Potential Molecular Mechanisms of Alcohol Use Disorder with Non-Coding RNAs and Gut Microbiota for the Development of Superior Therapeutic Application
Abstract
1. Introduction
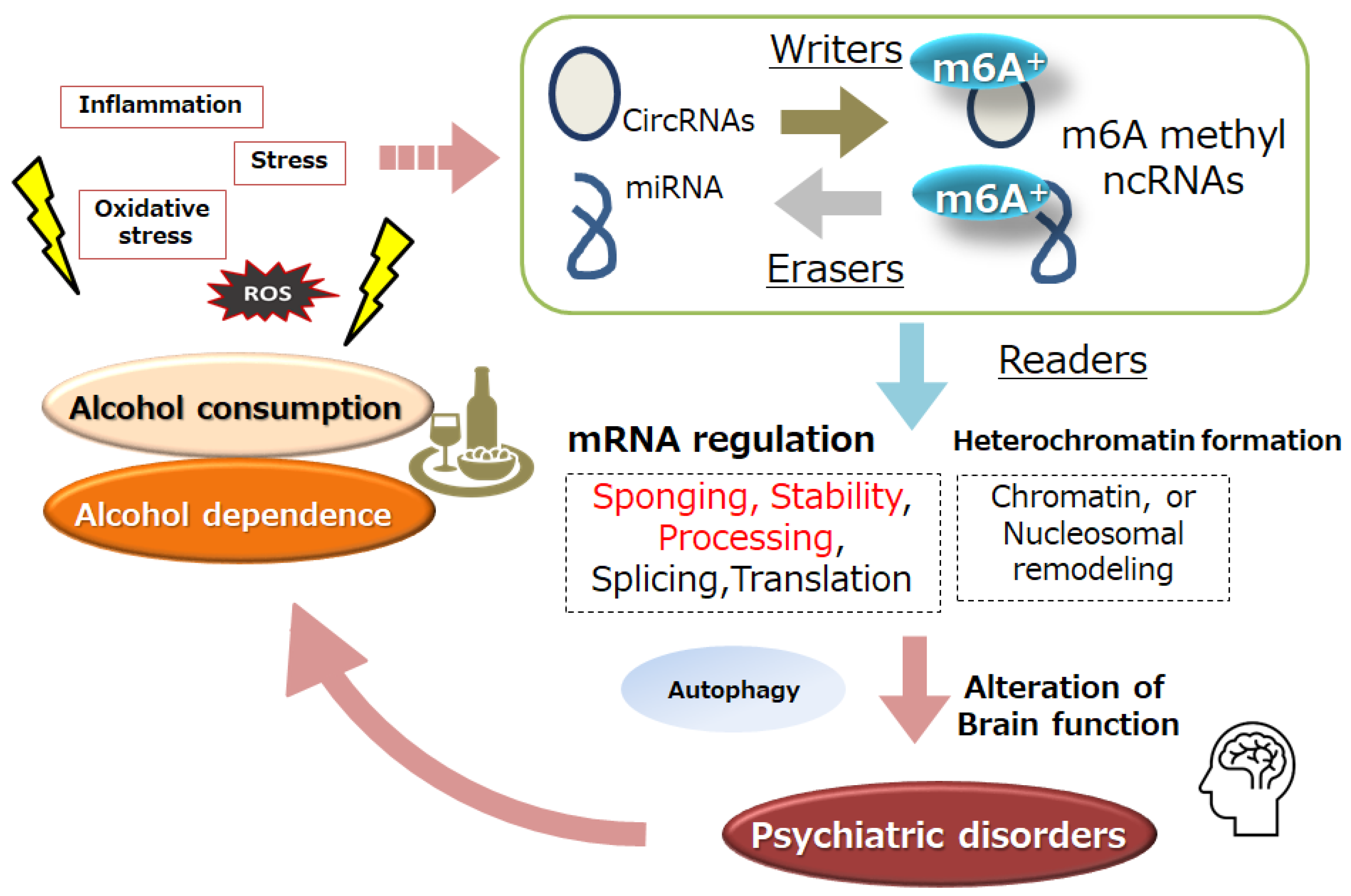
2. Alcohol Dependence and ncRNA
3. Alcohol Dependence and m6A Modification of RNAs
4. Individual Epigenetic Mechanisms for Alcohol Dependency
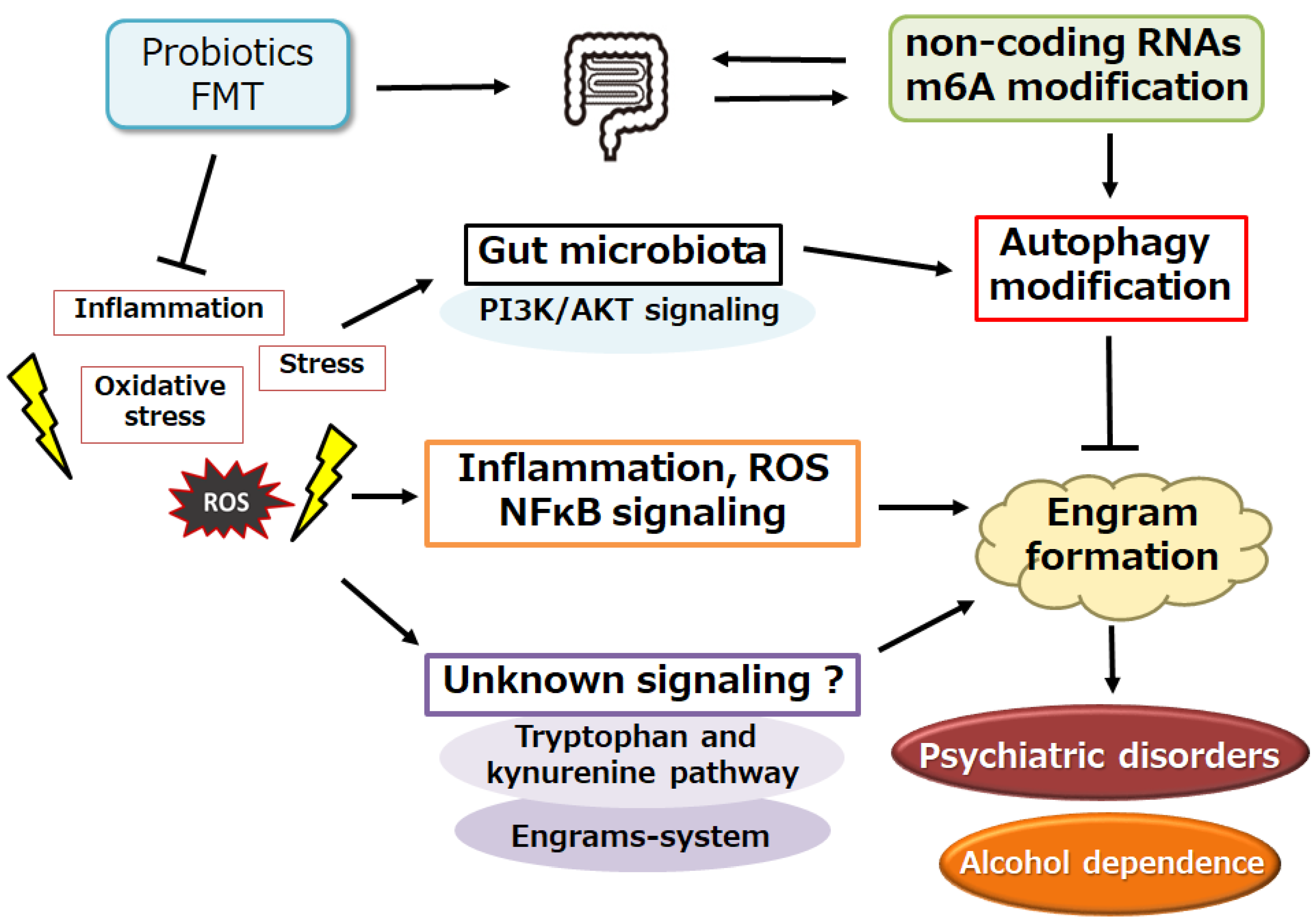
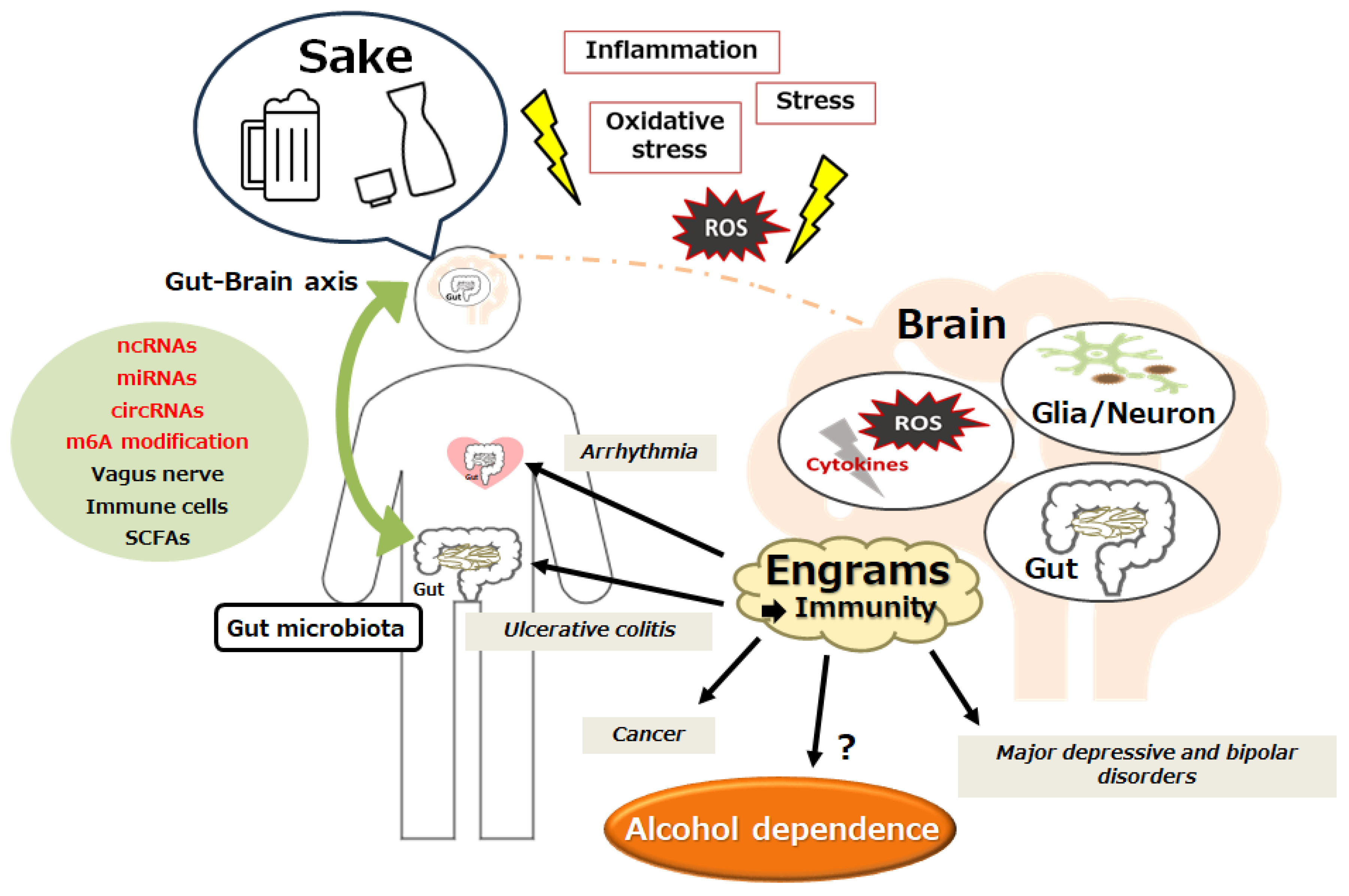
5. Connection between Gut Microbiota and Alcohol Dependency
6. A Possible Tactic with Alteration of Gut Microbiota against Alcohol Dependency
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| BBB | Blood–brain barrier |
| BDNF | brain-derived neurotrophic growth factor |
| CNS | central nervous system |
| circRNA | circular RNA |
| FMT | fecal microbiota transplantation |
| HDAC | histone deacetylase |
| lncRNA | long non-coding RNA |
| mRNA | messenger RNA |
| m6A | methylation of N6 adenosine |
| ncRNA | non-coding RNA |
| NF-kB | nuclear factor kappa B |
| NPY | neuropeptide Y |
| piRNAs | piwi interacting RNAs |
| siRNAs | small interfering RNAs |
| ROS | reactive oxygen species |
| SCFA | short-chain fatty acid |
| siRNA | short interference RNA |
| R-ketamine | arketamine |
| S-ketamine | esketamine |
References
- Ducci, F.; Goldman, D. Genetic approaches to addiction: Genes and alcohol. Addiction 2008, 103, 1414–1428. [Google Scholar] [CrossRef]
- Carvalho, A.F.; Heilig, M.; Perez, A.; Probst, C.; Rehm, J. Alcohol use disorders. Lancet 2019, 394, 781–792. [Google Scholar] [CrossRef]
- Rehm, J.; Shield, K.D. Global burden of disease and the impact of mental and addictive disorders. Curr. Psychiatry Rep. 2019, 21, 10. [Google Scholar] [CrossRef]
- Hasin, D.S.; Wall, M.; Witkiewitz, K.; Kranzler, H.R.; Falk, D.; Litten, R.; Mann, K.; O’Malley, S.S.; Scodes, J.; Robinson, R.L.; et al. Change in non-abstinent WHO drinking risk levels and alcohol dependence: A 3 year follow-up study in the US general population. Lancet Psychiatry 2017, 4, 469–476. [Google Scholar] [CrossRef]
- Mayfield, R.D. Emerging roles for ncRNAs in alcohol use disorders. Alcohol 2017, 60, 31–39. [Google Scholar] [CrossRef][Green Version]
- Slack, F.J.; Chinnaiyan, A.M. The role of non-coding RNAs in oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Kuo, H.C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 2020, 27, 49. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Wang, J.Q.; Liu, Y.R.; Xia, Q.R.; Liang, J.; Wang, J.L.; Li, J. Functional roles, regulatory mechanisms and theranostics applications of ncRNAs in alcohol use disorder. Int. J. Biol. Sci. 2023, 19, 1316–1335. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Berkel, T.D.; Pandey, S.C. Emerging Role of Epigenetic Mechanisms in Alcohol Addiction. Alcohol Clin. Exp. Res. 2017, 41, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Hu, R.; Pei, G.; Zhang, H.; Zhao, Z.; Jia, P. Diverse types of genomic evidence converge on alcohol use disorder risk genes. J. Med. Genet. 2020, 57, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wang, F.; Rosato, A.J.; Farrer, L.A.; Henderson, D.C.; Zhang, H. Prefrontal cortex eQTLs/mQTLs enriched in genetic variants associated with alcohol use disorder and other diseases. Epigenomics 2020, 12, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, Y.; Zhang, Y.; Wang, Q.; Miao, Q.; Xu, Y.; Soares, J.C.; Zhang, X.; Zhang, R. Correlation between the epigenetic modification of histone H3K9 acetylation of NR2B gene promoter in rat hippocampus and ethanol withdrawal syndrome. Mol. Biol. Rep. 2019, 46, 2867–2875. [Google Scholar] [CrossRef] [PubMed]
- Santos-Bezerra, D.P.; Cavaleiro, A.M.; Santos, A.S.; Suemoto, C.K.; Pasqualucci, C.A.; Jacob-Filho, W.; Leite, R.E.P.; Passarelli, M.; Marie, S.K.N.; Machado, U.F.; et al. Alcohol Use Disorder is Associated with Upregulation of MicroRNA-34a and MicroRNA-34c in Hippocampal Postmortem Tissue. Alcohol. Clin. Exp. Res. 2021, 45, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Karabegović, I.; Abozaid, Y.; Maas, S.C.E.; Labrecque, J.; Bos, D.; De Knegt, R.J.; Ikram, M.A.; Voortman, T.; Ghanbari, M. Plasma microRNA signature of alcohol consumption: The Rotterdam Study. J. Nutr. 2023, 152, 2677–2688. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.C. A critical role of brain-derived neurotrophic factor in alcohol consumption. Biol. Psychiatry 2016, 79, 427–429. [Google Scholar] [CrossRef] [PubMed]
- Van Booven, D.; Li, M.; Sunil Rao, J.; Blokhin, I.O.; Dayne Mayfield, R.; Barbier, E.; Heilig, M.; Wahlestedt, C. Alcohol use disorder causes global changes in splicing in the human brain. Transl. Psychiatry 2021, 11, 2. [Google Scholar] [CrossRef]
- Farris, S.P.; Mayfield, R.D. RNA-Seq reveals novel transcriptional reorganization in human alcoholic brain. Int. Rev. Neurobiol. 2014, 116, 275–300. [Google Scholar]
- Liu, Y.; Li, J.; Bu, H.; Wang, H.; Zhang, Y.; Shen, Q.; Li, M.; Lu, Z.; Rong, X.; Zheng, D.; et al. Circular RNA expression alteration identifies a novel circulating biomarker in serum exosomal for detection of alcohol dependence. Addict. Biol. 2021, 26, e13031. [Google Scholar] [CrossRef] [PubMed]
- Vornholt, E.; Drake, J.; Mamdani, M.; McMichael, G.; Taylor, Z.N.; Bacanu, S.; Miles, M.F.; Vladimirov, V.I. Identifying a novel biological mechanism for alcohol addiction associated with circRNA networks acting as potential miRNA sponges. Addict. Biol. 2021, 26, e13071. [Google Scholar] [CrossRef] [PubMed]
- Mohebbati, R.; Sadeghnia, H.R. The Role of microRNAs in Alcoholism: A Meta-analytic Review. Curr. Pharm. Des. 2022, 28, 1926–1931. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Goldman, D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 2018, 17, e12444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bai, W.; Sun, L.; Lin, Y.; Tian, M. Targeting Non-Coding RNA for CNS Injuries: Regulation of Blood-Brain Barrier Functions. Neurochem. Res. 2023, 48, 1997–2016. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Röh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The role of m(6)A/m-RNA methylation in stress Response regulation. Neuron 2018, 99, 389–403.e9. [Google Scholar] [CrossRef]
- Liu, Y.; Koo, J.S.; Zhang, H. Chronic intermittent ethanol exposure-induced m6A modifications around mRNA stop codons of opioid receptor genes. Epigenetics 2024, 19, 2294515. [Google Scholar] [CrossRef]
- Bohnsack, J.P.; Teppen, T.; Kyzar, E.J.; Dzitoyeva, S.; Pandey, S.C. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl. Psychiatry 2019, 9, 34. [Google Scholar] [CrossRef]
- Khani-Habibabadi, F.; Zare, L.; Sahraian, M.A.; Javan, M.; Behmanesh, M. Hotair and Malat1 Long Noncoding RNAs Regulate Bdnf Expression and Oligodendrocyte Precursor Cell Differentiation. Mol. Neurobiol. 2022, 59, 4209–4222. [Google Scholar] [CrossRef]
- Sendinc, E.; Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef]
- Du, T.; Rao, S.; Wu, L.; Ye, N.; Liu, Z.; Hu, H.; Xiu, J.; Shen, Y.; Xu, Q. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect Disord. 2015, 183, 279–286. [Google Scholar] [CrossRef]
- Joshi, K.; Wang, D.O.; Gururajan, A. The m6A-methylome in major depression: A bioinformatic analysis of publicly available datasets. Psychiatry Res. Commun. 2022, 2, 100089. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, Y.; Bai, Y.; Han, B.; Ju, M.; Chen, B.; Yan, L.; Wang, Y.; Zhang, H.; Zhang, H.; et al. N(6)-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1-Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol Psychiatry. 2020, 88, 392–404. [Google Scholar] [CrossRef]
- Lei, C.; Wang, Q. The Progression of N6-methyladenosine Study and Its Role in Neuropsychiatric Disorders. Int. J. Mol. Sci. 2022, 23, 5922. [Google Scholar] [CrossRef]
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 2020, 14, 98. [Google Scholar] [CrossRef]
- Daviet, R.; Aydogan, G.; Jagannathan, K.; Spilka, N.; Koellinger, P.D.; Koellinger, P.D.; Kranzler, H.R.; Nave, G.; Wetherill, R.R. Associations between alcohol consumption and gray and white matter volumes in the UK biobank. Nat. Commun. 2022, 13, 1175. [Google Scholar] [CrossRef]
- McCaul, M.E.; Hutton, H.E.; Stephens, M.A.; Xu, X.; Wand, G.S. Anxiety, anxiety sensitivity, and perceived stress as predictors of recent drinking, alcohol craving, and social stress response in heavy drinkers. Alcohol. Clin. Exp. Res. 2017, 41, 836–845. [Google Scholar] [CrossRef]
- Boden, J.M.; Fergusson, D.M. Alcohol and depression. Addiction 2011, 106, 906–914. [Google Scholar] [CrossRef]
- Ajoolabady, A.; Aslkhodapasandhokmabad, H.; Zhou, Y.; Ren, J. Epigenetic modification in alcohol-related liver diseases. Med. Res. Rev. 2022, 42, 1463–1491. [Google Scholar] [CrossRef]
- de Carvalho, L.M.; Chen, W.Y.; Lasek, A.W. Epigenetic mechanisms underlying stress-induced depression. Int. Rev. Neurobiol. 2021, 156, 87–126. [Google Scholar]
- Misztak, P.; Panczyszyn-Trzewik, P.; Sowa-Kucma, M. Histone deacetylases (HDACs) as therapeutic target for depressive disorders. Pharmacol. Rep. 2018, 70, 398–408. [Google Scholar] [CrossRef]
- Boers, R.; Boers, J.; de Hoon, B.; Kockx, C.; Ozgur, Z.; Molijn, A.; van Ijcken, W.; Laven, J.; Gribnau, J. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018, 28, 88–99. [Google Scholar] [CrossRef]
- Palmisano, M.; Pandey, S.C. Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol 2017, 60, 7–18. [Google Scholar] [CrossRef]
- Ciafrè, S.; Carito, V.; Ferraguti, G.; Greco, A.; Chaldakov, G.N.; Fiore, M.; Ceccanti, M. How alcohol drinking affects our genes: An epigenetic point of view. Biochem. Cell Biol. 2019, 97, 345–356. [Google Scholar] [CrossRef]
- Maier, H.B.; Neyazi, M.; Neyazi, A.; Hillemacher, T.; Pathak, H.; Rhein, M.; Bleich, S.; Goltseker, K.; Sadot-Sogrin, Y.; Even-Chen, O.; et al. Alcohol consumption alters Gdnf promoter methylation and expression in rats. J. Psychiatr. Res. 2020, 121, 1–9. [Google Scholar] [CrossRef]
- Mahna, D.; Puri, S.; Sharma, S. DNA methylation signatures: Biomarkers of drug and alcohol abuse. Mutat. Res. 2018, 777, 19–28. [Google Scholar] [CrossRef]
- Berkel, T.D.M.; Zhang, H.; Teppen, T.; Sakharkar, A.J.; Pandey, S.C. Essential role of histone methyltransferase G9a in rapid tolerance to the anxiolytic effects of ethanol. Int. J. Neuropsychopharmacol. 2019, 22, 292–302. [Google Scholar] [CrossRef]
- Jarmasz, J.S.; Stirton, H.; Basalah, D.; Davie, J.R.; Clarren, S.K.; Astley, S.J.; Del Bigio, M.R. Global DNA methylation and histone posttranslational modifications in human and nonhuman primate brain in association with prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 2019, 43, 1145–1162. [Google Scholar] [CrossRef]
- Chen, W.Y.; Zhang, H.; Gatta, E.; Glover, E.J.; Pandey, S.C.; Lasek, A.W. The histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) alleviates depression-like behavior and normalizes epigenetic changes in the hippocampus during ethanol withdrawal. Alcohol 2019, 78, 79–87. [Google Scholar] [CrossRef]
- Quagebeur, R.; Dalile, B.; Raes, J.; Van Oudenhove, L.; Verbeke, K.; Vrieze, E. The role of short-chain fatty acids (SCFAs) in regulating stress responses, eating behavior, and nutritional state in anorexia nervosa: Protocol for a randomized controlled trial. J. Eat. Disord. 2023, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Chachra, P.; Kennedy, P.; Pena, C.J.; Desouza, L.A.; Nestler, E.J.; Vaidya, V.A. Hippocampal HDAC4 contributes to postnatal fluoxetine-evoked depression-like behavior. Neuropsychopharmacology 2014, 39, 2221–2232. [Google Scholar] [CrossRef] [PubMed]
- Ardizzone, A.; Capra, A.P.; Repici, A.; Lanza, M.; Bova, V.; Palermo, N.; Paterniti, I.; Esposito, E. Rebalancing NOX2/Nrf2 to limit inflammation and oxidative stress across gut-brain axis in migraine. Free Radic Biol. Med. 2024, 213, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Zalar, B.; Haslberger, A.; Peterlin, B. The Role of Microbiota in Depression—A brief review. Psychiatr. Danub. 2018, 30, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Woo, V.; Alenghat, T. Host–microbiota interactions: Epigenomic regulation. Curr. Opin. Immunol. 2017, 44, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Kulyté, A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, P.; Luo, J.; Shen, L.; Zhang, S.; Gu, H.; He, J.; Wang, L.; Zhao, X.; Gan, M.; et al. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microbes 2021, 13, 1862612. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, Y.; Yan, R.; Wang, R.; Zhang, P.; Bai, Z.; Liu, Y.; Ren, Y.; Li, Y.; Jiang, X.; et al. Butyrate suppresses atherosclerotic inflammation by regulating macrophages and polarization via GPR43/HDAC-miRNAs axis in ApoE−/− mice. PLoS ONE 2023, 18, e0282685. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.T.; Abdullah, S.R.; Hussen, B.M.; Younis, Y.M.; Rasul, M.F.; Taheri, M. Role of circular RNAs and gut microbiome in gastrointestinal cancers and therapeutic targets. Noncoding RNA Res. 2023, 9, 236–252. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Chen, J.; Li, Y.; Kuang, Z.; Dende, C.; Raj, P.; Quinn, G.; Hu, Z.; Srinivasan, T.; et al. The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science 2023, 381, 851–857. [Google Scholar] [CrossRef]
- Fardi, F.; Khasraghi, L.B.; Shahbakhti, N.; Salami Naseriyan, A.; Najafi, S.; Sanaaee, S.; Alipourfard, I.; Zamany, M.; Karamipour, S.; Jahani, M.; et al. An interplay between non-coding RNAs and gut microbiota in human health. Diabetes Res. Clin. Pract. 2023, 201, 110739. [Google Scholar] [CrossRef]
- Jabs, S.; Biton, A.; Bécavin, C.; Nahori, M.A.; Ghozlane, A.; Pagliuso, A.; Spanò, G.; Guérineau, V.; Touboul, D.; Gianetto, Q.G.; et al. Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat. Commun. 2020, 11, 1344. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef]
- Kim, H.J.; Jeon, H.J.; Kim, J.Y.; Shim, J.J.; Lee, J.H. Lactiplantibacillus plantarum HY7718 Improves Intestinal Integrity in a DSS-Induced Ulcerative Colitis Mouse Model by Suppressing Inflammation through Modulation of the Gut Microbiota. Int. J. Mol. Sci. 2024, 25, 575. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Chen, W.; Shi, H.; Eren, A.M.; Morozov, A.; He, C.; Luo, G.-Z.; Pan, T. Transcriptome-wide reprogramming of N(6)-methyladenosine modification by the mouse microbiome. Cell Res. 2019, 29, 167–170. [Google Scholar] [CrossRef]
- Zhao, Y.; Qin, F.; Han, S.; Li, S.; Zhao, Y.; Wang, H.; Tian, J.; Cen, X. MicroRNAs in drug addiction: Current status and future perspectives. Pharmacol. Ther. 2022, 236, 108215. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H. RNA m6A Modification Changes in Postmortem Nucleus Accumbens of Subjects with Alcohol Use Disorder: A Pilot Study. Genes 2022, 13, 958. [Google Scholar] [CrossRef]
- Huang, P.; Liu, M.; Zhang, J.; Zhong, X.; Zhong, C. YTHDF1 Attenuates TBI-Induced Brain-Gut Axis Dysfunction in Mice. Int. J. Mol. Sci. 2023, 24, 4240. [Google Scholar] [CrossRef]
- Gonçalves, C.L.; Doifode, T.; Rezende, V.L.; Costa, M.A.; Rhoads, J.M.; Soutullo, C.A. The many faces of microbiota-gut-brain axis in autism spectrum disorder. Life Sci. 2024, 337, 122357. [Google Scholar] [CrossRef]
- Fuenzalida, C.; Dufeu, M.S.; Poniachik, J.; Roblero, J.P.; Valenzuela-Pérez, L.; Beltrán, C.J. Probiotics-based treatment as an integral approach for alcohol use disorder in alcoholic liver disease. Front. Pharmacol. 2021, 12, 729950. [Google Scholar] [CrossRef]
- Martino, C.; Zaramela, L.S.; Gao, B.; Embree, M.; Tarasova, J.; Parker, S.J.; Wang, Y.; Chu, H.; Chen, P.; Lee, K.C.; et al. Acetate reprograms gut microbiota during alcohol consumption. Nat. Commun. 2022, 13, 4630. [Google Scholar] [CrossRef]
- Jadhav, K.S.; Peterson, V.L.; Halfon, O.; Ahern, G.; Fouhy, F.; Stanton, C.; Dinan, T.G.; Cryan, J.F.; Boutrel, B. Gut microbiome correlates with altered striatal dopamine receptor expression in a model of compulsive alcohol seeking. Neuropharmacology 2018, 141, 249–259. [Google Scholar] [CrossRef]
- Flores-Bastías, O.; Karahanian, E. Neuroinflammation produced by heavy alcohol intake is due to loops of interactions between Toll-like 4 and TNF receptors, peroxisome proliferator-activated receptors and the central melanocortin system: A novel hypothesis and new therapeutic avenues. Neuropharmacology 2018, 128, 401–407. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Duong, N.K.; Brocato, E.R.; Bajaj, J.S. Gut-Liver-Brain Axis and Alcohol Use Disorder: Treatment Potential of Fecal Microbiota Transplantation. Alcohol Res. 2024, 44, 1. [Google Scholar] [CrossRef]
- Meng, Y.; Sun, J.; Zhang, G. Pick fecal microbiota transplantation to enhance therapy for major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 2024, 128, 110860. [Google Scholar] [CrossRef]
- Hashimoto, K. Neuroinflammation through the vagus nerve-dependent gut-microbiota-brain axis in treatment-resistant depression. Prog. Brain Res. 2023, 278, 61–77. [Google Scholar]
- Kalkman, H.O. Activation of σ1-Receptors by R-Ketamine May Enhance the Antidepressant Effect of S-Ketamine. Biomedicines 2023, 11, 2664. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, Y.; Zhou, H.; Lu, H.; Zhang Esketamine, Y.; Hua, J.; Liao, X. Esketamine is neuroprotective against traumatic brain injury through its modulation of autophagy and oxidative stress via AMPK/mTOR-dependent TFEB nuclear translocation. Exp. Neurol. 2023, 366, 114436. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, W.; Lin, H.; Gu, X.; Xie, H. The effects of esketamine on the intestinal microenvironment and intestinal microbiota in mice. Hum. Exp. Toxicol. 2023, 42, 9603271231211894. [Google Scholar] [CrossRef]
- Ma, L.; Wang, L.; Chang, L.; Shan, J.; Qu, Y.; Wang, X.; Fujita, Y.; Hashimoto, K. A role of microRNA-149 in the prefrontal cortex for prophylactic actions of (R)-ketamine in inflammation model. Neuropharmacology 2022, 219, 109250. [Google Scholar] [CrossRef]
- Delalle, I. MicroRNAs as Candidates for Bipolar Disorder Biomarkers. Psychiatr. Danub. 2021, 33, 451–455. [Google Scholar]
- Choi, J.L.; Kao, P.F.; Itriago, E.; Zhan, Y.; Kozubek, J.A.; Hoss, A.G.; Banigan, M.G.; Vanderburg, C.R.; Rezvani, A.H.; Latourelle, J.C.; et al. miR-149 and miR-29c as candidates for bipolar disorder biomarkers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017, 174, 315–323. [Google Scholar] [CrossRef]
- Suga, N.; Ikeda, Y.; Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Matsuda, S. Non-Coding RNAs and Gut Microbiota in the Pathogenesis of Cardiac Arrhythmias: The Latest Update. Genes 2023, 14, 1736. [Google Scholar] [CrossRef]
- Nakashima, M.; Suga, N.; Ikeda, Y.; Yoshikawa, S.; Matsuda, S. Circular RNAs, Noncoding RNAs, and N6-methyladenosine Involved in the Development of MAFLD. Non-Coding RNA 2024, 10, 11. [Google Scholar] [CrossRef]
- Ezquer, F.; Quintanilla, M.E.; Morales, P.; Santapau, D.; Munita, J.M.; Moya-Flores, F.; Ezquer, M.; Herrera-Marschitz, M.; Israel, Y. A dual treatment blocks alcohol binge-drinking relapse: Microbiota as a new player. Drug Alcohol. Depend. 2022, 236, 109466. [Google Scholar] [CrossRef]
- Cooper, T.E.; Khalid, R.; Chan, S.; Craig, J.C.; Hawley, C.M.; Howell, M.; Johnson, D.W.; Jaure, A.; Teixeira-Pinto, A.; Wong, G. Synbiotics, prebiotics and probiotics for people with chronic kidney disease. Cochrane Database Syst. Rev. 2023, 10, CD013631. [Google Scholar]
- Yacoub, R.; Nadkarni, G.N.; McSkimming, D.I.; Chaves, L.D.; Abyad, S.; Bryniarski, M.A.; Honan, A.M.; A Thomas, S.; Gowda, M.; He, J.C.; et al. Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study. Exp. Biol. Med. 2019, 244, 505–513. [Google Scholar] [CrossRef]
- Du, Y.; Zhu, Y.J.; Zhou, Y.X.; Ding, J.; Liu, J.Y. Metformin in therapeutic applications in human diseases: Its mechanism of action and clinical study. Mol. Biomed. 2022, 3, 41. [Google Scholar] [CrossRef]
- Tsuji, A.; Ikeda, Y.; Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Morikawa, S.; Nakashima, M.; Asai, T.; Matsuda, S. The Tryptophan and Kynurenine Pathway Involved in the Development of Immune-Related Diseases. Int. J. Mol. Sci. 2023, 24, 5742. [Google Scholar] [CrossRef] [PubMed]
- Håvik, B.; Røkke, H.; Dagyte, G.; Stavrum, A.K.; Bramham, C.R.; Steen, V.M. Synaptic activity-induced global gene expression patterns in the dentate gyrus of adult behaving rats: Induction of immunity-linked genes. Neuroscience 2007, 148, 925–936. [Google Scholar] [CrossRef]
- Ikeda, Y.; Taniguchi, K.; Yoshikawa, S.; Sawamura, H.; Tsuji, A.; Matsuda, S. A budding concept with certain microbiota, anti-proliferative family proteins, and engram theory for the innovative treatment of colon cancer. Explor. Med. 2022, 3, 468–478. [Google Scholar] [CrossRef]
- Suga, N.; Ikeda, Y.; Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Matsuda, S. In Search of a Function for the N6-Methyladenosine in Epitranscriptome, Autophagy and Neurodegenerative Diseases. Neurol. Int. 2023, 15, 967–979. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakashima, M.; Suga, N.; Yoshikawa, S.; Ikeda, Y.; Matsuda, S. Potential Molecular Mechanisms of Alcohol Use Disorder with Non-Coding RNAs and Gut Microbiota for the Development of Superior Therapeutic Application. Genes 2024, 15, 431. https://doi.org/10.3390/genes15040431
Nakashima M, Suga N, Yoshikawa S, Ikeda Y, Matsuda S. Potential Molecular Mechanisms of Alcohol Use Disorder with Non-Coding RNAs and Gut Microbiota for the Development of Superior Therapeutic Application. Genes. 2024; 15(4):431. https://doi.org/10.3390/genes15040431
Chicago/Turabian StyleNakashima, Moeka, Naoko Suga, Sayuri Yoshikawa, Yuka Ikeda, and Satoru Matsuda. 2024. "Potential Molecular Mechanisms of Alcohol Use Disorder with Non-Coding RNAs and Gut Microbiota for the Development of Superior Therapeutic Application" Genes 15, no. 4: 431. https://doi.org/10.3390/genes15040431
APA StyleNakashima, M., Suga, N., Yoshikawa, S., Ikeda, Y., & Matsuda, S. (2024). Potential Molecular Mechanisms of Alcohol Use Disorder with Non-Coding RNAs and Gut Microbiota for the Development of Superior Therapeutic Application. Genes, 15(4), 431. https://doi.org/10.3390/genes15040431





