.; Qi, S.S.; Dhandapani, V.; Chen, Q.; Rutherford, S.; Wan, J.S.; Jegadeesan, S.; Yang, H.Y.; Li, Q.; Li, J.; et al. Gene expression profiling reveals enhanced defense responses in an invasive weed compared to its native congener during pathogenesis. Int. J. Mol. Sci. 2019, 20, 4916. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Y.; Yin, T.F.; Liu, C.X.; Luo, F.L. The invasive wetland plant Alternanthera philoxeroides shows a higher tolerance to waterlogging than its native Congener Alternanthera sessilis. PLoS ONE 2013, 8, e81456. [Google Scholar] [CrossRef]
- Wang, T.; Hu, J.; Miao, L.; Yu, D.; Liu, C. The invasive stoloniferous clonal plant Alternanthera philoxeroides outperforms its co-occurring non-invasive functional counterparts in heterogeneous soil environments—Invasion implications. Sci. Rep. 2016, 6, 38036. [Google Scholar] [CrossRef] [PubMed]
- Gao, L. Comparisons of morphological variation and cellular osmotic potential adjustment between invasive species Alternanthera philoxeroides and its native congener A. sessilis under different water treatments. Plant Sci. 2015, 33, 195–202. [Google Scholar] [CrossRef]
- You, W.; Li, N.; Zhang, J.; Song, A.; Du, D. The plant invader Alternanthera philoxeroides benefits from clonal integration more than its native co-genus in response to patch contrast. Plants 2023, 12, 2371. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, J. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci. 2018, 19, 3900. [Google Scholar] [CrossRef] [PubMed]
- Zecca, G.; Abbott, J.R.; Sun, W.B.; Spada, A.; Sala, F.; Grassi, F. The timing and the mode of evolution of wild grapes (Vitis). Mol. Phylogenet Evol. 2012, 62, 736–747. [Google Scholar] [CrossRef]
- Li, C.; Cai, C.; Tao, Y.; Sun, Z.; Jiang, M.; Chen, L.; Li, J. Variation and evolution of the whole chloroplast genomes of Fragaria spp. (Rosaceae). Front. Plant Sci. 2021, 12, 754209. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.Z.; Li, Z.Y.; Jin, X.H. DNA barcoding of invasive plants in China: A resource for identifying invasive plants. Mol. Ecol. Resour. 2018, 18, 128–136. [Google Scholar] [CrossRef]
- Viljoen, E.; Odeny, D.A.; Coetzee, M.P.A.; Berger, D.K.; Rees, D.J.G. Application of chloroplast phylogenomics to resolve species relationships within the plant genus Amaranthus. J. Mol. Evol. 2018, 86, 216–239. [Google Scholar] [CrossRef]
- Doorduin, L.; Gravendeel, B.; Lammers, Y.; Ariyurek, Y.; Chin, A.W.T.; Vrieling, K. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011, 18, 93–105. [Google Scholar] [CrossRef]
- Su, Y.; Huang, L.; Wang, Z.; Wang, T. Comparative chloroplast genomics between the invasive weed Mikania micrantha and its indigenous congener Mikania cordata: Structure variation, identification of highly divergent regions, divergence time estimation, and phylogenetic analysis. Mol. Phylogenet. Evol. 2018, 126, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.S.; Kim, J.H.; Kim, C.S.; Mejias, J.A.; Kim, S.C. Sow thistle chloroplast genomes: Insights into the plastome evolution and relationship of two weedy species, Sonchus asper and Sonchus oleraceus (Asteraceae). Genes 2019, 10, 881. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.-Y.; Xiang, G.-H.; Luo, Y.-C. The complete chloroplast genome of the invasive alligator weed Alternanthera philoxeroides (Caryophyllales: Amaranthaceae). Mitochondrial DNA Part B 2019, 4, 1345–1346. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Subramanian, S. Nearly neutrality and the evolution of codon usage bias in eukaryotic genomes. Genetics 2008, 178, 2429–2432. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, W.; Xing, T.; Zhao, M.; Li, N.; Yan, L.; Xia, G.; Wang, M. Synonymous codon usage bias in the plastid genome is unrelated to gene structure and shows evolutionary heterogeneity. Evol. Bioinform. 2015, 11, 65–77. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Nugent, J.M.; Herbon, L.A. Unusual structure of geranium chloroplast DNA: A triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. USA 1987, 84, 769–773. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Morden, C.W.; Palmer, J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 1992, 89, 10648–10652. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-M.; Wang, J.; Feng, L.; Liu, S.; Pang, H.; Qi, L.; Li, J.; Sun, Y.; Qiao, W.; Zhang, L.; et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017, 7, 1555. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Bock, R.; Timmis, J.N. Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays 2008, 30, 556–566. [Google Scholar] [CrossRef]
- Yao, G.; Jin, J.J.; Li, H.T.; Yang, J.B.; Mandala, V.S.; Croley, M.; Mostow, R.; Douglas, N.A.; Chase, M.W.; Christenhusz, M.J.M.; et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenet. Evol. 2019, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, T.T.; Scharff, L.B.; Alkatib, S.; Hasdorf, S.; Schottler, M.A.; Bock, R. Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 2011, 23, 3137–3155. [Google Scholar] [CrossRef] [PubMed]
- Gantt, J.S.; Baldauf, S.L.; Calie, P.J.; Weeden, N.F.; Palmer, J.D. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991, 10, 3073–3078. [Google Scholar] [CrossRef] [PubMed]
- Ehrnthaler, M.; Scharff, L.B.; Fleischmann, T.T.; Hasse, C.; Ruf, S.; Bock, R. Synthetic lethality in the tobacco plastid ribosome and its rescue at elevated growth temperatures. Plant Cell 2014, 26, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Goulding, S.E.; Olmstead, R.G.; Morden, C.W.; Wolfe, K.H. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 1996, 252, 195–206. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.B.; Wang, H.; Song, Y.; Corlett, R.T.; Yao, X.; Li, D.Z.; Yu, W.B. Plastid NDH pseudogenization and gene loss in a recently derived lineage from the largest hemiparasitic plant genus Pedicularis (Orobanchaceae). Plant Cell Physiol. 2021, 62, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.; Stoebe, B.; Goremykin, V.; Hapsmann, S.; Hasegawa, M.; Kowallik, K.V. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 1998, 393, 162–165. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Maier, R.M.; Alcaraz, J.P.; Cottet, A.; Herrmann, R.G.; Mache, R. The plastid chromosome of spinach (Spinacia oleracea): Complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001, 45, 307–315. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Liu, H.; Wu, S.; Yuan, Y.; Li, H.; Dong, J.; Liu, Z.; An, C.; Su, Z.; Li, B. Species identification of Oaks (Quercus L., Fagaceae) from gene to genome. Int. J. Mol. Sci. 2019, 20, 5940. [Google Scholar] [CrossRef] [PubMed]
- Schwenkert, S.; Legen, J.; Takami, T.; Shikanai, T.; Herrmann, R.G.; Meurer, J. Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol. 2007, 144, 1924–1935. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, T.; Tsudzuki, T.; Sugiura, M. The genomics of land plant chloroplasts: Gene content and alteration of genomic information by RNA editing. Photosynth. Res. 2001, 70, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Kode, V.; Mudd, E.A.; Iamtham, S.; Day, A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005, 44, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-l.; Nie, L.-y.; Deng, S.-w.; Duan, L.; Wang, Z.-f.; Charboneau, J.L.M.; Ho, B.-C.; Chen, H.-f. Characterization of Firmiana danxiaensis plastomes and comparative analysis of Firmiana: Insight into its phylogeny and evolution. BMC Genom. 2024, 25, 203. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Merchant, S. The Plastid-encoded ccsA Gene is required for heme attachment to chloroplast c-type cytochromes (*). J. Biol. Chem. 1996, 271, 4632–4639. [Google Scholar] [CrossRef]
- Dong, W.-L.; Wang, R.-N.; Zhang, N.-Y.; Fan, W.-B.; Fang, M.-F.; Li, Z.-H. Molecular evolution of chloroplast genomes of orchid opecies: Insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 716. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Yang, J.; Cho, M.-S.; Stuessy, T.F.; Crawford, D.J.; Kim, S.-C. Chloroplast genome provides insights into molecular evolution and species relationship of fleabanes (Erigeron: Tribe Astereae, Asteraceae) in the Juan Fernández Islands, Chile. Plants 2024, 13, 612. [Google Scholar] [CrossRef]
- Sheikh-Assadi, M.; Naderi, R.; Kafi, M.; Fatahi, R.; Salami, S.A.; Shariati, V. Complete chloroplast genome of Lilium ledebourii (Baker) Boiss and its comparative analysis: Lights into selective pressure and adaptive evolution. Sci. Rep. 2022, 12, 9375. [Google Scholar] [CrossRef]
- Wang, J.-H.; Moore, M.J.; Wang, H.; Zhu, Z.-X.; Wang, H.-F. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies. Gene 2021, 765, 145103. [Google Scholar] [CrossRef] [PubMed]
- The Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Sánchez-Del Pino, I.; Motley, T.J.; Borsch, T. Molecular phylogenetics of Alternanthera (Gomphrenoideae, Amaranthaceae): Resolving a complex taxonomic history caused by different interpretations of morphological characters in a lineage with C4 and C3-C4 intermediate species. Bot. J. Linn. Soc. 2012, 169, 493–517. [Google Scholar] [CrossRef]
- Sage, R.F.; Sage, T.L.; Pearcy, R.W.; Borsch, T. The taxonomic distribution of C4 photosynthesis in Amaranthaceae sensu stricto. Am. J. Bot. 2007, 94, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).