Abstract
The starch synthase (SS) plays important roles in regulating plant growth and development and responding to adversity stresses. Although the SS family has been studied in many crops, it has not been fully identified in sweet potato and its two related species. In the present study, eight SSs were identified from Ipomoea batatas (I. batata), Ipomoea trifida (I. trifida), and Ipomoea trlioba (I. trlioba), respectively. According to the phylogenetic relationships, they were divided into five subgroups. The protein properties, chromosomal location, phylogenetic relationships, gene structure, cis-elements in the promoter, and interaction network of these proteins were also analyzed; stress expression patterns were systematically analyzed; and real-time polymerase chain reaction (qRT-PCR) analysis was performed. Ipomoea batatas starch synthase (IbSSs) were highly expressed in tuber roots, especially Ipomoea batatas starch synthase 1 (IbSS1) and Ipomoea batatas starch synthase 6 (IbSS6), which may play an important role in root development and starch biosynthesis. At the same time, the SS genes respond to potassium deficiency, hormones, cold, heat, salt, and drought stress. This study offers fresh perspectives for enhancing knowledge about the roles of SSs and potential genes to enhance productivity, starch levels, and resistance to environmental stresses in sweet potatoes.
1. Introduction
Starch is the second-most abundant carbohydrate on earth and an important energy source for human and animal nutrition. Starch mainly consists of amylose (AM) and branched-chain amylopectin (AP) [1,2,3]. AM/AP affects the pasting temperature, solubility, and palate of starch, while the composition of starch is mainly affected by the enzyme starch synthase [4,5,6]. Starch synthase (SS) contains soluble starch synthases (SSS) and granule-bound starch synthase (GBSS), which are highly similar in the C-terminal region and are key players in starch synthesis [7,8,9]. The SSSs are usually composed of a core region with a molecular weight of about 60 kDa, which is essential for catalytic activity, and consist of two conserved structural domains, including the Glyco_transf_5 structural domain (GT5, PF08323) and the Glycos_transf_1 structural domain (GT1, PF00534) [7,10,11,12,13]. The SSs contained Glyco_transf_1 (GT1; PF00534) and Glyco_transf_5 (GT5; PF08323) structural domains [7,11,13,14].
Starch synthesis is a complex biological process, requires the synergistic involvement of several enzymes, and is subject to environmental and developmental regulation [7]. GBSS is a key gene in AM synthesis, with GBSSI functioning mainly in endosperm and pollen tissues and GBSSII functioning mainly in leaves and photosynthetic tissues. At the same time, GBSS is also involved in AP synthesis. For example, by suppressing the expression of GBSSI, the content of AM was reduced, the long chains above degree of polymerization (DP) 100 were lacking in AP, and the chains of DP 6 and DP 7 were slightly reduced in AP; by up-regulating the expression of GBSSI, the content of AM was increased [1]. These were validated in potato, sweet potato, wheat, and rice [15,16]. SSSs play an important role in the process of AP synthesis, in which SSSI is mainly responsible for prolonging the short-branched side chains of DP 6 or DP 7 to DP 8–12 and is also involved in the synthesis of the external short chains of starch clusters [10,17]. The elongated branches of SSSI can be further extended to intermediate lengths by SSSII and alter the content of branched starch in the crop [17]; SSSIII produces longer chains that extend between short-chain polysaccharide clusters [8]; SSSIV was expressed predominantly in leaves, has low sequence identity to starch synthases expressed in the endosperm, and was involved in the process of starch granule initiation or at least controls the number of starch granules in the chloroplasts [18,19,20,21]; SSSV was expressed mainly in spike leaves and seeds, synthesizing transient and storage starch, respectively, and its transcript reaches its maximum at the seed filling stage, which was important for seed starch synthesis [22]. The ratio of AM/AP has an important role in crop quality, flavor, and quality, as well as stress tolerance, and is important for promoting crop yield and stress tolerance. Presently, the SS gene family members have been studied in several plants, such as Arabidopsis [23], rice [24], potato [25,26], and tomato [27].
Sweet potato (Ipomoea batatas (L.)), the world’s seventh largest food crop, is a dicotyledonous annual or perennial plant. It belongs to the family Convolvulaceae, which is grown on a large scale around the world because of its high yield and resistance to abiotic stresses. It is an important root crop, and the main component of its storage roots is starch, which accounts for 50–80% of the dry weight [6] and provides humans with abundant carbohydrates, dietary fiber, vitamins, and trace elements [5,28]. Some studies have shown that inhibition of IbGBSSI gene expression by RNA interference dramatically suppresses AM expression [15,16]. Overexpression of IbSSI affects sweet potato starch content, grain size, and proportion of branched-chain starch [29]. Inhibition of IbSSII gene expression by RNA interference dramatically alters the basic structure of starch and thus its functional properties [30]. Furthermore, IbGBSSI plays an important role in different drought-tolerant cultivars under drought stress [31]. Therefore, SSs may be involved in starch synthesis and abiotic stress responses in sweet potato. However, the SS gene family members have not been identified in sweet potato. Meanwhile, with the release of the genome assemblies of sweet potato and two diploid wild relatives, I. trifida and I. triloba, it will be possible to identify and analyze the SS gene family of sweet potato [28].
A total of 24 SS genes were successfully pinpointed in sweet potato as well as their two diploid wild counterparts. The results of the collinearity analysis showed that there was a strong correlation between sweet potato and its two relatives. The analysis of real-time fluorescence PCR and expression profiles revealed the significant involvement of SS genes in plant growth and response to various environmental challenges such as salt stress and heat stress. In conclusion, our results lay the groundwork for future exploration into the functionalities of sweet potato SS genes.
2. Results
2.1. Identification of SS in Sweet Potato and Two Closely Related Species
Eight SS genes were identified in I. batatas, I. trifida, and I. triloba, respectively, named after “IbSS1-8”, “ItfSS1-8”, and “ItbSS1-8”, respectively, according to their positions on the chromosomes (Table 1). Physicochemical properties analysis showed that the amino acid number of IbSSs ranged from 633 to 1402, the number of amino acids of ItfSSs ranged from 538 to 1391, and the number of amino acids of ItbSSs ranged from 608 to 1349. The molecular weights of IbSSs ranged from 70.69 to 157.43 kDa; the molecular weights of ItfSSs and ItbSSs ranged from 59.38 to 156.75 kDa and 66.70 to 152.42 kDa, respectively. The theoretical potentials of SSs in I. batatas ranged from 5.11 to 7.15, the theoretical potentials of ItfSSs and ItbSSs ranged from 4.96 to 6.56 and 5.13 to 8.31, respectively. In sweet potato, all IbSS genes were unstable except for IbSS2, IbSS6, and IbSS7. ItfSS2, ItfSS4, ItfSS5, and ItfSS8 were stable; the other ItfSS genes were unstable; and ItbSS2, ItbSS5, and ItbSS8 were stable ones. The hydrophilicity of IbSSs ranged from −0.561 to −0.023, the hydrophilicity of ItfSSs ranged from −0.571 to 0.035, and the hydrophilicity of ItbSSs ranged from −0.579 to 0.075. Overall, there was significant variability in the physicochemical properties of I. batatas, I. trifida, and I. triloba.

Table 1.
Characterization of SS in sweet potato, I. trifida, and I. triloba.
The chromosomal localization analysis showed that SSs in I. batatas, I. trifida, and I. triloba were separately distributed on 5 chromosomes (Figure 1A–C). In sweet potato, two SS genes were detected in LG5, LG7, and LG11, and one SS gene was detected in LG9 and LG10; I. trifida and I. triloba showed similar distributions in Chr01, Chr03, Chr08, Chr10, and Chr12, respectively, and it was hypothesized that there might be a one-to-one correspondence. In addition, a collinearity analysis of each species between I. batatas, I. trifida, I. triloba, Arabidopsis thaliana (A. thaliana), Solanum tuberosum L. (S. tuberosum), and Oryza sativaL (O. sativa) was generated. As shown in Figure 1D, a strong collinearity relationship occurred among I. batatas, I. trifida, and I. triloba, and a weak collinearity relationship occurred between I. batatas and other varieties. Among these genes, IbSS7 and IbSS8 were homologous to ItfSS1, ItSS2, ItbSS1, and ItbSS2, respectively; IbSS6 was homologous to ItfSS5 and ItbSS5; IbSS1 and IbSS2 were related to ItfSS7, ItfSS8, ItfSS3, ItfSS4, and ItbSS7, ItbSS8, ItbSS3, and ItbSS4, respectively; IbSS3 and IbSS4 were homologous to ItfSS7, ItfSS8, ItfSS3, ItfSS4, and ItbSS7, ItbSS8, ItbSS3, and ItbSS4, respectively; and IbSS5 was homologous to ItfSS6 and ItbSS6. These results suggested that segmental duplication was present in the evolutionary process from diploid to hexaploid.
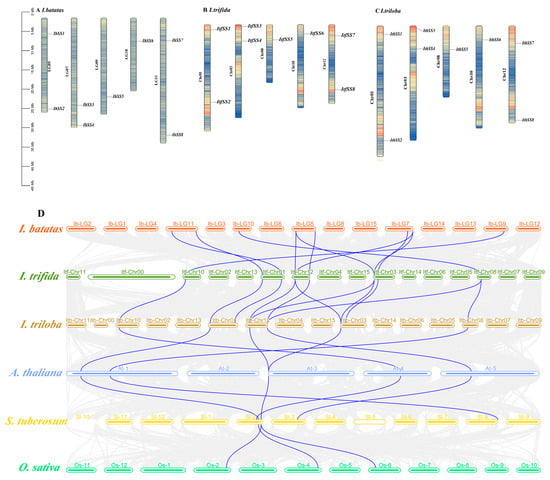
Figure 1.
Chromosome localization and distribution of SSs in I. batatas, I. trifida, and I. triloba (A–C). Chromosome numbers are shown on the left and gene names on the right. (D) Collinearity analyses of SSs in I. batatas, I. trifida, I. triloba, A. thaliana, S. lycopersicum, and O. sativa are indicated by different colors. Orange, light green, brown, light blue, yellow, and green, respectively, represent I. batatas, I. trifida, I. triloba, A. thaliana, S. lycopersicum, and O. sativa.
2.2. Analysis of Phylogenetic Relationships between Sweet Potato and Two Diploid Wild Relatives
In order to study the phylogenetic relationship of SSs among I. batatas, I. trifida, I. triloba, A. thaliana, S. bicolor, O. sativa, and S. lycopersicum, a phylogenetic tree was constructed using MEGA-X, and 55 SSs were classified into five groups (Figure 2). SSs were distributed in varying quantities in all groups; the specific distributions were as follows: (total: I. batatas, I. trifida, I. triloba, A. thaliana, S. bicolor, O. sativa, and S. lycopersicum): SSI (7: 1, 1, 1, 1, 1, 1, and 1); SSII: (14: 2, 2, 2, 1, 3, 3, and 1); SSIII (9: 1, 1, 1, 1, 2, 2, and 1); SSIV (17: 3, 3, 3, 1, 3, 3, and 1); GBSS (8: 1, 1, 1, 1, 2, 1, and 1). Additionally, each IbSS had a homologous gene in I. trifida and I. triloba, respectively.
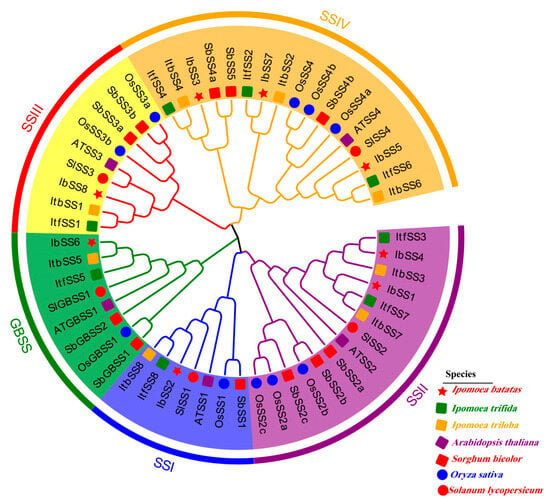
Figure 2.
Phylogenetic analyses of SSs in I. batatas, I. trifida, I. triloba, A. thaliana, S. bicolor, O. sativa, and S. lycopersicum. Based on the evolutionary analysis, the 24 SSs were categorized into five groups (I, II, III, and IV are indicated in blue, purple, red, and orange, respectively), with green indicating granule-bound starch synthases. Red pentagrams indicate eight IbSS in I. batatas, green squares indicate eight ItfSS in I. trifida, orange squares indicate eight ItbSS in I. triloba, purple squares indicate five ATSS in A. thaliana, red squares indicate 11 SbSS in S. bicolor, blue circles indicate 10 OSSs in O. sativa, and red squares indicate five SlSS in S. lycopersicum.
2.3. Conserved Motifs and Exon–Intron Structure Analysis of SS in Sweet Potato and Two Closely Related Species
The protein sequences of I. batatas, I. trifida, and I. triloba were analyzed for conserved motifs by the MEME online tool and Tbtools software, and 10 conserved motifs were identified (Figure 3). The analysis showed that most of the SSs contained these 10 conserved motifs; however, it varies in different subgroups; in group II, IbSS1 and IbSS4 lacked 1 and 2 motifs, respectively. In group III, IbSS8, ItfSS1, and ItbSS1 lacked 2, 1, and 1 motifs, respectively. In group IV, IbSS5 and ItbSS4 lack 2 and 3 motifs, respectively. ItbSS4, which contained 7 conserved motifs; IbSS4, IbSS5, and IbSS8, which contained 8 conserved motifs; ItfSS1, ItbSS1, and IbSS1, which contained 9 conserved motifs.
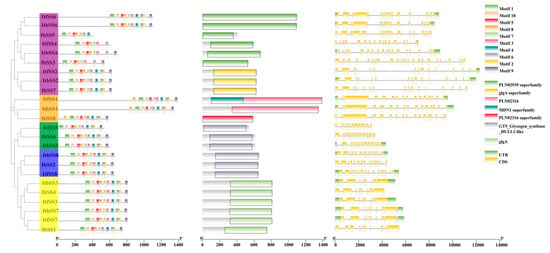
Figure 3.
Analysis of conserved motifs and exon–intron structure of starch synthase (SS) in three species, I. batatas, I. trifida, and I. triloba, revealed interesting findings. The phylogenetic tree indicated that SS can be classified into seven distinct subgroups, with the top ten most prevalent subgroups identified. The SS were divided into seven subgroups and the ten conserved motifs are shown in different colors, purple: SSIV, orange: SSIII, green: GBSS blue: SSI yellow: SSII. The far right represents the exon–intron structure of SS.
Analyzing the exon–intron distribution of IbSS, ItfSS, and ItbSS provides a better understanding of the evolution of the IbSS gene family. Overall, the number of exons and introns in SSs was highly variable (Figure 3). The number of exons ranged from 9 to 24. Additionally, the SSs in the same group have the same number of exons, such as ItfSS3, ItbSS3, IbSS1, and ItbSS7 in the SSII group (all containing 10 exons) and IbSS8, ItfSS1, ItbSS1, ItbSS1, ItfSS6, and ItbSS6 in the SSIII and SSI groups (all containing 18 exons). Interestingly, some exons were more abundant in sweet potato: IbSS5 in IV has 19 exons, while both ItfSS6 and ItbSS6 contain 18 exons; IbSS3 contains 24 exons, while ItfSS4 and ItbSS4 contain 18 and 22 exons, respectively; IbSS5 contains 19 exons, while both ItfSS6 and ItbSS6 contain 18 exons. These results suggested that the differences between exons and introns may lead to evolutionary and functional differences among SSs, and the SS gene family may have become more complex during evolution.
2.4. Analysis of Cis-Acting Elements of SS Promoters in Sweet Potato and Two Closely Related Species
Plant promoter cis-acting elements were closely related to plant development, hormone regulation, and response to adversity. A large number of core promoter elements and generic homeopathic elements were distributed in the promoter region of I. batatas, I. trifida, and I. triloba. Based on the functions of the components, we classified them as core promoter elements, light-responsive elements, hormone responsive elements, and stress response elements, respectively (Figure 4 and Table S1). SSs have a variety of hormonal response elements, such as abscisic acid response element ABRE; auxin response elements TGA and AUxR; and gibberellin response elements P-box, GARE, and TATC-box. In addition, there were abiotic stress response elements, for example, the low-temperature response element LTR; elements that were essential for anaerobic induction; drought response elements; and salt response elements MBS, MYB, and MYC. This indicates that plant growth and development, hormone action, light regulation, and abiotic/biotic stress responses in sweet potato and its two relatives were controlled by SSs.
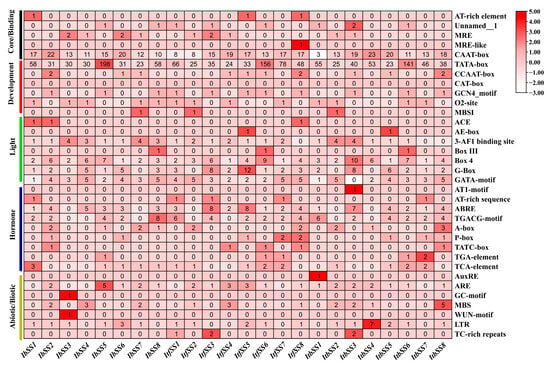
Figure 4.
Analysis of cis−elements in the promoters of SS from I. batatas, I. trifida, and I. triloba. The cis−elements were categorized into six groups. The intensity of the red hue indicates the quantity of cis-elements in the SS promoters.
2.5. Expression Analysis of SS Genes in Sweet Potato and Two Closely Related Species
2.5.1. Expression Analysis in Different Tissues
Based on the transcriptome data from sweet potato flower, fruit, leaf, stem, primary root, firewood root, fibrous root, and tuber root, a sweet potato SS gene expression heatmap was generated (Figure 5A). We found that these IbSSs had different levels of expression in the eight tissues of I. batatas. As can be seen from Figure 5A, seven IbSSs were highly expressed in tuber root, such as IbSS6, IbSS1, and IbSS8. The SSs were more expressed in stems and leaves, such as IbSS1, IbSS5, and IbSS6. Overall, the IbSS6 gene was highest expressed in all tissues of sweet potato except in leaves, where it was lower than the IbSS5 gene.
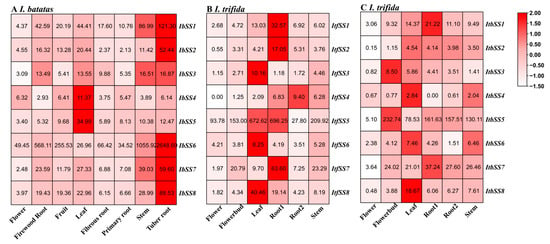
Figure 5.
Expression analysis of SS in different tissues of I. batatas, I. trifida, and I. triloba using RNA-seq. (A) Expression analysis of IbSSs in different tissues. (B) Expression patterns of ItfSSs in the flower, flowerbud, leaf, stem, root1, root2, and stem of I. trifida. (C) Expression patterns of ItbSSs in the flower, flowerbud, leaf, stem, root1, root2, and stem. The FPKM values were shown in the boxes.
We analyzed the expression of SSs in I. trifida and I. triloba (Figure 5B,C). In I. trifida, six genes were highly expressed in root1, especially ItfSS5 and ItfSS7; ItfSSs were also highly expressed in leaves, such as ItfSS5 and ItfSS8. Similarly, in I. triloba, four genes were highly expressed in flower buds, such as ItbSS5 and ItbSS7; ItbSSs were also expressed at a higher level in leaves and root1. Overall, SS5 and SS7 were highly expressed in I. trifida and I. triloba.
2.5.2. Expression Analysis under Potassium Deficiency in Sweet Potato
Figure 6 displays the expression patterns of the eight SS genes in sweet potato, which were identified through transcriptome analysis of the low-k-tolerant “Xushu 32” and low-k-sensitive “Ningzishu 1” varieties with contrasting. The data were retrieved from the PRJNA1013090 dataset in the NCBI repository. During a low-potassium treatment, with the exception of IbSS4, which showed decreased expression in the two cultivars, the remaining seven SSs exhibited increased expression in both varieties.
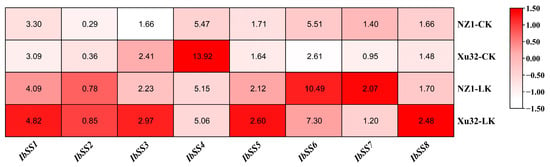
Figure 6.
Expression analysis of sweet potato SS genes under potassium deficiency as determined via RNA-seq. NZ1: “Ningzishu 1”; Xu32: “Xushu 32”. The FPKM values are shown in the color blocks.
2.5.3. Expression Analysis under Hormone Stress
Using the “Xushu 18” RNA-seq data retrieved from the NCBI database (PRJNA511028), the expression patterns of eight SS genes in sweet potato were analyzed in three different tissues following treatments with ABA, SA, and MeJA (Figure 7). In fibrous roots, the expression of eight SS genes was up-regulated after ABA treatment; among them, the expression level of the IbSS6 gene was particularly high; IbSS1 and IbSS6 genes were down-regulated after SA treatment; other genes showed no changes; IbSS1, IbSS4, and IbSS6 were up-regulated after MeJa treatment; among them, IbSS6 was significantly increased in the fibrous roots. In stems, IbSS3 and IbSS8 decreased after ABA treatment; there was no change before and after IbSS5, the other genes were up-regulated to varying degrees, among which IbSS1 and IbSS6 were significantly up-regulated; under SA treatment, except for IbSS3, all other genes were up-regulated, particularly IbSS1, IbSS2, IbSS6, and IbSS8 were significantly increased; after MeJa treatment, all IbSSs genes were up-regulated; among them, IbSS1, IbSS6 and IbSS7 have been significantly increased. In leaves, under ABA and SA treatment, IbSS6 was downregulated, and other genes were upregulated; after MeJa treatment, IbSS2, IbSS4, IbSS6, and IbSS8 were downregulated, and IbSS1 and IbSS7 were raised.
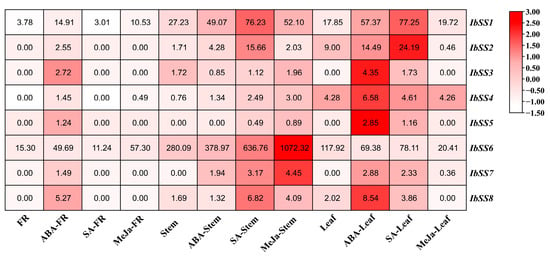
Figure 7.
Analysis of the expression of genes related to sweet potato SSs in fibrous roots (FR), stems, and leaves of sweet potato after hormone treatments using RNA-seq technology. The FPKM values are displayed in colored blocks.
We also utilized the expression levels of I. trifida and I. triloba under ABA, GA, and IAA treatments. After ABA treatment, ItfSS7, ItbSS3, and ItbSS7 were up-regulated. After GA treatment, ItfSS1, ItfSS8, ItbSS2, ItbSS4, and ItbSS6 were up-regulated. Under IAA treatment, ItfSS5, ItbSS1, ItbSS3, ItbSS4, ItbSS6, and ItbSS8 were up-regulated (Figure 8).
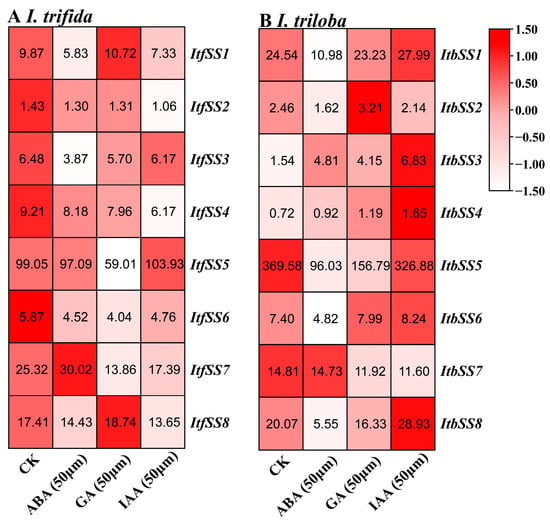
Figure 8.
Expression analysis of ItfSSs (A) and ItbSSs (B) in response to different hormones ABA (50 μm), GA (50 μm) and IAA (50 μm) as determined by RNA-seq. FPKM values are shown in the boxes.
2.5.4. Expression Analysis under Cold Stress
Figure 9 presents the detected expression profiles of the eight IbSSs, using transcriptome data from “Shenshu 28” (a cold-sensitive variety) and “Liaohanshu 21” (a cold-tolerant variety) under cold stress conditions. Under cold stress, in “Shenshu 28”, IbSS4 and IbSS6 were up-regulated, and in “Liaohanshu 21”, IbSS2, IbSS4, IbSS6, and IbSS8 were up-regulated.
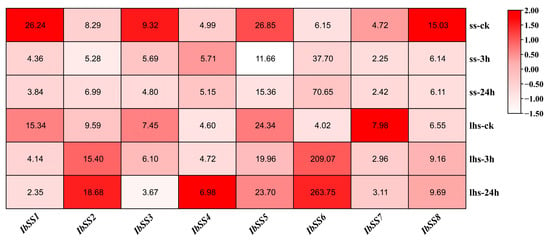
Figure 9.
RNA-seq analysis revealed the gene expression profiles of sweet potato SS genes under cold stress. The cold-sensitive variety “Shenshu 28” was compared to the cold-tolerant variety “Liaohanshu 21”. The color block displays the FPKM values for each sample.
Furthermore, the expression patterns of ItfSSs and ItbSSs were examined in response to cold stress (see Figure 10). With the exception of ItfSS5 and ItbSS2, which showed an increase in expression, the remaining genes demonstrated a decrease in expression levels when subjected to cold stress.
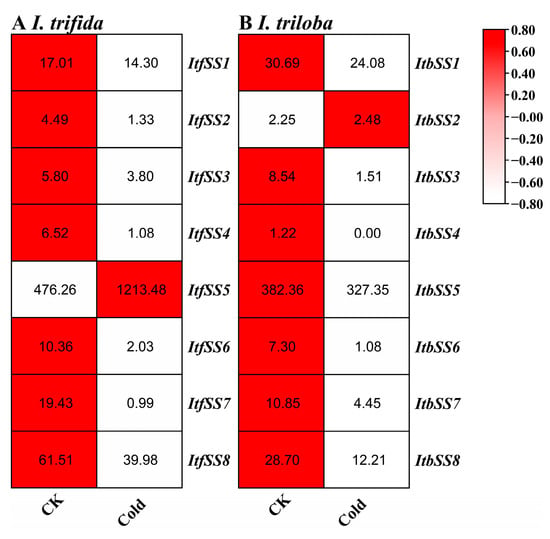
Figure 10.
Expression analysis of ItfSSs (A) and ItbSSs (B) in response to cold stress as determined by RNA-seq. FPKM values are shown in the boxes.
2.5.5. Expression Analysis under Heat Stress
We used the heat-tolerant variety “Guangshu 87” and the heat-sensitive material “Ziluolan” to detect the expression profiles of SS genes in sweet potatoes after heat stress (Figure 11). After heat treatment, the expression levels of the IbSS3, IbSS4, and IbSS5 genes in the fibrous roots of the “Ziluolan” cultivar were up-regulated, while the other genes were down-regulated. Except for IbSS3 and IbSS5, all other genes were down-regulated in the tuberous roots. IbSS3 and IbSS5 were up-regulated in the fibrous roots of the “Gunagshu 87” cultivar, and all other genes were down-regulated, except for IbSS3. The other genes were down-regulated in the tuber roots.
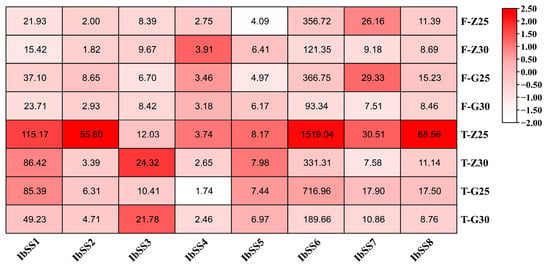
Figure 11.
Gene expression patterns of sweet potato SS genes under heat stress were determined using RNA-seq analysis. F represents fibrous roots, T represents tuberous roots, Z represents the heat-sensitive “Ziluolan”, and G represents the heat-tolerant “Guangshu 87”. The heatmap displays the FPKM values for each gene.
We also analyzed the expression levels of ItfSSs and ItbSSs under 35 °C heat stress (Figure 12). After heat treatment, except for ItfSS3, ItfSS4, ItfSS7, ItbSS3, ItbSS4, and ItbSS7, the transcript levels of the others were increased.
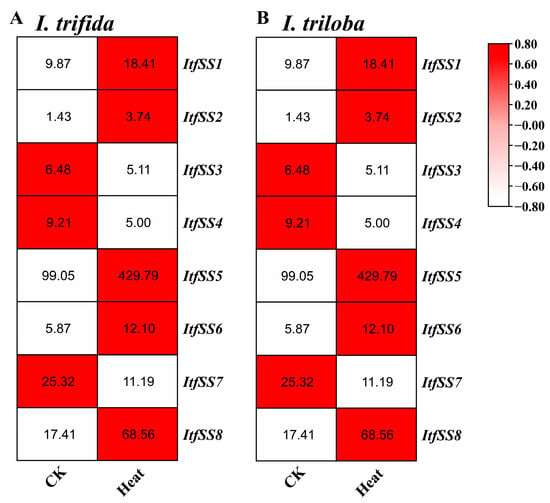
Figure 12.
Expression analysis of ItfSSs (A) and ItbSSs (B) in response to heat stress as determined by RNA-seq. FPKM values are shown in the boxes.
2.5.6. Expression Analysis under Salt and Drought Stresses
As shown in Figure 13, we analyzed the expression levels of IbSSs in the leaf, primary root, and stem under salt and drought treatments. In leaves, after salt treatment, IbSS1 and IbSS6 were up-regulated, and other IbSSs were down-regulated; under drought treatment, IbSS1 and IbSS2 were up-regulated, while other IbSSs were down-regulated. In the primary root, after salt treatment, IbSS4 and IbSS6 were up-regulated, and other genes were down-regulated; after drought treatment, IbSS4 was adjusted upward, while other IbSSs were downgraded. In stems, except for IbSS5, which was up-regulated in the drought treatment, the other genes were up-regulated in both salt and drought treatments.
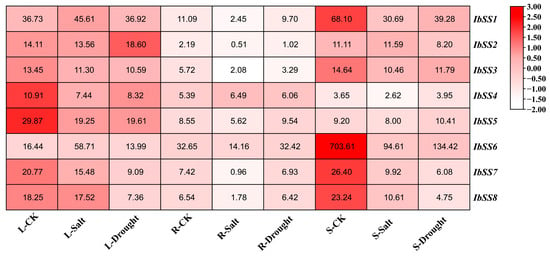
Figure 13.
Expression patterns of sweet potato SS genes under salt and drought stress as determined via RNA-seq. L: leaves; R: primary roots; S: stems. The FPKM values are shown in the color blocks.
We analyzed the expression levels of ItfSSs and ItbSSs under drought and salt stress to understand the role of ItfSSs and ItbSSs in two closely related species (Figure 14). In I. trifida (Figure 14A), after salt treatment, ItfSS1 and ItfSS5 were down-regulated, and other ItfSSs were up-regulated; under drought treatment, ItfSS1, ItfSS5, and ItfSS6 were down, and other ItfSSs were raised. In I. triloba (Figure 14B), under salt treatment, ItbSS2, ItbSS6, and ItbSS7 were regulated upward, ItbSS4 remained unchanged, and other ItbSSs were down-regulated; after drought treatment, ItbSS2, ItbSS4, ItbSS7, and ItbSS8 were adjusted upward, while other ItbSSs were downgraded.
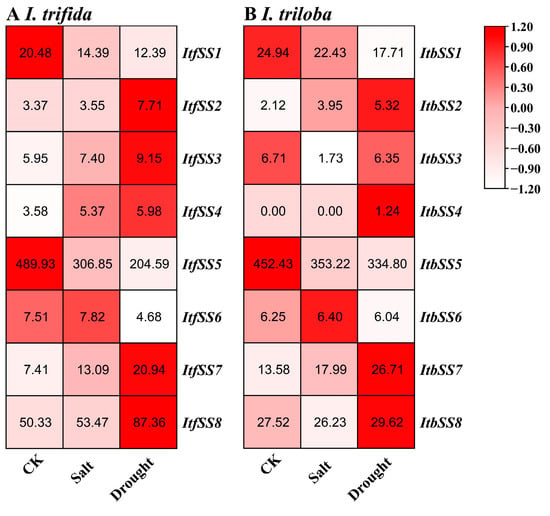
Figure 14.
Expression analysis of ItfSSs (A) and ItbSSs (B) in response to salt and drought stress as determined by RNA-seq. FPKM values are shown in the boxes.
2.6. Real-Time PCR Analysis of SS Genes in Sweet Potato
Two representative varieties of “J26” with high amylose content and “P32” with low amylose content were selected, and qRT-PCR was carried out to detect and analyze the expression of SSs in the stem, leaf, flower, seed, and root at four different stages (Figure 15). In root1, except that IbSS2 and IbSS6 were expressed higher in “J26” than in “P32”, the expression levels of other IbSSs in “P32” were higher than those in “J26”. In root2, except that IbSS6 was expressed higher in “J26” than in “P32”, the expression levels of other IbSSs in “P32” were higher than those in “J26”. In firewood root and flower, except that IbSS1 was expressed higher in “J26” than in “P32”, the expression of other IbSSs was approximately the same in both varieties. In fibrous roots, except that the expression of IbSS6 in both varieties was about the same, the expression levels of other IbSSs in “P32” were higher than those in “J26”. In stems, the expression of IbSSs in “P32” was higher than that in “J26”. In leaves, except that IbSS2, IbSS4, and IbSS5 were expressed higher in “J26” than in “P32”, the expression levels of other IbSSs in “J26” were lower than those in “P32”. In seed, except that IbSS2 and IbSS6 were expressed higher in “Jishu 26” than in “P32”, the expression levels of other IbSSs in “J26” were lower than those in “P32”. These results suggested that IbSSs may play an important role in different stages of root development. At the same time, SSs had a certain effect on the growth and development of stems and leaves, which may further affect the yield and quality of sweet potato by affecting photosynthesis.
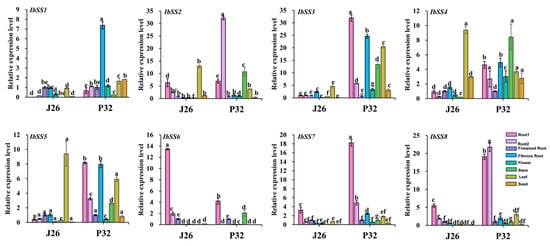
Figure 15.
Expression analysis of SS genes in sweet potato different tissue sites. J26: “Jishu 26”; P32: “Pushu 32”; root1: large tuber roots; root2: small tuber roots. Lowercase letters indicate a significant difference in each IbSS at p < 0.05 as determined by one-way ANOVA, with Tukey’s post hoc test conducted afterwards.
2.7. Sweet Potato IbSSs Protein Interactions
To investigate the potential regulatory network of SS, we constructed an IbSS interaction network map based on Arabidopsis homologous proteins (Figure 16). IbSS could interact with glucose-1-phosphate adenylyltransferases (ADG2, APS1, APL2, APL3, and APL4), dismutases (DPE1, DPE2), starch branching enzymes (SBE2.1, SBE2.2, SBE3), and UDP-glycosyltransferases (A0A1P8B9B9). Protein interaction predictions indicated that all eight IbSSs were homologous to Arabidopsis proteins, IbSS2 was homologous to SS1; IbSS1 and IbSS4 were homologous to SS2; IbSS3, IbSS5, and IbSS7 were homologous to SS4; IbSS6 was homologous to GBSS1; and the homology with SS4 was also up to 31.4%. These results suggest that IbSS plays an important role in the metabolic development of sweet potato.
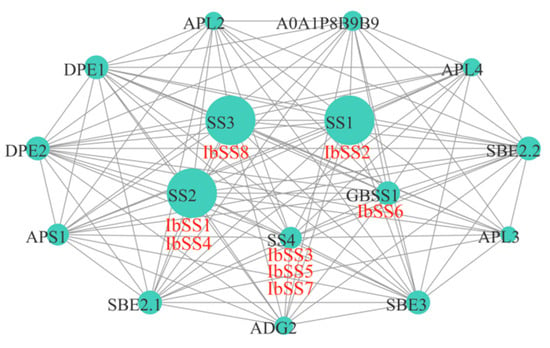
Figure 16.
Mapping the sweet potato SS protein–protein interaction network based on homolog proteins in Arabidopsis. The size of the circle and the thickness of the line represent the degree of interaction.
3. Discussion
3.1. Evolution of SS in Sweet Potato and Two Closely Related Species
Starch is widely found in green plants and is an important carbon and energy reserve in plants, which is essential for the yield and quality of sweet potato lakes. The SS gene family has been identified in many plants, such as Arabidopsis containing five SS genes [13,23], rice containing 10 SS genes [24], potato containing four SS genes [11], maize [11], and wheat [32,33], and tomato [27] containing six SS genes. In this study, 24 SS genes from I. batatas, I. trifida, and I. triloba were grouped into five subfamilies based on a phylogenetic tree. These genes had strong collinearity between I. batatas, I. trifida, and I. triloba; the homologs between the three species were located at similar points (Figure 1D and Figure 2). This suggests that sweet potato SS originated from its diploid relatives. As Figure 3 showed, except for IbSS3 and IbSS5, 10 conserved motifs were identified for all other IbSSs. IbSS3, ItfSS4, and ItbSS4 are homologous, but IbSS3 does not have motif 4 or motif 2. IbSS5, ItfSS6, and ItbSS6 are homologous, but IbSS5 does not have motif 2, motif 6, or motif 9. This may be since there may have been some changes in the function of the sweet potato SS genes during evolution.
The structures of gene exons and introns are generally preserved in homologous genes within a gene family [34]. Our research found that most homologous sequences in I. batatas, I. trifida, and I. triloba shared similar exon and intron counts. However, there was some difference; for example, in the SSIV group, IbSS3 contains 24 exons, ItfSS4 and ItbSS4 contain 18 and 22 exons, respectively; IbSS7 contains 19 exons, and ItfSS2 and ItbSS2 contain 18 exons. Other than that, in the SSI group, IbSS2 has two fewer exons than ItfSS8 and ItbSS8; in the SSII group, IbSS4 has two fewer exons than ItfSS3 and ItbSS3; and in the GBSS group, IbSS6 has one fewer exon than ItbSS5. This suggests that there may have been some changes in the function of SSs during the evolution of sweet potato and its relative.
3.2. Distinct Roles of SS Genes in Biological Processes
The distribution of SSs in various tissues, stages of development, and exposure to environmental stresses provided valuable insights into the elucidation of their inherent biological roles [35]. Overall, the expression of IbSSs was higher in the tuberous roots of two cultivars with different amylose contents, and the expression levels of IbSS1 and IbSS6 were the highest in the tuberous roots. This suggests that the IbSS1 and IbSS6 genes mainly acted in tuberous root development and starch biosynthesis. The IbSS1 and IbSS6 expression levels were also higher in the stems and leaves (Figure 5A). This suggested that the IbSS1 and IbSS6 genes may participate in transport, assimilation of carbohydrates, and leaf photosynthesis. Similarly, the ItfSS5 and ItbSS5 expression was highest in I. trifida and I. triloba root1 (Figure 5B,C); it also had higher expression in leaves and stems; the expression levels of SSs in storage roots and leaves were different from those in sweet potato; and interestingly, the relative expression levels of flower buds were higher in I. trifida and I. triloba. It is also worth noting that both the ItfSS5 and ItbSS5 genes were highly expressed in both I. trifida and I. triloba.
Starch plays an important role in balancing growth and carbon assimilation processes, especially under stress conditions, and plants can mobilize starch reserves to release energy, sugars, and derived metabolites to mitigate the effects of stress [36]. It accumulated during the photoperiod and was degraded at night to support respiration and growth. In maize, starch synthesis helped plants maintain leaf growth and facilitate carbon uptake under drought conditions [35]. Similar conclusions were also made in faba bean [37], tomato [38], reed [39], wheat [40,41], rice [42], and sorghum [43]. In our study, IbSS6 and IbSS1 showed high expression under both salt and drought stress (Figure 13), while ItfSS5, ItfSS8, ItbSS5, and ItbSS8 showed high expression under both salt and drought stress conditions (Figure 14). Thus, SS may play a crucial role in the salt and heat stress responses of I. batatas, I. trifida, and I. triloba.
In addition to salt and drought, cold stress and potassium deficiency also had important effects on sweet potato yield and quality. Using two varieties of cold-resistant “Liaohanshu 21” and cold-sensitive “Shenshu 28”, we found that IbSS4 and IbSS6 were upregulated in both varieties (Figure 9). High expression of ItfSS5 and ItbSS2 in two closely related species (Figure 10) suggests that SS may play an important role in the response of sweet potato and its relatives to cold stress in the evolutionary process. When two varieties of low-potassium-tolerant “Xushu 32” and low-potassium-sensitive “Ningzishu 1” were used, we found that all IbSSs genes were up-regulated, except for IbSS4, whose expression was down-regulated in both varieties (Figure 6). These results suggest that SS may have a positive effect on sweet potato under cold stress and potassium deficiency stress.
Finally, we examined the expression of different places in “P32” and “J26” (large storage roots, small storage roots, firewood root, and fibrous root) by qPCR. In large storage roots and small storage roots (root1 and root2), IbSS6 in the GBSS group had a higher expression in “J26” exhibiting more amylose, according to the results of other studies, i.e., when GBSS expression was higher, the content of rectilinear starch was also higher [1]. IbSSs showed higher expressions in “P32” than “J26”. IbSS3, IbSS4, and IbSS5 have higher expression in the stem and leaf (Figure 15), suggesting that these SS genes in sweet potato may affect carbohydrate transport, assimilation, and leaf photosynthesis. According to our cluster analysis and existing articles, it was known that SS genes not only play important roles in starch synthesis but also play key roles in abiotic stress response [44,45,46].
3.3. Involvement of SS in Hormonal Responses in Sweet Potato and Two Closely Related Species
Gene expression was usually regulated by cis-acting elements, and SS genes were involved in a number of physiological and biochemical processes such as plant growth and development, signal transduction, abiotic stress response, and hormone regulation [47]. A large number of cis-acting elements, including core promoter elements, light-responsive elements, hormone-responsive elements, stress-responsive elements, and unknown elements, have been identified in the promoter regions of I. batatas, I. trifida, and I. triloba (Figure 4). In this study, we found abundant ABA, GA, PHY, CAS, and MeJA response elements in the IbSS, ItfSS, and ItbSS promoters. Through expression analysis, we found that IbSSs showed different expression patterns. Among them, IbSS2, IbSS4, IbSS6, and IbSS7 were all up-regulated under ABA, SA, and MeJA treatments (Figure 7). After ABA, GA, and IAA treatments, ItfSS1, ItfSS5, ItfSS7, ItfSS8, ItbSS1, ItbSS2, ItbSS3, ItbSS4, ItbSS6, ItbSS7, and ItbSS8 show different degrees of up-regulation (Figure 8A,B). These results suggest that SS may be involved in hormone crosstalk in sweet potato and its diploid wild relatives.
Differences in expression patterns of SS genes in sweet potato and its two diploid relatives might provide potential candidate genes for further functional characterization and provide a certain reference significance for the molecular breeding of sweet potato in the future.
4. Materials and Methods
4.1. Identification of SS Genes Members in Sweet Potato
The protein sequences and GFF gene annotation files of I. batatas, I. trifida, and I. triloba were obtained from the Sweet Potato Genomics Resource (Sweetpotato Genomics Resource (uga.edu), accessed on 17 July 2023) and Ipomoea Genome Hub (https://ipomoea-genome.org/, accessed on 17 July 2023). The Cryptomarkov model (HMM) profile of the glycosyltransferase domain (PF08323 and PF00534) was downloaded from the Pfam (Pfam: Home page (xfam.org)) database and used to investigate all protein sequences in I. batatas, I. trifida, and I. triloba. The Arabidopsis, S. bicolor, and S. lycopersicum SS protein sequences were downloaded at Ensembl Plants (https://plants.ensembl.org/index.html, accessed 20 September 2023). The O. sativa SS protein sequences were obtained from the National Rice Data Center (RiceData = Rice Gene Database, accessed on 22 June 2023). The HMMER Search program in Tbtools software was used to preliminarily screen the SS candidate genes. All putative SS were further screened using SMART (SMART: Main page (embl.de), accessed on 22 June 2023) and finally further validated by the conserved database CDD (Welcome to NCBI Batch CD-search (nih.gov), accessed on 22 June 2023) [48]. Finally, with the full-length transcriptome of third-generation sequenced “Guangshu 87” as a reference, the candidate sweet potato SS family members were manually corrected using SnapGene software 6.0.2. Screened genes of IbSS, ItfSS, and ItbSS were named sequentially based on chromosomal location. The physicochemical properties of IbSS, ItfSS, and ItbSS proteins were analyzed based on the basic information of the protein sequences determined by our laboratory in the company and the Expasy online tool (SIB Swiss Institute of Bioinformatics Expasy, accessed on 22 June 2023) [49]. TBtools software Version 2.0 was used for visualization and mapping [50].
4.2. Phylogenetic Analysis of Sweet Potato SS Genes
Using ClustalW Muscle default parameters, we performed multiple sequence comparisons of I. batatas, I. trifida, I. triloba, A. thaliana, S. bicolor, O. sativa, and S. lycopersicum, constructed a phylogenetic tree using MEGA7[51], and grassed on the classification of SS in A. thaliana, S. bicolor, O. sativa, and S. lycopersicum to classify the sweet potato, triticale ragweed, and triticale yam SS genes. Finally, the evolutionary trees were landscaped using the EvoView online tool (EvolView: my projects and trees (evolgenius.info), accessed on 22 June 2023).
4.3. Conserved Motifs and Exon–Intron Structure Analysis of Sweet Potato SS
Using the MEME online tool (MEME—Submission form (meme-suite.org), accessed on 22 August 2023) analyzed conserved motifs of SS proteins, and the maximum number of motifs parameter set to 10. The required protein sequences were put into CDD (NCBI Conserved Domain Search (nih.gov), accessed on 15 October 2023) to obtain the corresponding files. Finally, TBtools software Version 2.0 was used to visualize and analyze the Motif predictions of IbSS, ItfSS, and ItbSS, as well as the NCBI results.
4.4. Analysis of the Promoter Cis-Acting Element of the Sweet Potato SS Genes
Based on the genome annotation information and whole genome sequences of I. batatas, I. trifida, and I. triloba, sequences 2000 bp upstream of the CDS of I. batatas, I. trifida, and I. triloba were extracted as promoter regions, and the promoter sequences were submitted to the PlantCare website (PlantCARE, a database of plant promoters and their cis-acting regulatory elements (ugent.be), accessed on 19 October 2023) for cis-acting element prediction. The PlantCare results were simplified, and the cis-acting elements of the SS promoter region were obtained using TBtools software Version 2.0.
4.5. Analysis of SS Genes Expression in Sweet Potato
For the analysis of sweet potato SSs expression profiles, a total of four transcriptome bio project datasets were selected. The data included three bio project datasets obtained from the NCBI database: PRJNA1013090 focusing on low potassium, PRJNA511028 on hormone, and PRJNA987163 on cold. Additionally, an in-house (unpublished) dataset was included, specifically addressing sweet potato heat treatment. Within these datasets, different sweet potato varieties were utilized for specific treatments. For hormonal treatment, “Xushu 18” was chosen, while cold-tolerant “Liaohanshu 21” and cold-sensitive “Shenshu 28” were used for cold treatment. Low-tolerant “Xushu 32” and low-k-sensitive “Ningzishu 1” were selected for low-potassium treatment, and heat-tolerant “Guangshu 87” and heat-sensitive “Ziluolan” were employed for heat treatment. Moreover, the gene expression data for I. trifida and I. triloba were obtained from the Sweet Potato Genomics Resource (http://sweetpotato.plantbiology.msu.edu/, accessed on 17 July 2023). The expression maps were created with TBtools software version 2.0.
4.6. qRT-PCR Detection of SS Genes in Sweet Potato
Eight different parts of two varieties, high amylose “J26” and low amylose “P32”, were used to analyze the expression levels. Total RNA was extracted using the TRIzol method (Invitrogen, Carlsbad, CA, USA). The qRT-PCR experiments were an improvement over previous studies [52]. The experiments were performed by fluorescence quantification kits, with 3 replicates per sample. For qRT-PCR analysis, the 20 µL total reaction quantity of each sample contained 2 µL cDNA template, 2 µL (10 µmol L−1) forward and reverse gene-specifc primers, 10 µL 2 × SYBR Green qPCR mix, and 4 µL sterile ddH2O. The qRT-PCR reaction was conducted using the Bio-Rad system with the following thermal cycle conditions: 3 min of pre-degeneration at 95 °C, followed by 40 cycles of denaturation at 95 °C for 10 s, and annealing at 60 °C for 30 s. The reaction was completed with a 5-s step at 65 °C and a cooling rate of 0.5 °C to reach 95 °C. The results were averaged for gene expression analysis. The relative expression of SSs in sweet potato was calculated by the 2−ΔΔCt method. Primers used in the study are listed in Supplemental Table S1, where the IbARF gene is an internal reference [53].
4.7. Protein–Protein Interaction Analysis of SS Protein in Sweet Potato
Using the default parameters, the online STRING database (STRING: functional protein association networks (string-db.org), accessed on 22 July 2023) was used to predict and execute protein–protein interaction networks based on the Arabidopsis homolog of the sweet potato SS protein. Finally, Cytoscape software was used for visualization and mapping.
5. Conclusions
Eight SSs were identified in I. batatas, I. trifida, and I. triloba, respectively. Their protein physicochemical properties, chromosomal localization, phylogenetic relationship, gene structure, promoter cis-elements, coercion expression, and protein interaction network and expression patterns were systematically analyzed. SS genes may be involved in various hormone and abiotic stress responses to regulate sweet potato growth and development. For example, IbSSs were highly expressed in sweet potato tuber root, suggesting that IbSSs may play an important role in sweet potato yield and quality. The IbSS6 gene was highly expressed in the face of various stress treatments; other genes are also expressed to varying degrees; among them, IbSS1 and IbSS8 are upward in the face of coercion. These candidate genes merit further investigation. The findings in this study will help us better understand the biological functions of SSs in coping with climate change and identify possible candidate genes for enhancing field and abiotic stress tolerance in sweet potato and its two diploid relatives.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15040400/s1, Table S1: The primer sequences for qPCR of sweet potato SS gene.
Author Contributions
Conceptualization, Z.S.; methodology, Z.S., H.Z., X.L., Z.H. and Z.L.; software, Z.S.; validation, Z.S. and H.Z.; formal analysis, Z.S.; investigation, M.J., B.T., Z.Z., M.X. and X.Y.; resources, H.Z.; data curation, Z.S. and Z.L.; writing—original draft preparation, Z.S.; writing—review and editing, Z.S., Z.L. and H.Z.; visualization, Z.S.; supervision, H.Z.; project administration, H.Z.; funding acquisition, H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China-Guangdong Joint Fund, China and Studies on Resistance Resources and Molecular Mechanisms of Sweet potato Weevil in South China (Grant No. U1701234).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare that there is no conflict of interest to report regarding this current study.
References
- Yang, Q.; Ding, J.; Feng, X.; Zhong, X.; Lan, J.; Tang, H.; Harwood, W.; Li, Z.; Guzmán, C.; Xu, Q.; et al. Editing of the starch synthase IIa gene led to transcriptomic and metabolomic changes and high amylose starch in barley. Carbohydr. Polym. 2022, 285, 119238. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Tan, H.; Zhang, C.; Li, Q.; Liu, Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2021, 2, 100237. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Höchstötter, A.; Jekle, M.; Arendt, E.; Becker, T. Physicochemical and morphological characterization of different starches with variable amylose/amylopectin ratio. Food Hydrocoll. 2013, 32, 52–63. [Google Scholar] [CrossRef]
- Gong, D.; Xu, X.; Wu, L.; Dai, G.; Zheng, W.; Xu, Z. Effect of biochar on rice starch properties and starch-related gene expression and enzyme activities. Sci. Rep. 2020, 10, 16917. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.C.; Wang, S.Y.; Gao, H.Y.; Nguyen, K.M.; Nguyen, C.H.; Shih, M.C.; Lin, K.H. Physicochemical properties of starches and expression and activity of starch biosynthesis-related genes in sweet potatoes. Food Chem. 2016, 199, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yang, J.; Hong, Y.; Liu, G.; Zheng, J.; Gu, Z.; Zhang, P. Impact of amylose content on starch physicochemical properties in transgenic sweet potato. Carbohydr. Polym. 2015, 122, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jang, S.; Lar, S.M.; Lee, A.R.; Cao, F.Y.; Seo, J.; Kwon, S.W. Genome-Wide Identification and Genetic Variations of the Starch Synthase Gene Family in Rice. Plants 2021, 10, 1154. [Google Scholar] [CrossRef]
- Mishra, B.P.; Kumar, R.; Mohan, A.; Gill, K.S. Conservation and divergence of Starch Synthase III genes of monocots and dicots. PLoS ONE 2017, 12, e189303. [Google Scholar] [CrossRef]
- Cao, H.; James, M.G.; Myers, A.M. Purification and Characterization of Soluble Starch Synthases from Maize Endosperm. Arch. Biochem. Biophys. 2000, 373, 135–146. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.; Zhang, C.; Jiang, L.; Jiang, M.; Zhong, M.; Fan, X.; Gu, M.; Liu, Q. Rice Soluble Starch Synthase I: Allelic Variation, Expression, Function, and Interaction With Waxy. Front. Plant Sci. 2018, 9, 1591. [Google Scholar] [CrossRef]
- Nazarian-Firouzabadi, F.; Visser, R.G.F. Potato starch synthases: Functions and relationships. Biochem. Biophys. Rep. 2017, 10, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Barchiesi, J.; Hedin, N.; Iglesias, A.A.; Gomez-Casati, D.F.; Ballicora, M.A.; Busi, M. Identification of a novel starch synthase III from the picoalgae Ostreococcus tauri. Biochimie 2017, 133, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Schwarte, S.; Brust, H.; Steup, M.; Tiedemann, R. Intraspecific sequence variation and differential expression in starch synthase genes of Arabidopsis thaliana. BMC Res. Notes 2013, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Yuan, Y.; Shen, Q.; Jiang, Q.; Hua, X.; Zhang, Q.; Zhang, M.; Ming, R.; Zhang, J. Evolution and Expression Analysis of Starch Synthase Gene Families in Saccharum spontaneum. Trop. Plant Biol. 2019, 12, 158–173. [Google Scholar] [CrossRef]
- Kitahara, K.; Hamasuna, K.; Nozuma, K.; Otani, M.; Hamada, T.; Shimada, T.; Fujita, K.; Suganuma, T. Physicochemical properties of amylose-free and high-amylose starches from transgenic sweet potatoes modified by RNA interference. Carbohydr. Polym. 2007, 69, 233–240. [Google Scholar] [CrossRef]
- Otani, M.; Hamada, T.; Katayama, K.; Kitahara, K.; Kim, S.H.; Takahata, Y.; Suganuma, T.; Shimada, T. Inhibition of the gene expression for granule-bound starch synthase I by RNA interference in sweet potato plants. Plant Cell Rep. 2007, 26, 1801–1807. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Iizuka, Y.; Abe, N.; Miura, S.; Kikuchi, K.; Matsushima, R.; Fujita, N. Rice Mutants Lacking Starch Synthase I or Branching Enzyme IIb Activity Altered Starch Biosynthetic Protein Complexes. Front. Plant Sci. 2018, 9, 1817. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, Y.; Li, X.; Yan, Z.; Xie, Y.; Xiong, H.; Zhao, L.; Gu, J.; Zhao, S.; Liu, L. Novel mutant alleles of the starch synthesis gene TaSSIVb-D result in the reduction of starch granule number per chloroplast in wheat. BMC Genom. 2017, 18, 358. [Google Scholar] [CrossRef]
- Malinova, I.; Alseekh, S.; Feil, R.; Fernie, A.R.; Baumann, O.; Schöttler, M.A.; Lunn, J.E.; Fettke, J. Starch Synthase 4 and Plastidal Phosphorylase Differentially Affect Starch Granule Number and Morphology. Plant Physiol. 2017, 174, 73–85. [Google Scholar] [CrossRef]
- Crumpton Taylor, M.; Pike, M.; Lu, K.J.; Hylton, C.M.; Feil, R.; Eicke, S.; Lunn, J.E.; Zeeman, S.C.; Smith, A.M. Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol. 2013, 200, 1064–1075. [Google Scholar] [CrossRef]
- Roldán, I.; Wattebled, F.; Mercedes Lucas, M.; Delvalle, D.; Planchot, V.; Jimenez, S.; Perez, R.; Ball, S.; d’Hulst, C.; Merida, A. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. Cell Mol. Biol. 2007, 49, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, G.; Wei, B.; Wang, Y.; Zhang, Z.; Hu, H.; Liu, Y.; Yu, G.; Zhang, H.; Huang, Y. Identification and Phylogenetic Analysis of a Novel Starch Synthase in Maize. Front. Plant Sci. 2015, 6, 1013. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, P.; Moreau, H.; Worden, A.Z.; Dauvillée, D.; Ball, S.G. Early gene duplication within Chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts. Genetics 2008, 178, 2373–2387. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zheng, Y.; Thi, K.M.; Lin, L.; Xie, X.; Khine, E.E.; Nyein, E.E.; Lin, M.H.W.; New, W.W.; Aye, S.S.; Wu, W. Genome-wide association study of cooking-caused grain expansion in rice (Oryza sativa L.). Front. Plant Sci. 2023, 14, 1250854. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.G.; Morell, M.K. From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003, 54, 207–233. [Google Scholar] [CrossRef]
- Imparl-Radosevich, J.M.; Keeling, P.L.; Guan, H. Essential arginine residues in maize starch synthase IIa are involved in both ADP-glucose and primer binding. FEBS Lett. 1999, 457, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Helle, S.; Bray, F.; Verbeke, J.; Courseaux, A.; Rolando, C.; Szydlowski, N. Proteome Analysis of Potato Starch Reveals the Presence of New Starch Metabolic Proteins as Well as Multiple Protease Inhibitors. Front. Plant Sci. 2018, 9, 746. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome sequences of two diploid wild relatives of cultivated sweet potato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Zhang, H.; Zhai, H.; Liu, Q.; He, S. A soluble starch synthase I gene, IbSSI, alters the content, composition, granule size and structure of starch in transgenic sweet potato. Sci. Rep. 2017, 7, 2315. [Google Scholar] [CrossRef]
- Kitahara, K.; Takahata, Y.; Otani, M.; Tanaka, M.; Katayama, K.; Yoshinaga, M.; Fujita, K.; Suganuma, T. Starch Properties of Transgenic Sweetpotato Plants Modified by RNA Interference of the Starch Synthase II Gene. J. Appl. Glycosci. 2011, 58, 85–90. [Google Scholar] [CrossRef]
- Sheng, M.; Xia, H.; Ding, H.; Pan, D.; He, J.; Li, Z.; Liu, J. Long-Term Soil Drought Limits Starch Accumulation by Altering Sucrose Transport and Starch Synthesis in Sweet Potato Tuberous Root. Int. J. Mol. Sci. 2023, 24, 3053. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Jiang, H.; Pan, X.; Li, M.; Chen, Y.; Wu, G. The gene encoding starch synthase IIc exists in maize and wheat. Plant Sci. 2009, 176, 51–57. [Google Scholar] [CrossRef]
- Li, Z.; Rahman, S.; Kosar-Hashemi, B.; Mouille, G.; Appels, R.; Morell, M.K. Cloning and characterization of a gene encoding wheat starch synthase I. Theor. Appl. Genet. 1999, 98, 1208–1216. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- AbdElgawad, H.; Avramova, V.; Baggerman, G.; Van Raemdonck, G.; Valkenborg, D.; Van Ostade, X.; Guisez, Y.; Prinsen, E.; Asard, H.; Van den Ende, W.; et al. Starch biosynthesis contributes to the maintenance of photosynthesis and leaf growth under drought stress in maize. Plant Cell Environ. 2020, 43, 2254–2271. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Ortiz, S.M.; De La Paz Arrieta-Montiel, M.; Acosta-Gallegos, J.; Covarrubias, A.A. Relationship between carbohydrate partitioning and drought resistance in common bean. Plant Cell Environ. 2008, 31, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- González-Cruz, J.; Pastenes, C. Water-stress-induced thermotolerance of photosynthesis in bean (Phaseolus vulgaris L.) plants: The possible involvement of lipid composition and xanthophyll cycle pigments. Environ. Exp. Bot. 2012, 77, 127–140. [Google Scholar] [CrossRef]
- Kanai, M.; Higuchi, K.; Hagihara, T.; Konishi, T.; Ishii, T.; Fujita, N.; Nakamura, Y.; Maeda, Y.; Yoshiba, M.; Tadano, T. Common reed produces starch granules at the shoot base in response to salt stress. New Phytol. 2007, 176, 572–580. [Google Scholar] [CrossRef]
- Tan, C.; Feng, C.; Guo, W.; Zhu, X.K.; Li, C.Y.; Peng, Y.X. Difference in Expression of Starch Synthase Gene and Starch Synthesis in the Grains of Different Wheat Cultivars. Mai Lei Zuo Wu Xue Bao 2011, 31, 1063–1070. [Google Scholar]
- Genschel, U.; Abel, G.; Lörz, H.; Lütticke, S. The sugary-type isoamylase in wheat: Tissue distribution and subcellular localisation. Planta 2002, 214, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ming, D.; Ma, W.; Xu, F. Changes in starch accumulation and activity of enzymes associated with starch synthesis under different N supplying date. Agric. Sci. China 2004, 3, 738–745. [Google Scholar]
- Bing, Y.I.; Zhou, Y.-F.; Gao, M.-Y.; Zhang, Z.; Yi, H.; Yang, G.-D.; Wenjuan, X.; Huang, R.-D. Effect of Drought Stress During Flowering Stage on Starch Accumulation and Starch Synthesis Enzymes in Sorghum Grains. J. Integr. Agric. 2014, 13, 2399–2406. [Google Scholar]
- Chen, Y.; Jiang, Y.; Chen, Y.; Feng, W.; Liu, G.; Yu, C.; Lian, B.; Zhong, F.; Zhang, J. Uncovering candidate genes responsive to salt stress in Salix matsudana (Koidz) by transcriptomic analysis. PLoS ONE 2020, 15, e236129. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hua, Y.; Wang, S.; Liu, X.; Zou, L.; Chen, C.; Zhao, H.; Yan, Y. Analysis of the Prunellae Spica transcriptome under salt stress. Plant Physiol. Biochem. 2020, 156, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Zhang, Z.; Yan, G.; Wang, F.; Zhao, L.; Liu, N.; Abudurezike, A.; Li, Y.; Wang, W.; Shi, S. Salt-responsive transcriptome analysis of triticale reveals candidate genes involved in the key metabolic pathway in response to salt stress. Sci. Rep. 2020, 10, 20669. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Meng, Y.; Bai, Y.; Yu, H.; Qian, Y.; Zhang, D.; Zhou, Y. Starch and Sucrose Metabolism and Plant Hormone Signaling Pathways Play Crucial Roles in Aquilegia Salt Stress Adaption. Int. J. Mol. Sci. 2023, 24, 3948. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Walker, J.M.J.; Walker, J.M. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Shao, Z.; He, M.; Zeng, Z.; Chen, Y.; Hanna, A.-D.; Zhu, H. Genome-Wide Identification and Expression Analysis of the MADS-Box Gene Family in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Genet. 2021, 12, 750137. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, T.; Yu, J.; Hong, H.; Liu, S.; Guo, F.; Ma, H.; Zhang, J.; Zhu, M.; Meng, X. Genome-wide survey and expression analysis of Dof transcription factor family in sweetpotato shed light on their promising functions in stress tolerance. Front. Plant Sci. 2023, 14, 1140727. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).