Highlights
What are the main findings?
- The introduction of PCR has revolutionised forensic DNA profiling.
- With increasing sample complexity and the demands of forensic science, the ability of PCR is being pushed to its limits.
- This paper reviews the different PCR applications and cycling conditions that have been adopted over the years for use in forensic science and suggests where the field may be heading.
Abstract
The polymerase chain reaction (PCR) has played a fundamental role in our understanding of the world, and has applications across a broad range of disciplines. The introduction of PCR into forensic science marked the beginning of a new era of DNA profiling. This era has pushed PCR to its limits and allowed genetic data to be generated from trace DNA. Trace samples contain very small amounts of degraded DNA associated with inhibitory compounds and ions. Despite significant development in the PCR process since it was first introduced, the challenges of profiling inhibited and degraded samples remain. This review examines the evolution of the PCR from its inception in the 1980s, through to its current application in forensic science. The driving factors behind PCR evolution for DNA profiling are discussed along with a critical comparison of cycling conditions used in commercial PCR kits. Newer PCR methods that are currently used in forensic practice and beyond are examined, and possible future directions of PCR for DNA profiling are evaluated.
1. The Polymerase Chain Reaction & Forensic Science
1.1. The General Principles of PCR
The polymerase chain reaction (PCR) is a fundamental technique that amplifies specific regions of deoxyribonucleic acid (DNA) via an enzymatic reaction. Amplification using PCR requires five key components: deoxynucleotide triphosphates (dNTPs), thermostable DNA polymerase, template DNA, primers, and a buffer containing potassium and magnesium [1,2,3]. The high specificity of PCR can be largely attributed to the sequence-specific primers present in the reaction and stringent cycling conditions employed. PCR programs involve the same three basic steps of denaturation: heating to dissociate the double strands of the DNA molecule; annealing to allow primers to bind to their complementary target sequence; and extension, where the sample is heated to a temperature slightly above annealing that is optimal for DNA polymerase to synthesise the new double-stranded molecule [2,3,4,5,6]. These three steps are cycled through until sufficient target DNA is produced that can be detected.
1.2. Historial Significance and Applications of PCR in Forensic Science
In 1981, the entire human mitochondrial genome was sequenced and published [7]. This sequence, known as the Cambridge Reference Sequence (CRS), was quickly adopted by geneticists and became a widely used reference point for many mitochondrial DNA (mtDNA) studies [7,8]. The development of DNA fingerprinting in 1985 [9] and the rise of the PCR in forensic biology during the early 1990s [10,11,12,13] revolutionized forensic casework and significantly increased the tissue types submitted for forensic analyses. Initially, forensic DNA profiling was conducted using restriction enzymes that targeted hypervariable regions within the human nuclear and mitochondrial genomes called restriction fragment length polymorphisms (RFLPs) [14]. However, the introduction of PCR to target and amplify specific hypervariable regions of the human genome marked the beginning of the DNA profiling era. The first instance of PCR being used in a criminal trial was in 1986 for the case of Pennsylvania v. Pestinikas, where the amplification of the Human Leukocyte Antigen (HLA) DQα was presented in court [15].
As a result, in 1991 the PCR was successfully used to amplify two regions of the mitochondrial genome, HV1 and HV2, from skeletal remains using oligonucleotide hybridization [16]. The remains were identified to belong to a 3-year-old child that had been reported missing by their parents in 1984, thus marking the first published instance of human identification using mtDNA analysis [16,17]. In 1992, the PCR was successfully used to amplify a single variable number tandem repeat (VNTR) locus within the human nuclear genome, called D1S80 [12], and in 1993 the same was done for an amplification restriction fragment polymorphism (ARFP) locus spanning the human major histocompatibility complex (MHC), also known as the HLA, called DQα [10]. Shortly after, in 1994, nine sets of human remains discovered in a Russian forest were identified to be the Romanov family, the Russian Imperial Family who had been assassinated in 1918, through extensive mtDNA and short tandem repeat (STR) analysis [18]. Later that same year, Kimpton et al. [19] published a quadruplex PCR method, which amplified four tetrameric STRs in a single reaction and produced DNA profiles that could provide a limited power of discrimination between individuals. For a significant amount of time, using restriction digests to isolate RFLPs was favored over STR PCR assays because the discrimination power afforded by RFLP analysis was significantly higher than that afforded by the four STR loci [20]. However, the advent of STR multiplexing PCR for DNA profiling allowed the introduction of additional hypervariable STR loci such that the technique of DNA profiling quickly superseded RFLP analysis to become the cornerstone of the forensic biology discipline. Importantly, the PCR cycling conditions used in these landmark publications are still largely the same as those used for DNA profiling today.
1.3. Driving Factors of PCR Evolution in Forensic Science
A phase of rapid change and discovery, driven by the need for standardization, reliability and reproducibility in forensic DNA analysis, occurred between 1990 and 1995. This resulted in the formation of the European DNA Profiling Group (EDNAP), the European Network of Forensic Science Institutes (ENFSI) and the Scientific Working Group on DNA Analysis Methods (SWGDAM) (originally TWGDAM), organizations that aimed to standardize techniques and ensure equal justice outcomes from biological examinations from different forensic laboratories. In the years following the first national DNA databases were established in the United Kingdom [21], other countries in Europe [22] and the United States [23]. DNA databases are compilations of DNA profiles obtained from references of individuals involved (or suspected of being involved, depending on the jurisdiction) in criminal activity (the exact criteria for inclusion on a database varying by country and state), or obtained as unknown profiles from casework. Such compilations allow DNA profiles from unknown sources, obtained from exhibits of forensic relevance, to be searched against the profiles in the database for a potential match. These databases required a set of standard STR markers to be used by all participating laboratories to allow profiles to be compared, and quality assurance so that all profiles uploaded would be reliable between laboratories [23,24,25]. To avoid coincidental matches, additional hypervariable loci were added to the multiplex STR systems [26,27], with separation initially on polyacrylamide gels and then capillary electrophoresis for DNA profiling becoming standard practice. However, despite the inherent value of using DNA databases in criminal investigations, the ethical and legal implications associated with their use posed considerable challenges for their implementation [28,29,30]. Over the last two decades, substantial progress has been made in this arena, with many countries now having their own national DNA databases. As a result of the continued development and expansion of standard STR marker sets and laboratory standardization, international databases, such as the INTERPOL DNA database, now exist and are routinely used in forensic investigation [31].
The rapid developments of PCR and DNA profiling in more recent years can largely be attributed to the need for high sensitivity, specificity and reproducibility in forensic techniques. The optimal amounts of DNA recommended by manufacturers to generate probative STR profiles have substantially decreased, from 2 ng in 1995 [19] to as little as 0.4 ng in current STR kits [32,33,34,35]. However, forensic samples are commonly submitted for PCR with far lower than optimal amounts of DNA, pushing the boundaries of PCR [36]. Importantly, as the sensitivity of these techniques has increased, the number of trace DNA samples submitted to operational laboratories for analysis has also increased dramatically [37,38,39,40]. Despite the improvements in machine and PCR assay sensitivity in recent years, the success when generating DNA profiles from these trace samples remains poor [41,42,43,44,45,46,47,48]. Given the volume of trace samples that are now routinely submitted for analysis, the time and monetary costs associated with processing these samples only to obtain very little genetic information is a strain on operational laboratories around the globe. Examples of the developments in forensic genetics are shown in Figure 1.

Figure 1.
A timeline of PCR evolution within forensic science from 1985 to the present day (2023). 1985–1990 [4,5,14,49], 1990–1995 [19,50,51], 1995–2000 [52,53,54,55,56], 2000–2005 [57,58,59], 2005–2010 [60,61,62], 2010–2015 [63,64,65,66], 2015–2020 [67], and 2020+ [68,69].
1.4. Current PCR Workflows in Forensic Science
The PCR process for DNA profiling, also known as STR PCR, utilizes a specialized form of chemistry where fluorophores attached to primers become incorporated into amplicons as the reaction progresses [33,54,70,71,72,73,74,75]. Once the STR PCR is complete, the amplified product is detected during capillary electrophoresis using the incorporated fluorescence tags. A visual representation of this traditional STR PCR workflow is shown below in pathway A (Figure 2).
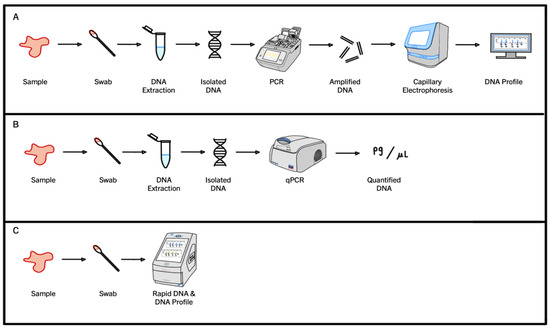
Figure 2.
A comparison of the steps required to go from the initial crime scene sample to the final product via three different PCR methodologies: traditional PCR (A), quantitative PCR (qPCR) (B), and Rapid DNA (C).
However, in accredited forensic laboratories, the DNA present in a sample must be quantified before it progresses to STR PCR for DNA profiling. The dynamics of quantitative PCR (qPCR) are substantially different to those of an STR PCR. Commercially available qPCR kits, such as QuantiFiler Trio (Thermo Fisher Scientific, Waltham, MA, USA) or Investigator Quantiplex Pro (QIAGEN, Hilden, Germany), use TaqMan® probes [75,76]. These probes are labeled with two fluorescent dyes: a reporter dye at the 5′-end and a quencher dye at the 3′end [23,77]. When the probe is intact (prior to polymerization), the two dyes are in close proximity due to the small size of the probe. The presence of the quencher dye so close to the reporter dye means the fluorescence of the reporter dye is suppressed due to an energy transfer occurring between the two. This energy transfer is based on fluorescent resonance energy transfer (FRET) principles, where the efficiency of FRET is dependent on the inverse sixth power of the intermolecular separation of the two dyes [77,78]. When a qPCR program is carried out, the probes bind to the target sequences during the annealing stage and the reporter dye fluorescence remains suppressed. However, during the extension stage, the 5′-exonuclease activity of DNA polymerase displaces the bound TaqMan probes, and the reporter dye attached to the 5′-end is separated from the quencher dye at the 3′-end [23,77,78]. The separation means the energy transfer that suppressed fluorescence when the two fluorophores were in close proximity is no longer occurring and the reporter molecule can now fluoresce [23,77,78]. The amount of fluorescence detected correlates directly with the amount of DNA present in the reaction, as the greater the amount of DNA, the more reporter dye molecules are cleaved, and the greater the fluorescence detected. Typically, as the qPCR program progresses, and the DNA increases exponentially, an amplification curve is generated using these fluorescence data, which (at the end of the qPCR program) are then used to calculate a DNA concentration. If a sample is found to contain a sufficient amount of DNA, such that useful genetic information could be obtained from it, then that sample can progress to STR PCR and a DNA profile can be generated. A visual representation of a standard qPCR workflow is shown below in pathway B (Figure 2).
The third PCR workflow that is often used in forensic science is Rapid PCR. Rapid PCR uses the same fluorescent primer chemistry as STR PCR, but has specialized instruments, PCR programs and reaction chemistries that allow DNA profiles to be generated in a much smaller time frame. Rapid PCR protocols skip the DNA quantification step but are capable of generating DNA profiles quickly using shortened PCR programs. Thus, this workflow allows informative DNA profiles to be generated quickly, but to do so samples must contain large amounts of high-quality template DNA, such as those obtained from reference swabs [73,79]. A visual representation of a standard Rapid PCR workflow is shown below in pathway C (Figure 2).
2. Fundamental Factors of the Polymerase Chain Reaction
2.1. PCR Variants—Uniplex and Multiplex PCR
While this basic formula for DNA amplification has been largely conserved since it was first conceptualized in the 1980s, some modifications have been made to the PCR process to allow it to be applied to a broad range of fields, such as microbiology, medicine and forensic science. Perhaps most notable of these developments is the advent of multiplex PCR. Initially, PCRs were only capable of amplifying one specific fragment of DNA in a single reaction [1,2,3], which was sufficient for many applications in clinical and medical diagnostics. However, in forensic science, PCR is used in an attempt to collect as much genetic information from a crime scene sample as possible. This is important because the more information that can be collected about DNA within an evidence sample, the more discrimination power there is to distinguish a donor of DNA from a non-donor, which then can be used to inform investigators about potential exclusions and contributors to a sample. To achieve this, multiplex PCR is conducted instead of uniplex PCR. Unlike uniplex PCR, multiplex PCR allows multiple regions of DNA to be amplified in a single reaction [80]. This helps reduce the amount of DNA extract needed to produce a DNA profile and the PCR reagents required to produce informative genetic data, while also increasing sample throughput when compared to uniplex PCR [81,82,83]. However, the stringency of the cycling conditions used for multiplex PCR is much higher than for those used for uniplex PCR.
The significance of multiplexing in the realm of DNA evidence is most apparent in the highly discriminatory DNA profiles obtained from the DNA amplified using multiplex PCR. The amplification of more than 20 hypervariable regions of the human genome, known as short-tandem repeats (or STRs), in a single reaction for DNA profiling imposes some restraints on the cycling conditions that can be used. As the number of regions targeted during multiplex PCR increases, so too does the number of primers needed in a reaction to ensure that the new target regions are amplified from all human populations around the world [70]. All these primers also need to be able to bind to their complementary sequence and avoid mismatching; thus, to ensure correct amplification, the annealing and extension temperatures used in multiplex PCR are highly stringent and well validated.
2.2. Factors Influencing PCR Cycling Conditions
There are two key factors that dictate the success of a PCR program: primer melting temperature and DNA polymerase processivity. The temperature at which the annealing step occurs is dictated by the primer melting temperature (Tm) [62]. The Tm value for a pair of primers is indicative of the stability of the double-stranded DNA (dsDNA), and is determined by the temperature at which one half of the dsDNA will dissociate. Longer primers and those that are rich in guanine and cytosine have been found to have higher Tm values due to greater amounts of energy being required to break the bonds between the primer and the target DNA [84]. The Tm value for a set of primers can be determined using specialized software programs [85,86,87]; however, the methods used to calculate Tm values differ slightly between programs, which means the calculated values differ slightly between programs. Importantly, for multiplex PCR (which involves many primers), all primers in a reaction must have similar annealing temperatures, and thus Tm values, but minimal overlap with each other to minimize primer interactions [23,88]. Primer–primer interactions are detrimental to PCR because primers with regions of high complementarity will preferentially bind to one another and form primer dimers or hairpin structures, rather than bind to the template DNA, which will significantly reduce the amount of target DNA amplified [23,89]. Due to the number of independent primer annealing events that must occur simultaneously in multiplex PCRs, the temperature and timing of the annealing step is often optimized and extensively validated for each specific set of primers. To ensure all STRs are amplified in approximately equal amounts to produce balanced DNA profiles, the primers used in commercially available STR PCR kits are often manipulated to ensure effective amplification. An example of this is primers being made smaller or larger, which will decrease or increase the binding specificity and alter the Tm value to align with the ideal annealing temperatures for a given STR kit. Therefore, the success of DNA amplification via PCR is largely dependent on the suitability of the annealing step for the set of primers present in the reaction.
The processivity of a DNA polymerase refers to the speed at which the polymerase can synthesize a complementary DNA strand, and is measured as the number of dNTPs incorporated in a single association/dissociation event [90]. The timing and temperature of the extension step of PCR is heavily influenced by the processivity of the DNA polymerase used. The importance of polymerase processivity in PCR cycling conditions is perhaps most evident in the context of DNA profiling. While the timely amplification of DNA is ideal for all applications of PCR, the most useful and informative DNA profiles are those that contain all the target STRs (alleles) in approximately equal amounts. To promote total and equal polymerization of the target STRs, the time allowed for the extension step of PCR needs to account for the size of the target sequences and the speed at which the DNA polymerase can synthesize the amplicons. However, it is well understood that the enzymatic activity of DNA polymerase does not remain constant across a PCR run [1,91]. In the early cycles of PCR, enzymatic activity is high as the enzyme is “fresh”, there are sufficient amounts of catalyst in the reaction vessel, and there are only a few copies of template DNA to be amplified. After a few cycles, the DNA polymerase will have started to lose enzymatic activity through the heat denaturation that is occurring during the denaturation steps of the PCR [91,92]. As the PCR progresses, the available catalysts in solution decrease, the amount of template DNA continues to increase exponentially, and the repeated heating to denature the dsDNA strands continues to reduce polymerase enzymatic activity. Therefore, accounting for enzyme processivity when PCR programs are designed is essential. This involves ensuring there is sufficient time for polymerization to occur, and that complete polymerization is encouraged as enzymatic activity decreases across the PCR.
3. Evolution of the Polymerase Chain Reaction
3.1. Evolution of PCR Cycling Conditions in Forensic Science
3.1.1. Short Tandem Repeat Profiling
While PCR’s cycling conditions have seen some minor variation since it was first used in forensic science in 1993, the process of amplifying STRs for DNA profiling has remained largely the same. This conservation can be largely attributed to the significant increase in STR loci targeted for DNA profiling. The introduction of more STR loci to the PCR multiplex mixtures meant the number of primers required for amplification of these highly polymorphic regions increased, and with them the stringency of the annealing stage [70]. With the addition of new STR loci came the addition of more primers to the PCR to ensure that the new loci would be amplified for all human populations around the world [70]. These primers also needed to be able to bind to their complementary sequence and avoid mismatching; thus, to ensure correct amplification, the PCR cycling conditions used today are highly stringent. As a result of this stringency and in order to further improve the quality of DNA profiles obtained from challenging samples (i.e., inhibited and trace material), changes were made to other elements of the amplification process (i.e., buffers, enzymes, instruments).
While there have been minor changes made to recommended cycling conditions between kits (Table 1), the same largely invariant conditions are often used in the validated commercial STR multiplex kits. In an effort to increase the success of DNA profiling for low-template DNA samples, the PCR cycle number can be increased by one to five cycles [57,58,59]. Importantly, increasing the cycle number in this manner has also been found to further amplify stochastic characteristics, which can make profile interpretation and deconvolution substantially more difficult [36,48,58]. Furthermore, while the addition of extra cycles of amplification has proven to be useful in obtaining more DNA profiles from low-template samples, it is not a change to the PCR cycling conditions, but rather an extra repeat of the same uniform amplification conditions.
The most significant change to PCR cycling conditions for DNA profiling came with the introduction of Rapid DNA. The temperatures of the fundamental stages of the PCR are largely the same in Rapid PCR as they are in traditional STR PCR programs; however, it is the timing of the steps that changes substantially. The general Rapid DNA kit has a specialized chemistry and a PCR protocol involving enzyme activation (initialization) at 96 °C for 60 s, followed by 28 cycles of denaturation at 96 °C for 5 s and a combined annealing and extension step at 60 °C for 40 s, before a final extension step (hold) at 60 °C for 8 min [73,74,79,93,94]. These timing differences are further accentuated when the cycling conditions of traditional kits and Rapid DNA kits are directly compared, as shown in Table 1. These Rapid PCR programs are valuable to forensic investigations as they allow informative DNA profiles to be generated quickly, in the field at remote locations, and by personnel with little training. However, it is important to highlight that Rapid DNA has a specific range of applications, and currently has only had varying success with the sub-optimal samples that are typically encountered in casework [95]. Thus, a major limitation of Rapid DNA cycling conditions is that they generally require high-quality samples (i.e., reference samples) to generate good-quality, informative DNA profiles [96].
While Rapid DNA has successfully proven that changes can be made to elements of a PCR program, it is important to highlight that the changes used remain non-variant across the entire program. While this non-variance is a common feature of PCR programs within forensic genetics, there have been variations in cycling conditions through the generations of traditional STR PCR kits, as can be seen in Table 1. This is perhaps most notable when looking at the recommended cycling conditions for PowerPlex® 2.1 and PowerPlex® 16 systems (supplied by the Promega Corporation, Madison, WI, USA) and the VeriFilerTM Plus system (available from ThermoFisher Scientific), which have a decreasing denaturation temperature and annealing/extension temperature after the first few cycles, respectively (Table 1).

Table 1.
PCR cycling conditions recommended by commercially available traditional and rapid STR kits and the total reaction volumes.
Table 1.
PCR cycling conditions recommended by commercially available traditional and rapid STR kits and the total reaction volumes.
| STR Kit | Type | Year | Cycling Conditions | Total Cycles | Reaction Volume | ||
|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | |||||
| PowerPlex1.1 [97] and PowerPlex 2.1 [98] | Traditional | 1997 | 94 °C 30 s (10 cycles) 90 °C 30 s (20 cycles) | 60 °C 30 s | 70 °C 45 s | 30 | 22.5 L |
| SGM Plus [99] | Traditional | 1999 | 94 °C 1 min | 59 °C 1 min | 72 °C 1 min | 28 | 50 L |
| PowerPlex 16 [100] | Traditional | 2001 | 94 °C 30 s (10 cycles) 90 °C 30 s (22 cycles) | 60 °C 30 s | 70 °C 45 s | 32 | 25 L |
| AmpFSTR Identifiler [101] | Traditional | 2001 | 94 °C 1 min | 59 °C 1 min | 72 °C 1 min | 28 | 26 L |
| MiniFiler [102] | Traditional | 2007 | 94 °C 20 s | 59 °C 2 min | 72 °C 1 min | 30 | 25 L |
| AmpFSTR Identifiler Plus [103] | Traditional | 2010 | 94 °C 20 s | 59 °C 3 min | 28–29 | 25 L | |
| AmpFSTR NGM Select Express [104] | Rapid | 2011 | 94 °C 3 s | 59 C 16 s | 65 °C 29 s | 25–28 | 25 L |
| PowerPlex 21 [32] | Traditional | 2012 | 94 °C 10 s | 59 °C 1 min | 72 °C 30 s | 30 | 25 L |
| GlobalFiler and GlobalFiler IQC [35] | Traditional | 2013 | 94 °C 10 s | 59 °C 90 s | 29–30 | 25 L | |
| GlobalFiler Express [105] | Rapid | 2015 | 94 °C 3 s | 60 °C 60 s | 25–28 | 15 L | |
| VeriFiler Plus [106] | Traditional | 2018 | 96 °C 10 s | 62 °C 90 s (2 cycles) 59 °C 90 s (27 cycles) | 29 | 25 L | |
| VeriFiler Express [107] | Rapid | 2021 | 96 °C 10 s | 59 C 16 s | 65 °C 29 s | 25–28 | 25 L |
The concept of unchanging PCR conditions makes little sense when the conditions within the tube at the start of the PCR are compared to the conditions at the end of the reaction. The variation in reaction conditions across a PCR run is based on the amount of initial DNA template compared to the number of amplicons present at the culmination of the PCR process and the processivity of the enzyme. At the first cycle of PCR, the amount of template may be small (if 10 cells, then 20 priming sites for each primer when amplifying autosomal STRs), yet the enzyme is at its most active. After 28 cycles of PCR, the number of amplicons will be in the billions, yet the enzyme will have lost much of its activity. The extension time in the initial cycles could be reduced given the activity of the enzyme and the small amount of template, but increased as the template increases and enzyme processivity decreases. Equally, the denaturation temperature could decrease if the amplicons are short in length and there are no longer sections of chromosomal DNA.
The evolution of PCR for DNA profiling has slowed in recent years, despite the characteristic issues of degraded and inhibited samples still being present. Many changes have been made to elements of DNA profiling beyond the PCR programs, and yet the issues of these challenging sample types still remain.
3.1.2. Mitochondrial DNA Testing
Parallel to the advent of STR amplification for DNA profiling, mitochondrial DNA (mtDNA) amplification for forensic analysis was also established in the early 1990s. The targeted amplification of three relatively small hypervariable regions in the human mitochondrial genome, HV1, HV2 and HV3 [108,109], provides an alternative means of identification when traditional sources of nuclear DNA (i.e., body fluids and tissues) are no longer available [17]. The power of mtDNA to provide information where standard methods fail comes from the fact that there can be thousands of mitochondria in each cell, each with 2 to 10 copies of mtDNA. However, unlike the nuclear DNA targeted for STR profiling, mtDNA is only inherited maternally. Thus, mtDNA analysis has the ability to identify maternally related individuals (i.e., siblings), as well as distinguish between individuals from different maternal lines [110]. This means the uses for mtDNA investigations are specialized, rather than being the primary tool for identification. The value of these markers is most evident where mtDNA amplification and analysis has been crucial to the identification of missing persons [18], unidentified human remains [111,112,113,114] and disaster victims [115].
As a result of mtDNA techniques developing alongside STR PCR methods, the PCR process used to amplify the mtDNA targets for forensic analysis is very similar. However, the reasoning behind the conservation of PCR cycling conditions for STR profiling and mtDNA analysis are not the same. While one of the main factors influencing PCR cycling conditions for STR profiling is the number of targets needed to generate highly discriminatory profiles, the cycling conditions used for mtDNA amplification do not face these same restrictions, as far fewer regions are targeted. This is because, prior to the use of massively parallel sequencing (MPS) platforms, all mtDNA amplifications were performed with one pair of primers, as it was not possible to multiplex if the amplicons were going to progress to Sanger sequencing. The smaller number of primers required for mtDNA analysis means there is a flexibility in the annealing stage that is not afforded in STR or SNP amplification. Most challenges of mtDNA analysis lie beyond the PCR process. The inherent variability in the mitochondrial genome of an individual, known as heteroplasmy [116,117,118], and the increased susceptibility for contamination to occur during the mtDNA extraction process [119] provides substantial challenges for mtDNA interpretation. However, the conservation of cycling conditions that can be seen in mtDNA PCR protocols used over the last three decades (Table 2) can instead be attributed to the push for reliability and reproducibility in the technique. Unlike STR PCR, the conservation of mtDNA amplification processes is likely a result of forensic laboratories adhering to the guidelines put forward by the International Society for Forensic Genetics for mtDNA typing [119,120] as a means of quality control. The need for quality control in mtDNA analysis is intensified due to of the lack of commercially available PCR kits. All commercially available STR PCR kits come with recommended cycling conditions that have been extensively validated by the manufacturer. This means laboratories only need to verify the kit prior to implementation in casework. The lack of commercial mtDNA kits means there is not the same pre-validation behind the PCR conditions used, allowing much more variability in the cycling conditions that can be used for mtDNA amplification between laboratories.

Table 2.
PCR cycling conditions used to amplify mitochondrial DNA and the total reaction volumes. “N/A” is used where the temperature of the annealing step is varied depending on the primers used in each reaction.
Due to the low-template and degraded nature of samples commonly submitted for mtDNA analysis (i.e., bone, teeth and hair), some changes have been made to the number of PCR cycles used to amplify mtDNA targets. The number of additional cycles ranges from 2–12 cycles higher than STR PCR in an effort to maximize the amount of mitochondrial data obtained from these challenging samples (Table 2). However, as mentioned earlier, the addition of extra PCR cycles is not actually a change to the cycling conditions themselves, but rather a repeat of the same uniform conditions. The shorter denaturation, annealing and extension steps used in current mtDNA PCR programs may be attributed to improved PCR instruments and reagents; however, it also provides some evidence to support the suggestion that the timing of the stages can be shortened without affecting target amplification. The challenges that heteroplasmy and contamination continue to present for mtDNA analysis are not dependent on PCR dynamics, and so cannot be remedied by altering the PCR process. Nevertheless, the potential to optimize mtDNA amplification methods by using non-uniform PCR programs that change as the conditions within the PCR tube change still exists.
3.1.3. Single Nucleotide Polymorphism Analysis
While the amplification of single nucleotide polymorphisms (SNPs) for forensic analysis was used in casework before STR profiling [15], the issues associated with creating SNP multiplexes and the substantially lower discrimination powers afforded by SNP profiles saw STR profiling become the primary technique for human identification [126]. However, SNP analysis continued to be developed in response to the need for faster and more reliable techniques for the amplification of highly degraded and trace samples. In addition, there are abundant SNPs on both nuclear and mtDNA, allowing profiling using this technology to target both DNA types [127,128].
In recent years, SNP analysis has emerged as a major player in the world of forensic genetics due to the broad range of information it can provide from small amplicons [129,130,131]. This is because the single nucleotide variations that are targeted by SNP analysis are highly abundant in the human genome as a result of mutagenesis [129]. As the number of SNPs identified to be highly variable in the human genome continues to increase, so too does the power of discrimination between genomes when it is applied to forensic investigation. Given the abundance of forensically relevant SNPs, they are divided into five main categories based on the type of information they can provide to investigators. These categories are identity-informative SNPs (iiSNPS), lineage-informative SNPs (liSNPs), ancestry-informative SNPs (aiSNPs), phenotype-informative SNPs (pISNPs) and pharmacogenetic SNPs [31,132,133]. The combination of these SNPs can give insight into the biogeographical ancestry (BGA) and externally visible characteristics (EVCs) of an individual, in addition to the human identification capabilities that are afforded by STR and mtDNA assays [134], which highlights the value of SNP analysis to forensic investigations.
There are clear similarities between the previously mentioned mtDNA and STR PCR programs and the cycling conditions used for SNP analysis (Table 3). However, this is not surprising, given that SNP multiplex was developed in an attempt to overcome some of the challenges associated with the analysis of degraded and low-template mtDNA and STR samples. The variation in PCR cycling conditions between SNP assays (Table 3) can mostly be attributed to the different combinations of SNP targets that are used, and thus the different combinations of primers that require different annealing conditions to ensure all targets are amplified equally and efficiently. While early assays targeted 12 individual SNPs [135], more recently developed SNP panels, such as those used for modern forensic investigative genetic genealogy (FIGG) applications, target up to 1 million individual SNPs in a single assay [136]. The addition of large numbers of new SNP targets is also the reason why the cycling conditions have been highly conserved, as all targets need to be amplified in a balanced manner, and thus, have highly stringent annealing conditions. The increased number of PCR cycles can once again be attributed to the push for increased amounts of genetic data to be obtained from SNP analysis, with many samples submitted for SNP analysis containing low amounts of DNA or highly degraded material [127,128]. However, as previously noted, the variation in cycle number only signifies extra repeats of the same uniform PCR conditions and not actual changes to the cycling conditions themselves. As with STR profiling, changes have been made to other elements of the PCR process (i.e., commercial buffers, enzymes, instruments) to help further improve the amplification efficiency, and thus the overall quality, of the SNP data obtained [137].

Table 3.
PCR cycling conditions used to amplify single nucleotide polymorphisms (SNPs) and the total reaction volumes.
Despite the success of SNP analysis in recent years, the technique has begun to move away from traditional target amplification methods, such as PCR, to new technologies that sequence the DNA molecules directly without any need for PCR. Such technologies, which include the Oxford Nanopore minION [142], Illumina NovaSeq 6000 [143] and silicon microchips [67], provide higher-throughput methods for SNP analysis using MPS platforms. Therefore, while the potential does exist for SNP PCR programs to be further optimized by employing gradually changing cycling conditions that account for the changes in enzyme activity across a run, the shift in focus from traditional Sanger sequencing platforms that require PCR to next generation technologies that do not indicates that perhaps the continued evolution of SNP analysis lies beyond the PCR process.
While the PCR processes used for STR profiling, mtDNA testing and SNP analysis have all evolved substantially since they were first introduced to forensic science, the once-rapid evolution has slowed substantially in recent years. Though this is not necessarily an issue for mtDNA and SNP analysis, the characteristic issues of interpreting degraded and inhibited samples for STR profiling still present significant limitations for DNA profiling. However, other disciplines, such as medical science, have continued to see rapid evolution of the PCR process, and as a result, highly successful PCR variants have been developed and integrated into their workflows.
3.2. Evolution of PCR Cycling Conditions in Other Disciplines
Touchdown PCR differs from the traditional PCR method as it involves a stepwise decrease in annealing temperature in each cycle of PCR [144]. This process developed in response to a demand for increased primer binding specificity in clinical research [144,145,146]. The annealing temperature in the first PCR cycle is substantially higher than the temperature at which the primers will melt, which is approximately 60–66 °C [145,146]. This reduces the amount of off-target primer binding due to their stringent binding requirements only allowing them to bind to exactly complementary regions on the target DNA at such high temperatures [145,146]. With each cycle, the temperature of denaturation decreases (typically by a standard amount such as 1 °C) as the highly specific regions amplified in early cycles become template DNA strands of only the target regions [145,146]. Importantly, touchdown PCR has been developed for both multiplex and uniplex reactions, but it has yet to be integrated into forensic casework, due in part to laboratories following the PCR cycling conditions provided by the manufacturers of the DNA profiling kits for standard PCR setups [145,146], and the fact that primers within these profiling kits are all designed to work optimally within a small range of temperature.
Similarly, gradient PCR was developed to aid in the determination of the optimal annealing temperature to increase primer specificity during PCR [147]. However, it is not technically a modification of the traditional PCR program; rather, it relies on changing the annealing temperatures by using a heating block that possesses a temperature gradient across its surface during the annealing stages. The determination of the optimal annealing temperature has played an important role in clinical pathology, where it has aided in the development of PCR protocols for SARS-CoV-2 (COVID-19) [148]. Additionally, the technique can also be applied to optimize the denaturation and extension phases of PCR, but it has only been tested for a single amplification, rather than a multiplex [84,149]. Thus, research into the application of gradient PCR to multiplex systems must be conducted to determine the viability of integrating this technique into forensic practice.
4. Recent Developments in PCR for DNA Profiling
4.1. Increased Speed
The significant increases in reaction speed and sample throughput that have been seen over the last three decades can largely be attributed to the improved technology within PCR instruments. In the early days of PCR, the process was highly labor-intensive, requiring the manual movement of individual tubes between water baths and the addition of DNA polymerase at the beginning of each cycle (as the enzymes were not yet thermostable) [2]. The production of “Mr. Cycle” in 1987 [150], the first automated thermal cycler that heated and cooled using a metal block, and the use of a thermostable DNA polymerase (Thermus aquaticus) in 1988 [5], are the two features that pushed PCR into a new era. As significant advancements in technology were made, the PCR process became faster, with mineral oil being replaced by heated lids (preventing sample evaporation and condensation) and bulky plumbing compressors traded for Peltier blocks with temperature control algorithms that can heat and cool rapidly [150,151]. The significant increase in machine ramp rates and heat dispersal rates within the PCR process in recent years has allowed the speed and throughput of PCR to increase substantially.
Technological advancements in the thermal cyclers resulted in improved PCR instrument ramp rates and faster sample heating and cooling, which have helped speed up the time required to generate DNA profiles considerably. In conjunction, the identification of mutant DNA polymerases with improved processivity (amplification efficiency) and their application to forensic analysis has helped to speed up PCR [60,152,153,154]. Zhang et al. [154] developed a PCR enhancer cocktail containing non-ionic detergent, l-carnitine and d-(+)-trehalose, which worked to improve the performances of both commercially available Taq polymerases (i.e., AmpliTaq Gold and HotStarTaq Plus) and mutant DNA polymerases. While there has been substantial evidence that mutant polymerases can overcome inhibition more effectively than AmpliTaq Gold and amplify DNA more efficiently [60,152,153,154], they are yet to be adopted into commercially available STR kits. However, it is important to note that while DNA polymerases with increased processivity have successfully been used for DNA profiling, the challenges associated with sub-optimal samples still have not been overcome using these methods.
The largest alteration to PCR cycling conditions in forensic science came with the introduction of Rapid DNA instruments to laboratory workflows. While the features and importance of Rapid DNA have already been discussed, it is important to emphasize that Rapid DNA was designed for high-quality samples (i.e., reference samples) and has only had varying success when used with sub-optimal samples [95]. This highlights an important limitation of Rapid DNA: it requires good-quality DNA samples to generate informative DNA profiles [96].
4.2. Increased Sensitivity and Discrimination Power
The five-fold decrease in the amount of starting DNA required to generate probative STR profiles [19,32,35,99,103,106] in the last 30 years can be attributed to a number of factors: improved commercial DNA extraction processes ensuring inhibitors are removed, better buffer components in the STR PCR kits, and the increased number of STRs targeted for DNA profiling. The early multiplex PCRs for DNA profiling targeted only three loci, but quickly increased to seven loci, resulting in match probabilities of 1 in 50 million [54]. The identification and designation of the 13 core CODIS loci (USA) and 12 European Standard Set (ESS) loci then increased the possible match probability for informative DNA profiles to exceed 1 in 1 trillion [55,155]. The substantial increase in discrimination power of the widely used STR kits can be directly attributed to the increased number of loci targeted in recent years (see Table 4). The introduction of new, highly polymorphic loci, such as SE33, which has a high mutation rate of 0.64% [156,157,158], to the core CODIS and ESS loci in STR kits, as well as the increase in discrimination power that they afforded, have been key driving factors of DNA profiling in the last three decades. However, this push for increased sensitivity and discrimination powers has diminished in recent years as match probabilities now considerably exceed the total global population by many orders of magnitude. While the amount of DNA required to generate informative DNA profiles has decreased and the discrimination powers from good-quality genetic material has increased, the match probabilities (and likelihood ratios) of sub-optimal STR profiles generated from low-template and degraded samples are still limited by poor success rates.

Table 4.
Number of loci targeted by commercially available STR Kits.
4.3. Optimization of Commercially Available Kits
With the introduction of the new STR kit AmpFlSTR MinifilerTM by Applied Biosystems (Foster City, USA) in 2007 came a new and improved PCR buffer [23,102]. This new buffer allowed the pH of the amplification reaction to be maintained through the presence of potassium (K+) and ammonium (NH4+) ions, which work to optimize and sustain the processivity of the DNA polymerase [159,160]. The identification of inhibitor-tolerant DNA polymerases [60,152,153] and their application to STR PCR saw a substantial increase in the number of complete DNA profiles generated from samples that contain PCR inhibitors [66]. Through the use of improved buffers, DNA polymerases and primers in commercially available STR kits, the amount of PCR product generated from samples has increased, and thus, the chances of producing informative DNA profiles from trace, inhibited or degraded samples have also increased.
While the success of generating DNA profiles from challenging samples has increased substantially in recent years, the quality of the profiles generated can still be less than ideal. The characteristic features of sub-optimal samples, such as small peak heights, allelic drop-in and drop-out, inhibited locus amplification, and heterozygote imbalance, are still present in the DNA profiles generated from these samples [23,36,57,161,162]. Therefore, while the optimization of commercial STR kit components has improved the ability to generate DNA profiles from trace materials, the presence of stochastic effects and the challenges they pose during profile interpretation still have not been overcome [23,36,52,57,161].
4.4. PCR Amplification Kinetics
The advent of fully quantitative PCR (qPCR) provided insight into how the environment within a PCR tube changes across the course of the reaction [51]. This insight has allowed researchers to identify how different components of the reaction, such as the primer concentration, magnesium (Mg2+) concentration, pH, DNA polymerase processivity and polymerase inhibitor tolerance, affect the kinetic behavior of the amplification [64,68,163]. Furthermore, the amount of template DNA present, and the quality of this DNA, have been found to affect PCR kinetics [68]. In addition to furthering our understanding of the amplification process, qPCR has allowed a range of PCR inhibitors to be identified, such as hemoglobin [65,164,165,166], proteases [167], calcium [65,167] and ethylenediaminetetraacetic acid (EDTA) [168], along with the mechanisms through which they inhibit DNA amplification [169].
The significant developments in our understanding of PCR kinetics and how they change as a reaction progresses have provided critical insight into the mechanisms of DNA amplification. Importantly, this level of understanding was not something that was known when PCR was developed as a tool for DNA profiling, and while the process has been effectively validated over the last few decades [170], there are likely ways that it can be made more efficient. Due to dramatic improvements in qPCR system sensitivity and our deeper understanding of how the composition within a PCR tube changes across the course of a PCR program, the opportunity now exists to monitor how amplification kinetics change as specific elements of the PCR process are altered. The effects of changes to PCR cycling conditions, such as small changes to timing and temperature, can be carefully monitored, and their influence on amplification efficiency can be clearly defined. Furthermore, the cycling conditions that are found to increase amplification efficiency could then be used to improve the amplification of degraded and/or inhibited samples. The concept here would be to revisit the PCR cycling conditions and adjust the first cycles to account for low template DNA, and then alter the cycling conditions in stages through to the last cycle where enzyme processivity has slowed dramatically.
5. Recent Developments in PCR beyond Forensics
Lab-on-a-Chip devices for biological analyses outside laboratories and their integration with intelligent computer systems in recent years have significantly improved the ability to manipulate and monitor the PCR process [171]. The development of electrowetting-on-dielectric (EWOD) digital microfluidics devices came as an alternative to the time-, labor- and cost-intensive procedures required to study biomolecular interactions for clinical diagnostics and pathogen detection [171,172]. EWOD devices utilize fluorescent feedback detected through optical sensors to monitor reactions in real-time. Since they were first pioneered, EWOD systems have continued to evolve, with detectors becoming more sensitive, and only picolitres of sample are required for successful quantitative analysis [173,174]. The rapid heating and cooling rates, low reagent consumption, high sensitivity, high throughput, portability, and short run times of the EWOD platforms makes them ideal for PCR assays [175,176,177]. Additionally, the multidisciplinary collaboration in recent years between biochemists, engineers and computer scientists has enabled the development of high-level biological programming languages that can be integrated into microfluidic devices to detect and monitor reactions in real-time, and control the devices using feedback loops [177,178]. Importantly, multiplex PCR procedures have been designed and optimized for these microfluidic systems, and they have been proven to have high amplification efficiencies [175,179,180]. These high amplification efficiencies can be attributed to the intelligent computer software developed to make decisions in real time based on the fluorescence feedback obtained [177,178]. Multiple different DNA samples have also successfully been amplified concurrently and monitored in real-time using a digital microfluidic platform [179]. The development of microfluidic multiplex PCR assays highlights the viability and value of integrating intelligent computer systems into traditional biochemical processes to optimize them based on real-time feedback. The potential exists for such a system to be developed for forensic DNA profiling that could monitor amplification efficiency in real time and alter the PCR cycling conditions to generate an ideal amount of amplified product for DNA profiling.
6. Remaining Challenges in PCR for DNA Profiling
While the PCR programs used for STR PCR have been shown over the years to be reliable and robust, their largest pitfall is that they require large quantities of good-quality genetic material to produce informative DNA profiles. The development and improvement of commercially available PCR kits and instruments in recent years has significantly improved the speed and sensitivity of the technique, but the once-strong drive for improvement has slowed despite the characteristic issues of degraded and inhibited samples still being present. Many changes have been made to elements of DNA profiling beyond the PCR programs; however, they have not been overly successful in improving our ability to profile challenging DNA samples. With trace DNA samples being one of the most commonly submitted sample types in many jurisdictions, any improvement in the success rate of DNA profiling these samples would be valuable to forensic investigations. Rapid DNA and commercial STR kits have provided evidence to suggest that changes to the timing and temperatures used for PCR (either in a univariant or gradient form) could be made to increase amplification efficiency and improve the quality of DNA profiles produced. This begs the question: what changes can be made to PCR cycling conditions to finally improve DNA profiling success rates from challenging samples?
Gradual changes have been successfully made to the denaturation and polymerization steps of a PCR without compromising the evidentiary value of the DNA profiles generated in a recent proof-of-concept study [181]. While this was done using ideal amounts of pristine DNA, it does suggest that the PCR process could be altered to improve the DNA profiles obtained from characteristically challenging samples. Given the success of incorporating machine learning into biological processes in microfluidics [177,178], an exciting future direction would be to integrate such a system into the process of setting PCR cycling conditions for DNA profiling. Such a system would require a means to monitor the PCR as it progresses through real-time fluorescence feedback. A method to obtain this real-time fluorescence feedback was recently suggested that involved combining a standard STR reaction and a qPCR into a single tube [182]. Furthermore, the smart system would also require a means to use the fluorescence information in a machine learning algorithm to adjust the cycling conditions on a per-cycle basis, and a means to do this on a per-sample basis. Such work is in its inception, but may provide exciting avenues in the future.
Author Contributions
Conceptualization, C.M., D.T. and A.L.; writing—original draft preparation, C.M. and A.L.; writing—review and editing, C.M., D.T. and A.L.; visualization, C.M. and A.L.; supervision, D.T. and A.L.; project administration, A.L.; funding acquisition, C.M., D.T. and A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a Flinders University Research Scholarship and funding was provided from grants from the Attorney General’s Department through Forensic Science SA and the Ross Vining Research Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to acknowledge Oliva Handt for the valuable feedback she provided in relation to this work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lorenz, T.C. Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. J. Vis. Exp. 2012, 63, e3998. [Google Scholar] [CrossRef]
- Microbiology, B.O. The polymerase chain reaction: An overview and development of diagnostic PCR protocols at the LCDC. Can. J. Infect. Dis. 1991, 2, 89–91. [Google Scholar] [CrossRef]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51 Pt 1, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Jeffreys, A.J.; Wilson, V.; Neumann, R.; Keyte, J. Amplification of human minisatellites by the polymerase chain reaction: Towards DNA fingerprinting of single cells. Nucleic Acids Res. 1988, 16, 10953–10971. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5’----3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Kloss-Brandstätter, A.; Richards, M.B.; Yao, Y.-G.; Logan, I. The case for the continuing use of the revised Cambridge Reference Sequence (rCRS) and the standardization of notation in human mitochondrial DNA studies. J. Hum. Genet. 2014, 59, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Jeffreys, A.J.; Werrett, D.J. Forensic application of DNA ‘fingerprints’. Nature 1985, 318, 577–579. [Google Scholar] [CrossRef]
- Comey, C.T.; Budowle, B.; Adams, D.E.; Baumstark, A.L.; Lindsey, J.A.; Presley, L.A. PCR amplification and typing of the HLA DQ α gene in forensic samples. J. Forensic Sci. 1993, 38, 239–249. [Google Scholar] [CrossRef]
- Harrington, C.S.; Dunaiski, V.; Williams, K.E.; Fowler, C. HLA DQ α typing of forensic specimens by amplification restriction fragment polymorphism (ARFP) analysis. Forensic Sci. Int. 1991, 51, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, A.D.; Budowle, B.; Daselaar, P. PCR-amplification and detection of the human D1S80 VNTR locus. Amplification conditions, population genetics and application in forensic analysis. Int. J. Leg. Med. 1993, 105, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Sajantila, A.; Budowle, B.; Ström, M.; Johnsson, V.; Lukka, M.; Peltonen, L.; Ehnholm, C. PCR amplification of alleles at the DIS80 locus: Comparison of a Finnish and a North American Caucasian population sample, and forensic casework evaluation. Am. J. Hum. Genet. 1992, 50, 816–825. [Google Scholar] [PubMed]
- Jeffreys, A.J.; Wilson, V.; Thein, S.L. Individual-specific ‘fingerprints’ of human DNA. Nature 1985, 316, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Erlich, H. Part 1.1 in the Beginning: Forensic Applications of DNA Technologies in Silent Witness: Forensic DNA Evidence in Criminal Investigations and Humanitarian Disasters; Oxford University Press: Oxford, UK, 2020. [Google Scholar]
- Stoneking, M.; Hedgecock, D.; Higuchi, R.G.; Vigilant, L.; Erlich, H.A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am. J. Hum. Genet. 1991, 48, 370–382. [Google Scholar] [PubMed]
- Amorim, A.; Fernandes, T.; Taveira, N. Mitochondrial DNA in human identification: A review. PeerJ 2019, 7, e7314. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Ivanov, P.L.; Kimpton, C.; Piercy, R.; Benson, N.; Tully, G.; Evett, I.; Hagelberg, E.; Sullivan, K. Identification of the remains of the Romanov family by DNA analysis. Nat. Genet. 1994, 6, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Kimpton, C.; Fisher, D.; Watson, S.; Adams, M.; Urquhart, A.; Lygo, J.; Gill, P. Evaluation of an automated DNA profiling system employing multiplex amplification of four tetrameric STR loci. Int. J. Leg. Med. 1994, 106, 302–311. [Google Scholar] [CrossRef]
- Butler, J.M. The future of forensic DNA analysis. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140252. [Google Scholar] [CrossRef]
- Werrett, D.J. The National DNA Database. Forensic Sci. Int. 1997, 88, 33–42. [Google Scholar] [CrossRef]
- Martin, P.D.; Schmitter, H.; Schneider, P.M. A brief history of the formation of DNA databases in forensic science within Europe. Forensic Sci. Int. 2001, 119, 225–231. [Google Scholar] [CrossRef]
- Butler, J.M. Advanced Topics in Forensic DNA Typing: Methodology; Elsevier Science: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Panneerchelvam, S.; Norazmi, M.N. Forensic DNA profiling and database. Malays. J. Med. Sci. 2003, 10, 20–26. [Google Scholar] [PubMed]
- Jakovski, Z.; Ajanovska, R.J.; Stankov, A.; Poposka, V.; Bitoljanu, N.; Belakaposka, V. The power of forensic DNA data bases in solving crime cases. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e275–e276. [Google Scholar] [CrossRef]
- Ge, J.; Eisenberg, A.; Budowle, B. Developing criteria and data to determine best options for expanding the core CODIS loci. Investig. Genet. 2012, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Fereday, L.; Morling, N.; Schneider, P.M. The evolution of DNA databases—Recommendations for new European STR loci. Forensic Sci. Int. 2006, 156, 242–244. [Google Scholar] [CrossRef]
- Margarita, G.; María Victoria, L.; Carmela, P.; Antonio, S.; Angel, C. Ethical-legal problems of DNA databases in criminal investigation. J. Med. Ethics 2000, 26, 266. [Google Scholar] [CrossRef]
- Williams, R.; Johnson, P. Inclusiveness, effectiveness and intrusiveness: Issues in the developing uses of DNA profiling in support of criminal investigations. J. Law. Med. Ethics 2005, 33, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Wallace, H.M.; Jackson, A.R.; Gruber, J.; Thibedeau, A.D. Forensic DNA databases–Ethical and legal standards: A global review. Egypt. J. Forensic Sci. 2014, 4, 57–63. [Google Scholar] [CrossRef]
- Butler, J.M. Recent advances in forensic biology and forensic DNA typing: INTERPOL review 2019–2022. Forensic Sci. Int. Synerg. 2023, 6, 100311. [Google Scholar] [CrossRef]
- Corporation, P. PowerPlex® 21 System for Use on the Applied Biosystems® Genetic Analyzers. Available online: https://www.promega.com/-/media/files/resources/protocols/technical-manuals/tmd/powerplex-21-system-protocol.pdf?rev=7db853167600419eb6ddb19c0e88d4ab&sc_lang=en (accessed on 13 April 2023).
- Ludeman, M.J.; Zhong, C.; Mulero, J.J.; Lagacé, R.E.; Hennessy, L.K.; Short, M.L.; Wang, D.Y. Developmental validation of GlobalFiler™ PCR amplification kit: A 6-dye multiplex assay designed for amplification of casework samples. Int. J. Leg. Med. 2018, 132, 1555–1573. [Google Scholar] [CrossRef]
- Technologies, L. AmpFlSTR® Identifiler® Direct PCR Amplification Kit User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_065522.pdf (accessed on 22 May 2023).
- Technologies, L. GlobalFiler™ and GlobalFiler™ IQC PCR Amplification Kits: User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/4477604.pdf (accessed on 22 May 2023).
- Gill, P. Application of low copy number DNA profiling. Croat. Med. J. 2001, 42, 229–232. [Google Scholar] [PubMed]
- Bonsu, D.O.M.; Higgins, D.; Austin, J.J. Forensic touch DNA recovery from metal surfaces—A review. Sci. Justice 2020, 60, 206–215. [Google Scholar] [CrossRef]
- Burrill, J.; Daniel, B.; Frascione, N. A review of trace “Touch DNA” deposits: Variability factors and an exploration of cellular composition. Forensic Sci. Int. Genet. 2019, 39, 8–18. [Google Scholar] [CrossRef]
- Tozzo, P.; Mazzobel, E.; Marcante, B.; Delicati, A.; Caenazzo, L. Touch DNA Sampling Methods: Efficacy Evaluation and Systematic Review. Int. J. Mol. Sci. 2022, 23, 15541. [Google Scholar] [CrossRef]
- van Oorschot, R.A.; Ballantyne, K.N.; Mitchell, R.J. Forensic trace DNA: A review. Investig. Genet. 2010, 1, 14. [Google Scholar] [CrossRef]
- Cook, R.; Mitchell, N.; Henry, J. Assessment of Diamond™ Nucleic Acid Dye for the identification and targeted sampling of latent DNA in operational casework. Forensic Sci. Int. Genet. 2021, 55, 102579. [Google Scholar] [CrossRef] [PubMed]
- Dziak, R.; Peneder, A.; Buetter, A.; Hageman, C. Trace DNA Sampling Success from Evidence Items Commonly Encountered in Forensic Casework. J. Forensic Sci. 2018, 63, 835–841. [Google Scholar] [CrossRef]
- Castella, V.; Mangin, P. DNA profiling success and relevance of 1739 contact stains from caseworks. Forensic Sci. Int. Genet. Suppl. Ser. 2008, 1, 405–407. [Google Scholar] [CrossRef]
- Wong, H.Y.; Tan, J.; Lim, Z.G.; Kwok, R.; Lim, W.; Syn, C.K.-C. DNA profiling success rates of commonly submitted crime scene items. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 597–599. [Google Scholar] [CrossRef]
- Mapes, A.A.; Kloosterman, A.D.; de Poot, C.J. DNA in the Criminal Justice System: The DNA Success Story in Perspective. J. Forensic Sci. 2015, 60, 851–856. [Google Scholar] [CrossRef]
- Mapes, A.A.; Kloosterman, A.D.; van Marion, V.; de Poot, C.J. Knowledge on DNA Success Rates to Optimize the DNA Analysis Process: From Crime Scene to Laboratory. J. Forensic Sci. 2016, 61, 1055–1061. [Google Scholar] [CrossRef]
- Raymond, J.J.; van Oorschot, R.A.H.; Gunn, P.R.; Walsh, S.J.; Roux, C. Trace DNA success rates relating to volume crime offences. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 136–137. [Google Scholar] [CrossRef]
- Raymond, J.J.; Walsh, S.J.; Van Oorschot, R.A.; Gunn, P.R.; Roux, C. Trace DNA: An Underutilized Resource or Pandora’s Box? A Review of the use of Trace DNA Analysis in the Investigation of Volume Crime. J. Forensic Identif. 2004, 54, 668–686. [Google Scholar]
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef]
- Edwards, A.; Civitello, A.; Hammond, H.A.; Caskey, C.T. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am. J. Hum. Genet. 1991, 49, 746–756. [Google Scholar] [PubMed]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions. Bio/Technology 1993, 11, 1026–1030. [Google Scholar] [CrossRef]
- van Oorschot, R.A.H.; Jones, M.K. DNA fingerprints from fingerprints. Nature 1997, 387, 767. [Google Scholar] [CrossRef]
- Findlay, I.; Taylor, A.; Quirke, P.; Frazier, R.; Urquhart, A. DNA fingerprinting from single cells. Nature 1997, 389, 555–556. [Google Scholar] [CrossRef]
- Kimpton, C.P.; Oldroyd, N.J.; Watson, S.K.; Frazier, R.R.; Johnson, P.E.; Millican, E.S.; Urquhart, A.; Sparkes, B.L.; Gill, P. Validation of highly discriminating multiplex short tandem repeat amplification systems for individual identification. Electrophoresis 1996, 17, 1283–1293. [Google Scholar] [CrossRef]
- Chakraborty, R.; Stivers, D.N.; Su, B.; Zhong, Y.; Budowle, B. The utility of short tandem repeat loci beyond human identification: Implications for development of new DNA typing systems. Electrophoresis 1999, 20, 1682–1696. [Google Scholar] [CrossRef]
- Linacre, A. Review of low template DNA typing. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 549–550. [Google Scholar] [CrossRef]
- Gill, P.; Whitaker, J.; Flaxman, C.; Brown, N.; Buckleton, J. An investigation of the rigor of interpretation rules for STRs derived from less than 100 pg of DNA. Forensic Sci. Int. 2000, 112, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, J.P.; Cotton, E.A.; Gill, P. A comparison of the characteristics of profiles produced with the AMPFlSTR® SGM Plus™ multiplex system for both standard and low copy number (LCN) STR DNA analysis. Forensic Sci. Int. 2001, 123, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, A.D.; Kersbergen, P. Efficacy and limits of genotyping low copy number (LCN) DNA samples by multiplex PCR of STR loci. J. Soc. Biol. 2003, 197, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kermekchiev, M.B.; Kirilova, L.I.; Vail, E.E.; Barnes, W.M. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 2009, 37, e40. [Google Scholar] [CrossRef]
- Niederstätter, H.; Köchl, S.; Grubwieser, P.; Pavlic, M.; Steinlechner, M.; Parson, W. A modular real-time PCR concept for determining the quantity and quality of human nuclear and mitochondrial DNA. Forensic Sci. Int. Genet. 2007, 1, 29–34. [Google Scholar] [CrossRef]
- Yang, I.; Kim, Y.-H.; Byun, J.-Y.; Park, S.-R. Use of multiplex polymerase chain reactions to indicate the accuracy of the annealing temperature of thermal cycling. Anal. Biochem. 2005, 338, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Linacre, A.; Pekarek, V.; Swaran, Y.C.; Tobe, S.S. Generation of DNA profiles from fabrics without DNA extraction. Forensic Sci. Int. Genet. 2010, 4, 137–141. [Google Scholar] [CrossRef]
- Booth, C.S.; Pienaar, E.; Termaat, J.R.; Whitney, S.E.; Louw, T.M.; Viljoen, H.J. Efficiency of the Polymerase Chain Reaction. Chem. Eng. Sci. 2010, 65, 4996–5006. [Google Scholar] [CrossRef]
- Opel, K.L.; Chung, D.; McCord, B.R. A study of PCR inhibition mechanisms using real time PCR. J. Forensic Sci. 2010, 55, 25–33. [Google Scholar] [CrossRef]
- Hedman, J.; Dufva, C.; Norén, L.; Ansell, C.; Albinsson, L.; Ansell, R. Applying a PCR inhibitor tolerant DNA polymerase blend in forensic DNA profiling. Forensic Sci. Int. Genet. Suppl. Series 2011, 3, e349–e350. [Google Scholar] [CrossRef]
- Cornelis, S.; Fauvart, M.; Gansemans, Y.; Vander Plaetsen, A.-S.; Colle, F.; Wiederkehr, R.S.; Deforce, D.; Stakenborg, T.; Van Nieuwerburgh, F. Multiplex STR amplification sensitivity in a silicon microchip. Sci. Rep. 2018, 8, 9853. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- Kanokwongnuwut, P.; Martin, B.; Taylor, D.; Kirkbride, P.; Linacre, A. How many cells are required for successful DNA profiling? Forensic Sci. Int. Genet. 2021, 51, 102453. [Google Scholar] [CrossRef] [PubMed]
- Moretti, T.R.; Baumstark, A.L.; Defenbaugh, D.A.; Keys, K.M.; Smerick, J.B.; Budowle, B. Validation of short tandem repeats (STRs) for forensic usage: Performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. J. Forensic Sci. 2001, 46, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Chang, C.-W.; Lagacé, R.E.; Calandro, L.M.; Hennessy, L.K. Developmental Validation of the AmpFℓSTR® Identifiler® Plus PCR Amplification Kit: An Established Multiplex Assay with Improved Performance. J. Forensic Sci. 2012, 57, 453–465. [Google Scholar] [CrossRef]
- Ensenberger, M.G.; Hill, C.R.; McLaren, R.S.; Sprecher, C.J.; Storts, D.R. Developmental validation of the PowerPlex(®) 21 System. Forensic Sci. Int. Genet. 2014, 9, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, L.K.; Mehendale, N.; Chear, K.; Jovanovich, S.; Williams, S.; Park, C.; Gangano, S. Developmental validation of the GlobalFiler® express kit, a 24-marker STR assay, on the RapidHIT® System. Forensic Sci. Int. Genet. 2014, 13, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Gopinath, S.; Lagacé, R.E.; Norona, W.; Hennessy, L.K.; Short, M.L.; Mulero, J.J. Developmental validation of the GlobalFiler® Express PCR Amplification Kit: A 6-dye multiplex assay for the direct amplification of reference samples. Forensic Sci. Int. Genet. 2015, 19, 148–155. [Google Scholar] [CrossRef]
- Vraneš, M.; Scherer, M.; Elliott, K. Development and validation of the Investigator® Quantiplex Pro Kit for qPCR-based examination of the quantity and quality of human DNA in forensic samples. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e518–e519. [Google Scholar] [CrossRef]
- Holt, A.; Wootton, S.C.; Mulero, J.J.; Brzoska, P.M.; Langit, E.; Green, R.L. Developmental validation of the Quantifiler(®) HP and Trio Kits for human DNA quantification in forensic samples. Forensic Sci. Int. Genet. 2016, 21, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, P.; Hill, V.; Narayanan, J. Design of FRET-TaqMan probes for multiplex real-time PCR using an internal positive control. Biotechniques 2009, 46, 519–524. [Google Scholar] [CrossRef]
- Didenko, V.V. DNA probes using fluorescence resonance energy transfer (FRET): Designs and applications. Biotechniques 2001, 31, 1106–1121. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.; Laurin, N. Development of a fast PCR protocol enabling rapid generation of AmpFℓSTR® Identifiler® profiles for genotyping of human DNA. Investig. Genet. 2012, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-H. Chapter 9—Amplification of Nucleic Acids. In Diagnostic Molecular Biology; Shen, C.-H., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 215–247. [Google Scholar]
- Hao, L.; Xie, J.; Chen, S.; Wang, S.; Gong, Z.; Ling, K.S.; Guo, L.; Fan, Z.; Zhou, T. A multiple RT-PCR assay for simultaneous detection and differentiation of latent viruses and apscarviroids in apple trees. J. Virol. Methods 2016, 234, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.; Dorneles, E.M.S.; Diniz, C.; Abreu, V.; Sousa, C.; Alves, J.; Carneiro, A.; Bagano, P.; Spier, S.; Barh, D.; et al. Quadruplex PCR assay for identification of Corynebacterium pseudotuberculosis differentiating biovar Ovis and Equi. BMC Vet. Res. 2017, 13, 290. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Zhang, X.; Gao, Y.; Song, S.; Xu, D.; Yan, L. Development and application of multiplex PCR method for simultaneous detection of seven viruses in ducks. BMC Vet. Res. 2019, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, J.; Jurisic, V.; Tosic, N.; Mrdjanovic, J.; Perin, B.; Pavlovic, S.; Djordjevic, N. Optimization of PCR conditions for amplification of GC-Rich EGFR promoter sequence. J. Clin. Lab. Anal. 2013, 27, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Mann, T.; Humbert, R.; Dorschner, M.; Stamatoyannopoulos, J.; Noble, W.S. A thermodynamic approach to PCR primer design. Nucleic Acids Res. 2009, 37, e95. [Google Scholar] [CrossRef]
- Vallone, P.M.; Butler, J.M. AutoDimer: A screening tool for primer-dimer and hairpin structures. Biotechniques 2004, 37, 226–231. [Google Scholar] [CrossRef]
- Holleley, C.E.; Geerts, P.G. Multiplex Manager 1.0: A cross-platform computer program that plans and optimizes multiplex PCR. Biotechniques 2009, 46, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M.; Devaney, J.M.; Marino, M.A.; Vallone, P.M. Quality control of PCR primers used in multiplex STR amplification reactions. Forensic Sci. Int. 2001, 119, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.G.; Wang, M.X.; Song, P.; Mao, S.; Wang, Y.; Yang, Y.; Luo, J.; Ren, S.; Zhang, D.Y. Designing highly multiplex PCR primer sets with Simulated Annealing Design using Dimer Likelihood Estimation (SADDLE). Nat. Commun. 2022, 13, 1881. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Ai, Y. Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis. Biochim. Biophys. Acta 2010, 1804, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Wages, J.M., Jr. Polymerase Chain Reaction. Encycl. Anal. Sci. (Second. Ed.) 2005, 243–250. [Google Scholar] [CrossRef]
- Karantzeni, I.; Ruiz, C.; Liu, C.C.; Licata, V.J. Comparative thermal denaturation of Thermus aquaticus and Escherichia coli type 1 DNA polymerases. Biochem. J. 2003, 374, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, A.J.; Hurth, C.; Yang, J.; Cai, Z.; Moran, N.; Lee-Edghill, J.G.; Nordquist, A.; Lenigk, R.; Estes, M.D.; Haley, J.P.; et al. Integrated Microfluidic System for Rapid Forensic DNA Analysis: Sample Collection to DNA Profile. Anal. Chem. 2010, 82, 6991–6999. [Google Scholar] [CrossRef]
- LaRue, B.L.; Moore, A.; King, J.L.; Marshall, P.L.; Budowle, B. An evaluation of the RapidHIT(®) system for reliably genotyping reference samples. Forensic Sci. Int. Genet. 2014, 13, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.; Henry, J.; Taylor, D. Analysis of mixed DNA profiles from the RapidHIT™ ID platform using probabilistic genotyping software STRmix™. Forensic Sci. Int. Genet. 2022, 58, 102664. [Google Scholar] [CrossRef]
- Hennessy, L.K.; Franklin, H.; Li, Y.; Buscaino, J.; Chear, K.; Gass, J.; Mehendale, N.; Williams, S.; Jovanovich, S.; Harris, D.; et al. Developmental validation studies on the RapidHIT™ Human DNA Identification System. Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e7–e8. [Google Scholar] [CrossRef]
- Greenspoon, S.A.; Lytle, P.J.; Turek, S.A.; Rolands, J.M.; Scarpetta, M.A.; Carr, C.D. Validation of the PowerPlex 1.1 loci for use in human identification. J. Forensic Sci. 2000, 45, 677–683. [Google Scholar] [CrossRef]
- Corporation, P. PowerPlex® 2.1 System Technical Manual. Available online: https://www.promega.com/-/media/files/resources/profiles-in-dna/302/the-geneprint-powerplex-system-for-the-fbi-selection-of-thirteen-codis-core-str-loci.pdf?la=en (accessed on 2 June 2023).
- ThermoFischer Scientific. AmpFlSTR® SGM Plus® PCR Amplification Kit. Available online: https://tools.thermofisher.com/content/sfs/manuals/cms_041049.pdf (accessed on 2 June 2023).
- Corporation, P. PowerPlex® 16 System. Available online: https://www.promega.com/-/media/files/resources/protocols/technical-manuals/tmd/powerplex-16-system-protocol.pdf?rev=390c9836b26044b19ce1f709fc245352&sc_lang=en (accessed on 2 June 2023).
- ThermoFischer Scientific. AmpFlSTR® Identifiler® PCR Amplification Kit. Available online: https://tools.thermofisher.com/content/sfs/manuals/cms_041201.pdf (accessed on 2 June 2023).
- Technologies, L. AmpFlSTR™ MiniFiler™ PCR Amplification Kit User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_042748.pdf (accessed on 2 June 2023).
- ThermoFischer Scientific. AmpFlSTR® Identifiler® Plus PCR Amplification Kit User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/4440211_AmpFlSTR_IdentifilerPlus_UG.pdf (accessed on 22 May 2023).
- Technologies, L. AmpFlSTR® NGM SElect™ Express PCR Amplification Kit User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_104061.pdf (accessed on 2 June 2023).
- Technologies, L. GlobalFiler™ Express PCR Amplification Kit User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/4477672_GlobalFilerExpress_UG.pdf (accessed on 22 May 2023).
- ThermoFischer Scientific. VeriFiler™ Plus PCR Amplification Kit. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0017493_VeriFilerPlusPCRAmpKit_UG.pdf (accessed on 2 June 2023).
- Technologies, L. VeriFiler™ Express PCR Amplification Kit User Guide. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/100043588_VFE_UG.pdf (accessed on 2 June 2023).
- Greenberg, B.D.; Newbold, J.E.; Sugino, A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene 1983, 21, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Lutz, S.; Wittig, H.; Weisser, H.J.; Heizmann, J.; Junge, A.; Dimo-Simonin, N.; Parson, W.; Edelmann, J.; Anslinger, K.; Jung, S.; et al. Is it possible to differentiate mtDNA by means of HVIII in samples that cannot be distinguished by sequencing the HVI and HVII regions? Forensic Sci. Int. 2000, 113, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Budowle, B.; Allard, M.W.; Wilson, M.R.; Chakraborty, R. Forensics and Mitochondrial DNA: Applications, Debates, and Foundations. Annu. Rev. Genom. Hum. Genet. 2003, 4, 119–141. [Google Scholar] [CrossRef]
- Ríos, L.; García-Rubio, A.; Martínez, B.; Alonso, A.; Puente, J. Identification process in mass graves from the Spanish Civil War II. Forensic Sci. Int. 2012, 219, e4–e9. [Google Scholar] [CrossRef] [PubMed]
- Piccinini, A.; Coco, S.; Parson, W.; Cattaneo, C.; Gaudio, D.; Barbazza, R.; Galassi, A. World War One Italian and Austrian soldier identification project: DNA results of the first case. Forensic Sci. Int. Genet. 2010, 4, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Ossowski, A.; Diepenbroek, M.; Kupiec, T.; Bykowska-Witowska, M.; Zielińska, G.; Dembińska, T.; Ciechanowicz, A. Genetic Identification of Communist Crimes’ Victims (1944–1956) Based on the Analysis of One of Many Mass Graves Discovered on the Powazki Military Cemetery in Warsaw, Poland. J. Forensic Sci. 2016, 61, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Handt, O.; Richards, M.; Trommsdorff, M.; Kilger, C.; Simanainen, J.; Georgiev, O.; Bauer, K.; Stone, A.; Hedges, R.; Schaffner, W.; et al. Molecular genetic analyses of the Tyrolean Ice Man. Science 1994, 264, 1775–1778. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.J.; Li, Y.Z.; Yu, X.G.; Li, L.; Wu, D.Y.; Zhou, J.; Man, T.Y.; Yang, G.; Yan, J.W.; Cai, D.Q.; et al. Preliminary DNA identification for the tsunami victims in Thailand. Genom. Proteom. Bioinform. 2005, 3, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Montiel, R.; Sierra, B.; Bettencourt, C.; Fernandez, E.; Alvarez, L.; Lima, M.; Abade, A.; Aluja, M.P. Understanding Differences Between Phylogenetic and Pedigree-Derived mtDNA Mutation Rate: A Model Using Families from the Azores Islands (Portugal). Mol. Biol. Evol. 2005, 22, 1490–1505. [Google Scholar] [CrossRef]
- Sekiguchi, K.; Kasai, K.; Levin, B.C. Inter-and intragenerational transmission of a human mitochondrial DNA heteroplasmy among 13 maternally-related individuals and differences between and within tissues in two family members. Mitochondrion 2003, 2, 401–414. [Google Scholar] [CrossRef]
- Calloway, C.D.; Reynolds, R.L.; Herrin, G.L., Jr.; Anderson, W.W. The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. Am. J. Hum. Genet. 2000, 66, 1384–1397. [Google Scholar] [CrossRef] [PubMed]
- Bär, W.; Brinkmann, B.; Budowle, B.; Carracedo, A.; Gill, P.; Holland, M.; Lincoln, P.J.; Mayr, W.; Morling, N.; Olaisen, B.; et al. Guidelines for Mitochondrial DNA Typing. Vox Sang. 2000, 79, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Parson, W.; Gusmão, L.; Hares, D.R.; Irwin, J.A.; Mayr, W.R.; Morling, N.; Pokorak, E.; Prinz, M.; Salas, A.; Schneider, P.M.; et al. DNA Commission of the International Society for Forensic Genetics: Revised and extended guidelines for mitochondrial DNA typing. Forensic Sci. Int. Genet. 2014, 13, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.M.; Hopgood, R.; Gill, P. Identification of human remains by amplification and automated sequencing of mitochondrial DNA. Int. J. Legal Med. 1992, 105, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Handt, O.; Krings, M.; Ward, R.H.; Pääbo, S. The retrieval of ancient human DNA sequences. Am. J. Hum. Genet. 1996, 59, 368–376. [Google Scholar] [PubMed]
- Berger, C.; Parson, W. Mini-midi-mito: Adapting the amplification and sequencing strategy of mtDNA to the degradation state of crime scene samples. Forensic Sci. Int. Genet. 2009, 3, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Lee, H.Y.; Park, S.J.; Yang, W.I.; Shin, K.-J. Modified Midi- and Mini-Multiplex PCR Systems for Mitochondrial DNA Control Region Sequence Analysis in Degraded Samples. J. Forensic Sci. 2013, 58, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Cooley, A.M. Mitochondrial DNA Analysis. In Forensic DNA Analysis: Methods and Protocols; Cupples Connon, C., Ed.; Springer: New York, NY, USA, 2023; pp. 331–349. [Google Scholar]
- Butler, J.M.; Coble, M.D.; Vallone, P.M. STRs vs. SNPs: Thoughts on the future of forensic DNA testing. Forensic Sci. Med. Pathol. 2007, 3, 200–205. [Google Scholar] [CrossRef]
- Andréasson, H.; Asp, A.; Alderborn, A.; Gyllensten, U.; Allen, M. Mitochondrial Sequence Analysis for Forensic Identification Using Pyrosequencing Technology. Biotechniques 2002, 32, 124–133. [Google Scholar] [CrossRef]
- Budowle, B. SNP Typing Strategies. Forensic Sci. Int. 2004, 146, S139–S142. [Google Scholar] [CrossRef]
- Novroski, N.M.M.; Cihlar, J.C. Evolution of single-nucleotide polymorphism use in forensic genetics. WIREs Forensic Sci. 2022, 4, e1459. [Google Scholar] [CrossRef]
- Sobrino, B.; Carracedo, A. SNP Typing in Forensic Genetics. In Forensic DNA Typing Protocols; Carracedo, A., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 107–126. [Google Scholar]
- Sanghavi, H. Recent Advancements in SNP Typing Methods Used in Forensic Science. In Advances in Genetic Polymorphisms; Nouha Bouayed, A., Balkiss, A., Eds.; IntechOpen: Rijeka, Yugoslavia, 2023; pp. 13–30. [Google Scholar]
- Budowle, B.; van Daal, A. Forensically relevant SNP classes. Biotechniques 2008, 44, 603–608. [Google Scholar] [CrossRef]
- Wendt, F.R.; Budowle, B. Pharmacogenetics and the postmortem molecular autopsy. WIREs Forensic Sci. 2020, 2, e1361. [Google Scholar] [CrossRef]
- Mehta, B.; Daniel, R.; Phillips, C.; McNevin, D. Forensically relevant SNaPshot® assays for human DNA SNP analysis: A review. Int. J. Leg. Med. 2017, 131, 21–37. [Google Scholar] [CrossRef]
- Tully, G.; Sullivan, K.M.; Nixon, P.; Stones, R.E.; Gill, P. Rapid Detection of Mitochondrial Sequence Polymorphisms Using Multiplex Solid-Phase Fluorescent Minisequencing. Genomics 1996, 34, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Glynn, C.L. Bridging Disciplines to Form a New One: The Emergence of Forensic Genetic Genealogy. Genes 2022, 13, 1381. [Google Scholar] [CrossRef]
- Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001, 11, 3–11. [Google Scholar] [CrossRef]
- Ahmadian, A.; Gharizadeh, B.; Gustafsson, A.C.; Sterky, F.; Nyrén, P.; Uhlén, M.; Lundeberg, J. Single-Nucleotide Polymorphism Analysis by Pyrosequencing. Anal. Biochem. 2000, 280, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Yamamoto, Y.; Doi, Y.; Takata, T.; Ishikawa, T.; Imabayashi, K.; Yoshitome, K.; Miyaishi, S.; Ishizu, H. A new 39-plex analysis method for SNPs including 15 blood group loci. Forensic Sci. Int. 2004, 144, 45–57. [Google Scholar] [CrossRef]
- Divne, A.-M.; Allen, M. A DNA microarray system for forensic SNP analysis. Forensic Sci. Int. 2005, 154, 111–121. [Google Scholar] [CrossRef]
- McNevin, D.; Bate, A.; Daniel, R.; Walsh, S.J. A preliminary mitochondrial DNA SNP genotyping assay for inferring genealogy. Aust. J. Forensic Sci. 2011, 43, 39–51. [Google Scholar] [CrossRef]
- Cornelis, S.; Gansemans, Y.; Deleye, L.; Deforce, D.; Van Nieuwerburgh, F. Forensic SNP Genotyping using Nanopore MinION Sequencing. Sci. Rep. 2017, 7, 41759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xue, J.; Tan, M.; Chen, D.; Xiao, Y.; Liu, G.; Zheng, Y.; Wu, Q.; Liao, M.; Lv, M.; et al. An MPS-Based 50plex Microhaplotype Assay for Forensic DNA Analysis. Genes 2023, 14, 865. [Google Scholar] [CrossRef] [PubMed]
- Don, R.H.; Cox, P.T.; Wainwright, B.J.; Baker, K.; Mattick, J.S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991, 19, 4008. [Google Scholar] [CrossRef]
- Coyle, P.V.; Ong, G.M.; O’Neill, H.J.; McCaughey, C.; De Ornellas, D.; Mitchell, F.; Mitchell, S.J.; Feeney, S.A.; Wyatt, D.E.; Forde, M.; et al. A touchdown nucleic acid amplification protocol as an alternative to culture backup for immunofluorescence in the routine diagnosis of acute viral respiratory tract infections. BMC Microbiol. 2004, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Touchdown Polymerase Chain Reaction (PCR). Cold Spring Harb. Protoc. 2018, 2018, pdb-prot095133. [Google Scholar] [CrossRef] [PubMed]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Won, J.; Choi, B.Y.; Lee, C.J. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp. Mol. Med. 2020, 52, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Prezioso, V.R.; Jahns, A. Using Gradient PCR to Determine the Optimum Annealing Temperature; Eppendorf Scientific: Hamburg, Germany, 2000. [Google Scholar]
- ThermoFischer Scientific. PCR Thermal Cyclers Education. Available online: https://www.thermofisher.com/au/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-thermal-cyclers.html#:~:text=Peltier%20block%20elements%20are%20a,and%20better%20predict%20sample%20temperatures (accessed on 18 September 2023).
- Liu, H.; Fang, Y.; Su, X.; Wang, Y.; Ji, M.; Xing, H.; Gao, Y.; Zhang, Y.; He, N. Temperature control algorithm for polymerase chain reaction (PCR) instrumentation based upon improved hybrid fuzzy proportional integral derivative (PID) control. Instrum. Sci. Technol. 2023, 51, 109–131. [Google Scholar] [CrossRef]
- Nilsson, M.; Grånemo, J.; Buś, M.M.; Havsjö, M.; Allen, M. Comparison of DNA polymerases for improved forensic analysis of challenging samples. Forensic Sci. Int. Genet. 2016, 24, 55–59. [Google Scholar] [CrossRef]
- Yamagami, T.; Ishino, S.; Kawarabayasi, Y.; Ishino, Y. Mutant Taq DNA polymerases with improved elongation ability as a useful reagent for genetic engineering. Front. Microbiol. 2014, 5, 108670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kermekchiev, M.B.; Barnes, W.M. Direct DNA amplification from crude clinical samples using a PCR enhancer cocktail and novel mutants of Taq. J. Mol. Diagn. 2010, 12, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Dognaux, S.; Larmuseau, M.H.D.; Jansen, L.; Heylen, T.; Vanderheyden, N.; Bekaert, B.; Noel, F.; Decorte, R. Allele frequencies for the new European Standard Set (ESS) loci and D1S1677 in the Belgian population. Forensic Sci. Int. Genet. 2012, 6, e75–e77. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M.; Hill, C.R.; Kline, M.C.; Bastisch, I.; Weirich, V.; McLaren, R.S.; Storts, D.R. SE33 variant alleles: Sequences and implications. Forensic Sci. Int. Genet. Suppl. Ser. 2011, 3, e502–e503. [Google Scholar] [CrossRef]
- Davis, C.; Ge, J.; King, J.; Malik, N.; Weirich, V.; Eisenberg, A.J.; Budowle, B. Variants observed for STR locus SE33: A concordance study. Forensic Sci. Int. Genet. 2012, 6, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.; Shrivastava, P.; Dash, H.R.; Kaitholia, K.; Sahajpal, V.; Sahoo, S.; Srivastava, V.; Surekha Rani, H.; Mishra, A.; Choudhary, S.K.; et al. Assessment of significance and forensic relevance of SE33 (ACTBP2) locus in five Indian populations. Gene Rep. 2021, 24, 101293. [Google Scholar] [CrossRef]
- Karunanathie, H.; Kee, P.S.; Ng, S.F.; Kennedy, M.A.; Chua, E.W. PCR enhancers: Types, mechanisms, and applications in long-range PCR. Biochimie 2022, 197, 130–143. [Google Scholar] [CrossRef]
- Klenow, H.; Henningsen, I. Effect of monovalent cations on the activity of the DNA polymerase of Escherichia coli B. Eur. J. Biochem. 1969, 9, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Balding, D.J.; Buckleton, J. Interpreting low template DNA profiles. Forensic Sci. Int. Genet. 2009, 4, 1–10. [Google Scholar] [CrossRef]
- Balding, D.J. Evaluation of mixed-source, low-template DNA profiles in forensic science. Proc. Natl. Acad. Sci. USA 2013, 110, 12241–12246. [Google Scholar] [CrossRef]
- Sidstedt, M.; Steffen, C.R.; Kiesler, K.M.; Vallone, P.M.; Rådström, P.; Hedman, J. The impact of common PCR inhibitors on forensic MPS analysis. Forensic Sci. Int. Genet. 2019, 40, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Akane, A.; Matsubara, K.; Nakamura, H.; Takahashi, S.; Kimura, K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 1994, 39, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Al-Soud, W.A.; Rådström, P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001, 39, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Hedman, J.; Romsos, E.L.; Waitara, L.; Wadsö, L.; Steffen, C.R.; Vallone, P.M.; Rådström, P. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 2018, 410, 2569–2583. [Google Scholar] [CrossRef] [PubMed]
- Bickley, J.; Short, J.K.; McDowell, D.G.; Parkes, H.C. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett. Appl. Microbiol. 1996, 22, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Rossen, L.; Nørskov, P.; Holmstrøm, K.; Rasmussen, O.F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 1992, 17, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. App. Microb. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR past, present and future. Biotechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Berthier, J. Chapter 8—Biological Applications of EWOD. In Micro-Drops and Digital Microfluidics, 2nd ed.; Berthier, J., Ed.; William Andrew Publishing: Norwich, NY, USA, 2013; pp. 339–366. [Google Scholar]
- Brassard, D.; Malic, L.; Miville-Godin, C.; Normandin, F.; Veres, T. Advanced EWOD-based digital microfluidic system for multiplexed analysis of biomolecular interactions. In Proceedings of the 2011 IEEE 24th International Conference on Micro Electro Mechanical Systems, Cancun, Mexico, 23–27 January 2011; pp. 153–156. [Google Scholar]
- Schwarz, M.A.; Hauser, P.C. Recent developments in detection methods for microfabricated analytical devices. Lab A Chip 2001, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, P.S.; Manz, A. Single-molecule fluorescence detection in microfluidic channels-the Holy Grail in μtAS? Anal. Bioanal. Chem. 2005, 382, 1771–1782. [Google Scholar] [CrossRef]
- Luo, Y.; Bhattacharya, B.B.; Ho, T.Y.; Chakrabarty, K. Design and Optimization of a Cyberphysical Digital-Microfluidic Biochip for the Polymerase Chain Reaction. IEEE Trans. Comput. Aided Des. Integr. Circuits Syst. 2015, 34, 29–42. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, B.; Wu, W. Application of automatic feedback photographing by portable smartphone in PCR. Sens. Actuators B Chem. 2019, 298, 126782. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, L.; Guo, P.; Zhang, Y.; Lan, X.; Xu, W. Research progress of All-in-One PCR tube biosensors based on functional modification and intelligent fabrication. Biosens. Bioelectron. 2024, 246, 115824. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Brisk, P. Simulation of feedback-driven PCR assays on a 2D electrowetting array using a domain-specific high-level biological programming language. Microelectron. Eng. 2015, 148, 110–116. [Google Scholar] [CrossRef]
- Hua, Z.; Rouse, J.L.; Eckhardt, A.E.; Srinivasan, V.; Pamula, V.K.; Schell, W.A.; Benton, J.L.; Mitchell, T.G.; Pollack, M.G. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal. Chem. 2010, 82, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yu, C.; Li, S.; Meng, J.; Li, T.; Cheng, J.; Li, J. A droplet-based multivolume microfluidic device for digital polymerase chain reaction. Sens. Actuators B Chem. 2022, 371, 132473. [Google Scholar] [CrossRef]
- McDonald, C.; Taylor, D.; Linacre, A. Towards a smart PCR process. Aust. J. Forensic Sci. 2023, 1–12. [Google Scholar] [CrossRef]
- McDonald, C.; Taylor, D.; Linacre, A. Smart PCR leading to improved DNA profiles. Aust. J. Forensic Sci. 2023. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).