Wasabi Gone Wild? Origin and Characterization of the Complete Plastomes of Ulleung Island Wasabi (Eutrema japonicum; Brassicaceae) and Other Cultivars in Korea
Abstract
1. Introduction
2. Results
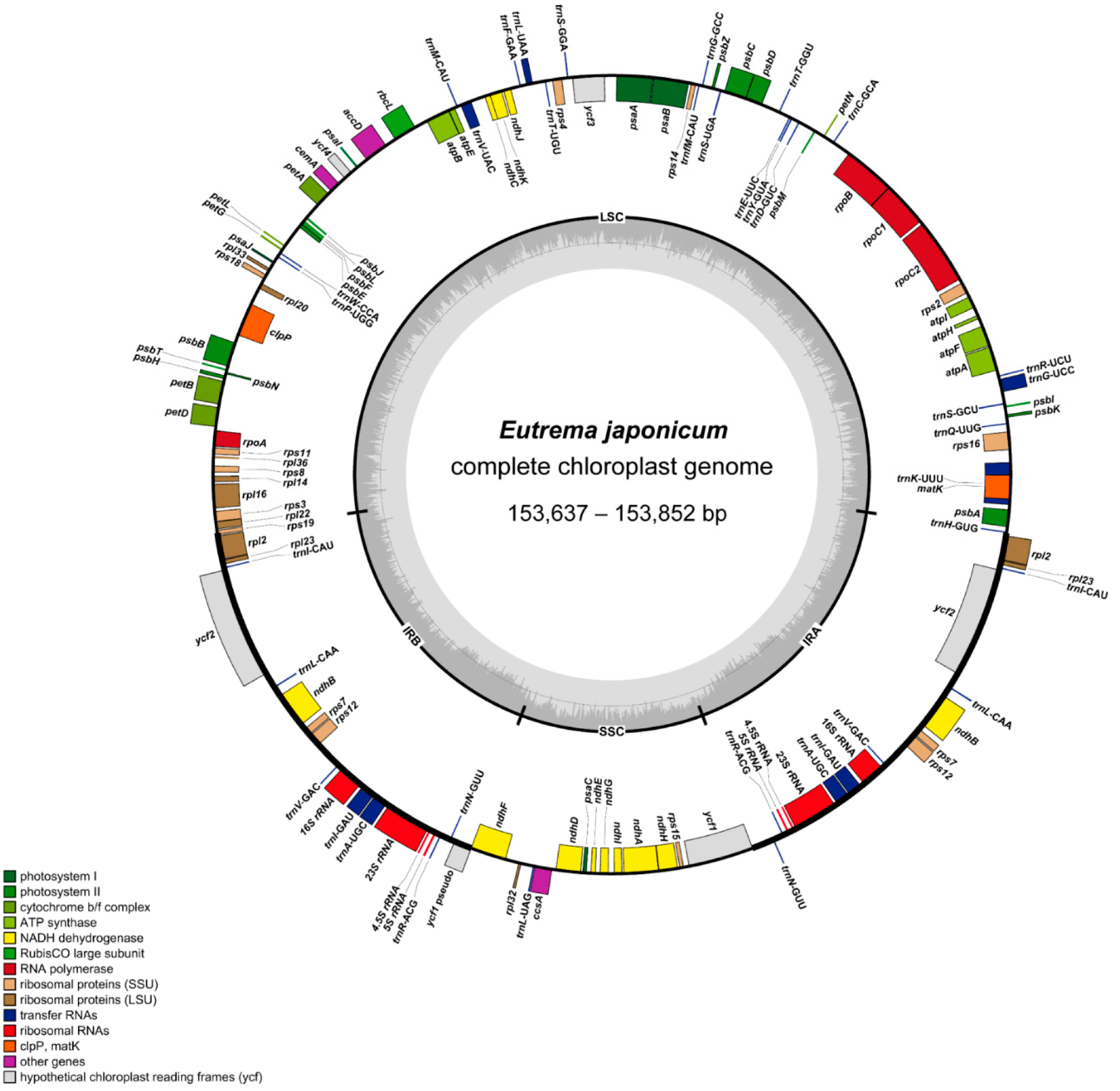
2.1. Genome Size and Features
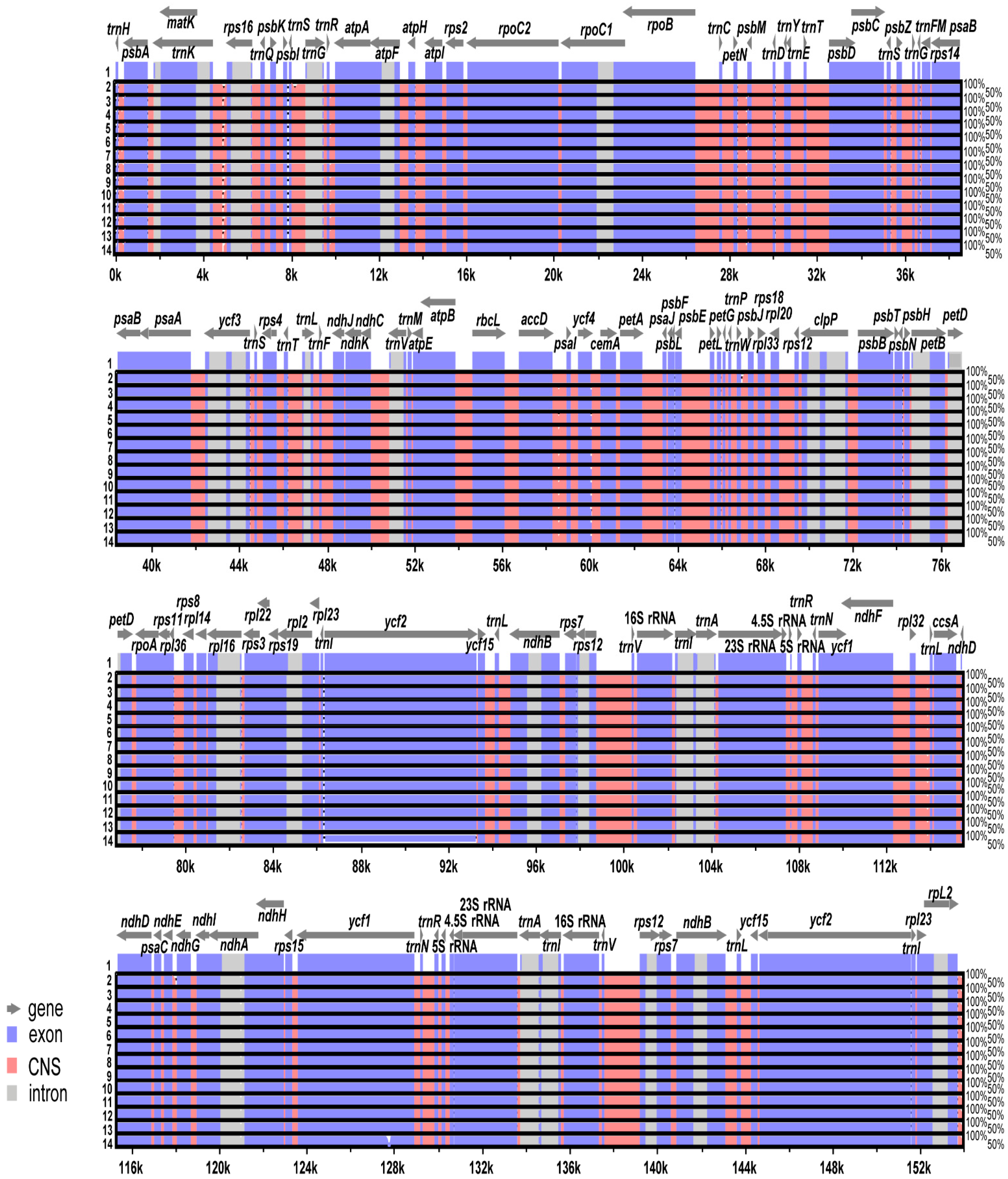
2.2. Comparative Analysis of Genome Structure
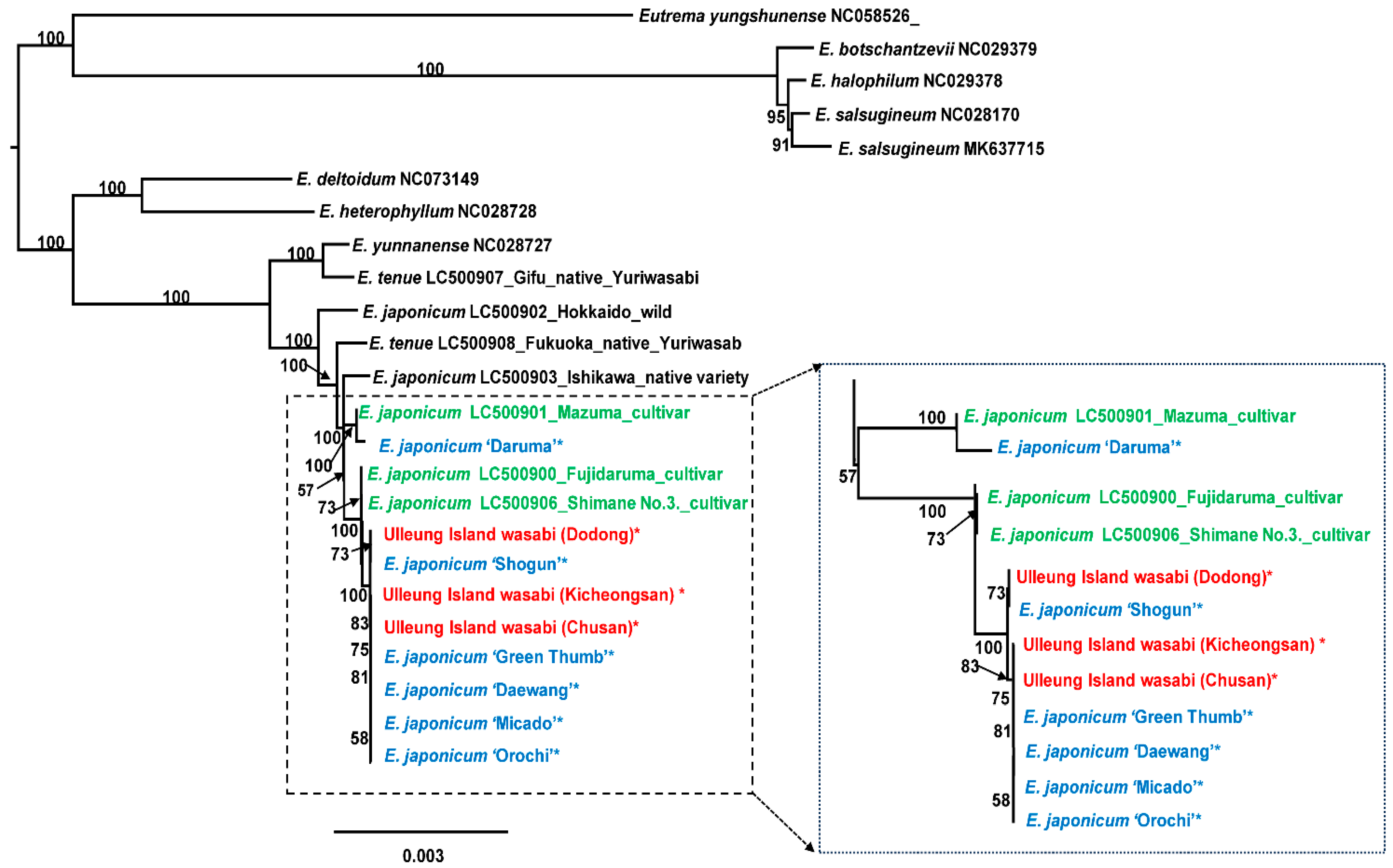
2.3. Phylogenetic Analysis
3. Discussion
3.1. Taxonomic Identity of Ulleung Island Wasabi and Its Relationships to Wild and Cultivated Wasabis Inferred by Complete Chloroplast Genomes
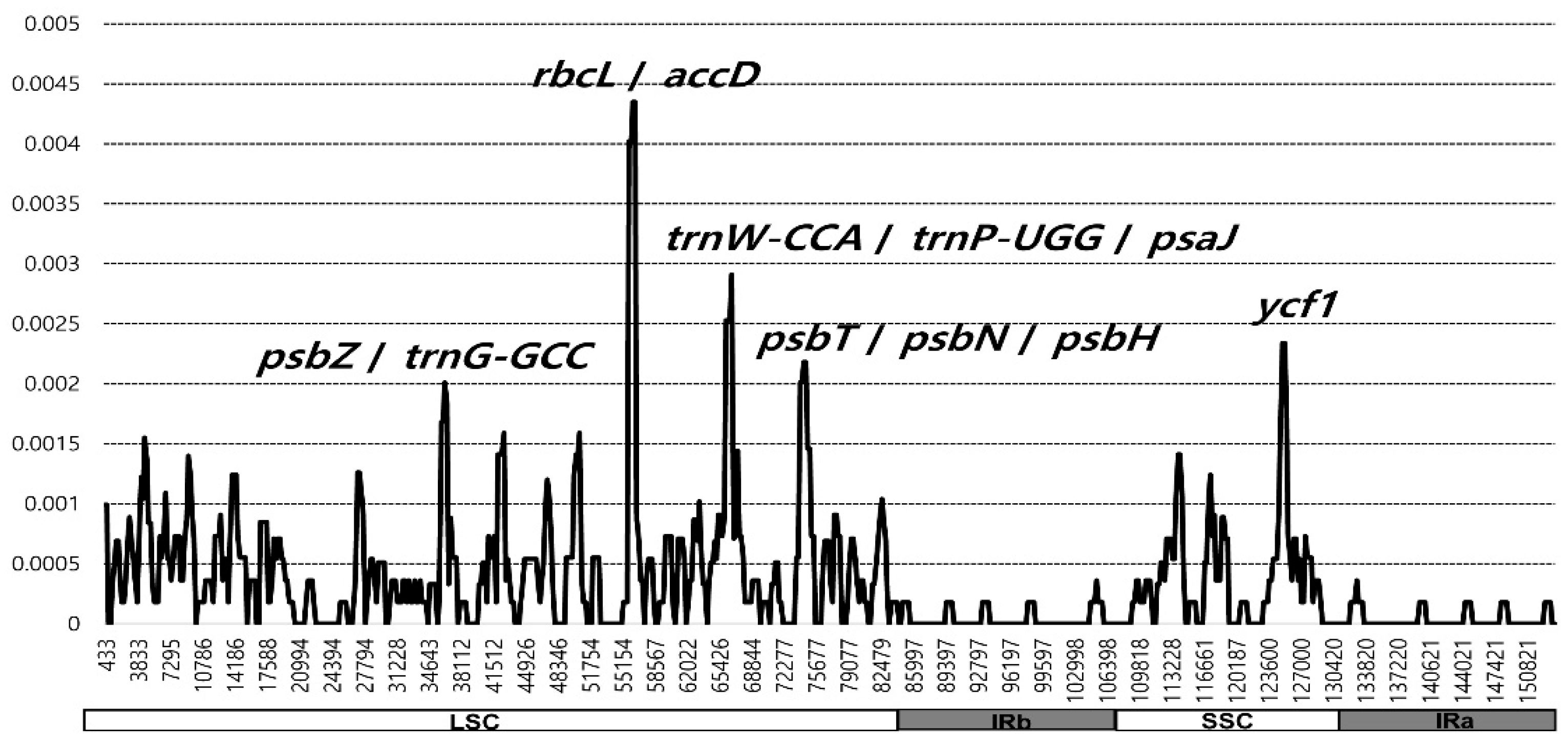
3.2. Plastome Divergence Hotspots in Wild and Cultivated Wasabi E. japonicum
4. Materials and Methods
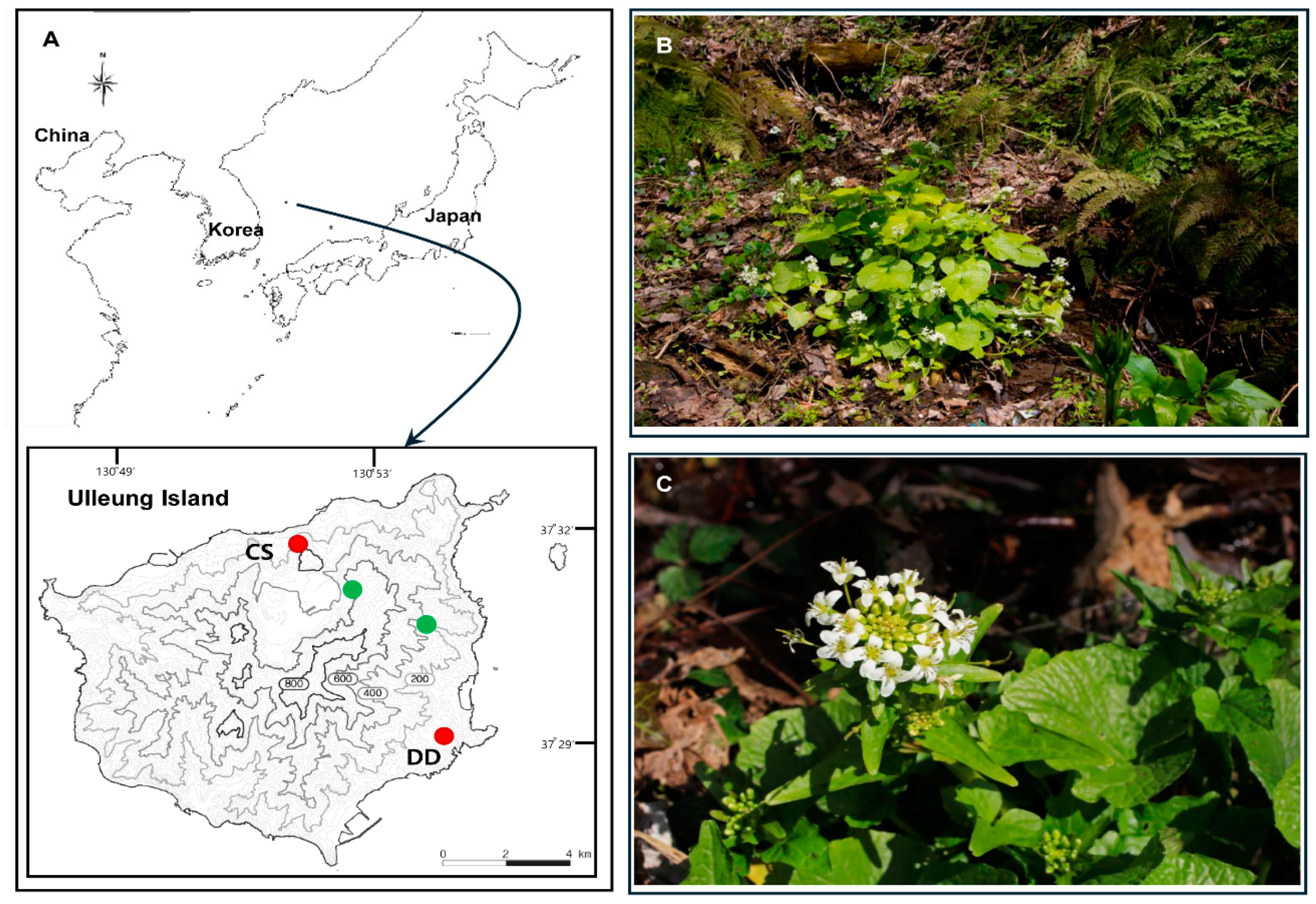
4.1. Plant Sampling, DNA Extraction, and Plastome Sequencing: Assembly and Annotation
4.2. Comparative Plastome Analysis
4.3. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodge, W.H. Wasabi, native condiment plant of Japan. Econ. Bot. 1974, 28, 118–129. [Google Scholar] [CrossRef]
- Yamane, K.; Sugiyama, Y.; Lu, Y.X.; Lü, N.; Tanno, K.; Kimura, E.; Yamaguchi, H. Genetic differentiation, molecular phylogenetic analysis, and ethnobotanical study of Eutrema japonicum and E. tenue in Japan and E. yunnanense in China. Hortic. J. 2016, 85, 46–54. [Google Scholar] [CrossRef]
- Haga, N.; Kobayashi, M.; Michiki, N.; Takano, T.; Baba, F.; Kobayashi, K.; Ohyanagi, H.; Ohgane, J.; Yano, K.; Yamane, K. Complete chloroplast genome sequence and phylogenetic analysis of wasabi (Eutrema japonicum) and its relatives. Sci. Rep. 2019, 9, 14377. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, D.; Abellán-Victorio, A.; Beretta, V.; Camargo, A.; Moreno, D.A. Functional ingredients from Brassicaceae species: Overview and perspectives. Int. J. Mol. Sci. 2020, 21, 1998. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T. Archaeological report of “Asuka Kyo Ato Enchi iko”, the site of a large pond in the garden located in the Asuka capital site. In Archaeological Report of “Asuka Kyo Ato Enchi iko”; Kashihara Archaeological Research Institute, Ed.; Gakuseisha Press: Tokyo, Japan, 2002; p. 55. [Google Scholar]
- Al-Shehbaz, I.A.; Warwick, S.I. A synopsis of Eutrema (Brassicaceae). Harv. Pap. Bot. 2005, 10, 129–135. [Google Scholar] [CrossRef]
- Yoshida, M. Cytological studies on Wasabia. Part 1. Wasabia tenuis. Bull. Fac. Agric. Shimane Univ. 1974, 8, 47–48. [Google Scholar]
- Yoshida, M. Cytological studies on Wasabia. Part 2. Wasabia japonica cultivar. Bull. Fac. Agric. Shimane Univ. 1974, 8, 49–50. [Google Scholar]
- Chadwick, C.I.; Lumpkin, T.A.; Elberson, L.R. The botany, uses and production of Wasabia japonica (Miq.) (Cruciferae) Matsum. Econ. Bot. 1993, 47, 113–135. [Google Scholar] [CrossRef]
- Warwick, S.I.; Al-Shehbaz, A.; Sauder, C.A. Phylogenetic position of Arabis arenicola and generic limits of Aphragmus and Eutrema (Brassicaceae) based on sequences of nuclear ribosomal DNA. Can. J. Bot. 2006, 84, 269–281. [Google Scholar] [CrossRef]
- Adachi, S. Wasabi Saibai; Shizuoka Prefecture Agricultural Experimental Station. Pub.: Shizuoka, Japan, 1987. [Google Scholar]
- Suzuki, H.; Adachi, S.; Nakamura, S. Control of the main disease of wasabi: Eutrema wasabi Maxim by the adaptation of seeding culture. Bull. Shizuoka Pref. Agric. Exp. Stn. 1976, 21, 59–66. [Google Scholar]
- Yokogi, K.; Ueno, R. Wasabi: A profitable side business in the mountains. In Special Products Series No. 34, 3rd ed.; Noosangyoson Bunkkyo (Agricultural and Fishing Villages Assoc.) Pub.: Tokyo, Japan, 1990. [Google Scholar]
- Toda, M. Simple Root Crops, Wasabi; Tokita Seed Company: Saitama, Japan, 1987; Volume 6, pp. 795–799. [Google Scholar]
- Yamane, K. Origin, production, and consumption of ordinary vegetable and fruit-Wasabi (part I). Food Preserv. Sci. 2010, 36, 189–196. [Google Scholar]
- Kim, Y.K. Petrology of Ulreung volcanic island, Korea Part 1. Geology. J. Jpn. Assoc. Mineral. Petrol. Econ. Geol. 1985, 80, 128–135. [Google Scholar] [CrossRef]
- Yim, Y.-J.; Lee, E.-B.; Kim, S.-H. Vegetation of Ulreung and Dogdo Islands. In The Report on the KSCN No. 19: A Report on Scientific Survey of Ulreung and Dogdo Islands; The Korean Association for Conservation of Nature: Seoul, Republic of Korea, 1981. [Google Scholar]
- Sun, B.-Y.; Shin, H.; Hyun, J.-O.; Kim, Y.-D.; Oh, S.-H. Vascular Plants of Dokdo and Ulleungdo Islands in Korea; The National Institute of Biological Resources, GeoBook Publishing Co.: Seoul, Republic of Korea, 2014. [Google Scholar]
- Sun, B.-Y.; Stuessy, T.F. Preliminary observations on the evolution of endemic angiosperms of Ullung Island, Korea. In Evolution and Speciation of Island Plants; Stuessy, T.F., Ono, M., Eds.; Cambridge University Press: New York, NY, USA, 1998; pp. 181–202. [Google Scholar]
- Stuessy, T.F.; Jakubowsky, G.; Gömez, R.S.; Pfosser, M.; Schlüter, P.M.; Fer, T.; Sun, B.-Y.; Kato, H. Anagenetic evolution in island plants. J. Biogeogr. 2006, 33, 1259–1265. [Google Scholar] [CrossRef]
- Yang, J.; Pak, J.-H.; Maki, M.; Kim, S.-C. Multiple origins and the population genetic structure of Rubus takesimensis (Rosaceae) on Ulleung Island: Implications for the genetic consequences of anagenetic speciation. PLoS ONE 2019, 14, e0222707. [Google Scholar] [CrossRef]
- Cheong, W.Y.; Kim, S.-H.; Yang, J.Y.; Lee, W.; Pak, J.-H.; Kim, S.-C. Insights from chloroplast DNA into the progenitor-derivative relationship between Campanula punctata and C. takesimana (Campanulaceae) in Korea. J. Plant Biol. 2020, 63, 431–444. [Google Scholar] [CrossRef]
- Seo, H.S.; Kim, S.-H.; Kim, S.-C. Chloroplast DNA insights into the phylogenetic position and anagenetic speciation of Phedimus takesimensis (Crassulaceae) on Ulleung and Dokdo Islands, Korea. PLoS ONE 2020, 15, e0239734. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.-S.; Takayama, K.; Yang, J.Y.; Maki, M.; Kim, S.-C. Genome-wide single nucleotide polymorphism analysis elucidates the evolution of Prunus takesimensis in Ulleung Island: The genetic consequences of anagenetic speciation. Front. Plant Sci. 2021, 12, 706196. [Google Scholar] [CrossRef]
- Jung, S.-Y.; Park, S.-H.; Nam, C.-H.; Lee, H.-J.; Lee, Y.-M.; Chang, K.-S. The distribution of vascular plants in Ulleungdo and nearby island regions (Gwaneumdo, Jukdo), Korea. J. Asia-Pac. Biodivers. 2013, 6, 123–156. [Google Scholar] [CrossRef][Green Version]
- Yoon, J.W.; Shin, H.T.; Yi, M.H. Distribution of vascular plants in the Ulleung forest trail area, Korea. J. Asia-Pac. Biodivers. 2013, 6, 1–30. [Google Scholar] [CrossRef][Green Version]
- Yang, S.; Jang, H.-D.; Nam, B.M.; Chung, G.Y.; Lee, R.-Y.; Lee, J.-H.; Oh, B.-U. A floristic study of Ulleungdo Island in Korea. Korean J. Plant Taxon. 2015, 45, 192–212. [Google Scholar] [CrossRef]
- Lee, W.C.; Yang, I.S. The flora of the Ulreung Is. and Dogdo Island. Rep. Korean Assoc. Conserv. Nat. 1981, 19, 61–95. (In Korean) [Google Scholar]
- Shin, H.; Kim, Y.-D. A short note on the taxonomic identity of Wasabia koreana Nakai (Brassicaceae). Korean J. Plant Taxon. 2008, 38, 223–231. [Google Scholar] [CrossRef]
- Chung, J.-M.; Shin, J.-K.; Kim, H.-M. Diversity of vascular plants native to the Ulleungdo and Dokdo Islands in Korea. J. Asia-Pac. Biodivers. 2020, 13, 701–708. [Google Scholar] [CrossRef]
- Heo, S.J.; Kwon, S.B.; Byeon, H.S.; Seo, J.S.; Yoo, G.O. Intraspecific genetic relation of Wasabia japonica Matsum. Korean J. Med. Crop Sci. 2004, 12, 31–35. [Google Scholar]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E. DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.H. Korean Flora. In Woody Plants; Shinjisa: Seoul, Republic of Korea, 1957; Volume 1. [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Suyama, Y.; Matsuki, Y. MIG-seq: An effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci. Rep. 2015, 5, 16963. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Blaxter, M.L. RADSeq: Next-generation population genetics. Brief. Funct. Genom. 2010, 9, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Q.; Zhang, C.; Huang, H.; He, H.; Wang, M.; Li, M.; Huang, Z.; Tang, Y.; Chen, Q.; et al. Sequencing and analysis of complete chloroplast genomes provide insights into the evolution and phylogeny of Chinese kale (Brassica oleracea var. alboglabra). Int. J. Mol. Sci. 2023, 24, 10287. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Hu, Q.; Al-Schbaz, I.A.; Luo, X.; Zeng, T.; Guo, X.; Liu, J. Species delimitation and interspecific relationships of the genus Orychophragmus (Brassicaceae) inferred from whole chloroplast genomes. Front. Plant Sci. 2016, 7, 1826. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Amar, M.H. ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica. J. Genet. Eng. Biotechnol. 2020, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-N.; Milne, R.I.; Du, X.-Y.; Liu, J.; Wu, Z.-Y. Characteristics and mutational hotspots of plastomes in Debregeasia (Urticaceae). Front. Genet. 2020, 11, 729. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hu, H.; Zhang, D. DNA barcoding and phylogenomic analysis of the genus Fritillaria in China based on complete chloroplast genomes. Front. Plant Sci. 2022, 13, 764255. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, T.-J. Genetic species identification using ycf1b, rbcL, and trnH-psbA in the genus Pinus as a complementary method for anatomical wood species identification. Forests 2023, 14, 1095. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. Organellar genome DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Brundo, M.; Malde, S.; Poliakov, A.; Do, C.B.; Couronne, O.; Dubchak, I. Global alignment: Finding rearrangements during alignment. Bioinformatics 2003, 19 (Suppl. S1), i54–i62. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software v7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. 1. The codon adaptation index-A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 1983, 47, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]

| Taxa | Ulleung Island | Major Cultivars in Korea (GARES *) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Kicheongsan | Chusan | Dodong | ‘Daewang’ | ‘Daruma’ | ‘Green Thumb’ | ‘Micado’ | ‘Orochi’ | ‘Shogun’ | |
| Total cpDNA size (bp) | 153,851 | 153,851 | 153,852 | 153,851 | 153,794 | 153,851 | 153,849 | 153,849 | 153,852 |
| GC content (%) | 36.4% | 36.4% | 36.4% | 36.4% | 36.4% | 36.4% | 36.4% | 36.4% | 36.4% |
| LSC size (bp) /GC content (%) | 83,889/34.1% | 84,006/34.1% | 84,006/34.1% | 84,006/34.1% | 83,990/34.0% | 84,006/34.1% | 84,006/34.1% | 84,006/34.1% | 84,006/34.1% |
| IR size (bp) /GC content (%) | 26,017/42.5% | 26,017/42.5% | 26,017/42.5% | 26,017/42.5% | 26,007/42.5% | 26,017/42.5% | 26,016/42.5% | 26,016/42.5% | 26,017/42.5% |
| SSC size (bp) /GC content (%) | 17,811/29.4% | 17,811/29.4% | 17,812/29.4% | 17,811/29.4% | 17,790/29.4% | 17,811/29.4% | 17,811/29.4% | 17,811/29.4% | 17,812/29.4% |
| Number of genes | 131 | 131 | 131 | 131 | 131 | 131 | 131 | 131 | 131 |
| Number of protein-coding genes | 84 | 84 | 84 | 84 | 84 | 84 | 84 | 84 | 84 |
| Number of tRNA genes | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 37 |
| Number of rRNA genes | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of duplicated genes | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 |
| Accession Number | PP413704 | PP413705 | PP413706 | PP413707 | PP413708 | PP413709 | PP413710 | PP413711 | PP413712 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Park, C.G.; Cho, M.-S.; Kim, S.-C. Wasabi Gone Wild? Origin and Characterization of the Complete Plastomes of Ulleung Island Wasabi (Eutrema japonicum; Brassicaceae) and Other Cultivars in Korea. Genes 2024, 15, 457. https://doi.org/10.3390/genes15040457
Yang J, Park CG, Cho M-S, Kim S-C. Wasabi Gone Wild? Origin and Characterization of the Complete Plastomes of Ulleung Island Wasabi (Eutrema japonicum; Brassicaceae) and Other Cultivars in Korea. Genes. 2024; 15(4):457. https://doi.org/10.3390/genes15040457
Chicago/Turabian StyleYang, JiYoung, Cheon Gyoo Park, Myong-Suk Cho, and Seung-Chul Kim. 2024. "Wasabi Gone Wild? Origin and Characterization of the Complete Plastomes of Ulleung Island Wasabi (Eutrema japonicum; Brassicaceae) and Other Cultivars in Korea" Genes 15, no. 4: 457. https://doi.org/10.3390/genes15040457
APA StyleYang, J., Park, C. G., Cho, M.-S., & Kim, S.-C. (2024). Wasabi Gone Wild? Origin and Characterization of the Complete Plastomes of Ulleung Island Wasabi (Eutrema japonicum; Brassicaceae) and Other Cultivars in Korea. Genes, 15(4), 457. https://doi.org/10.3390/genes15040457







