Unveiling the Impact of ApoF Deficiency on Liver and Lipid Metabolism: Insights from Transcriptome-Wide m6A Methylome Analysis in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Lipid Metabolism Disorder Mice with ApoF Gene Knockout
2.2. Histopathological Examination of Liver Tissue
2.3. CT Image Acquisition
2.4. Measurement of Serum Transaminase Enzymes and Blood Lipids
2.5. RNA Isolation and Extraction
2.6. High-Throughput m6A MeRIP-seq and mRNA-seq
2.7. RT-qPCR
2.8. Bioinformatic Analysis
2.9. Western Blotting
2.10. Statistical Analysis
3. Results
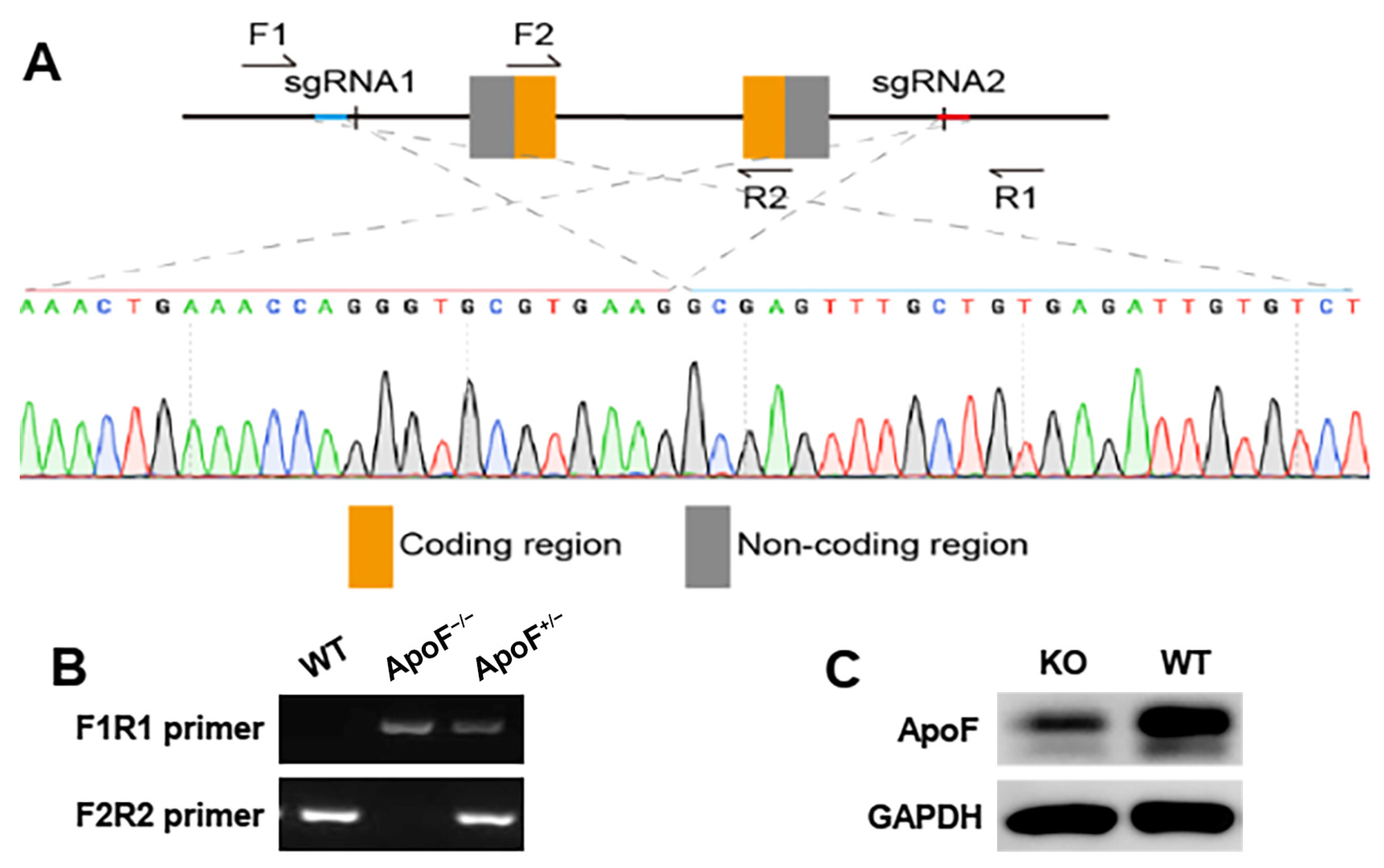
3.1. Genotype Identification
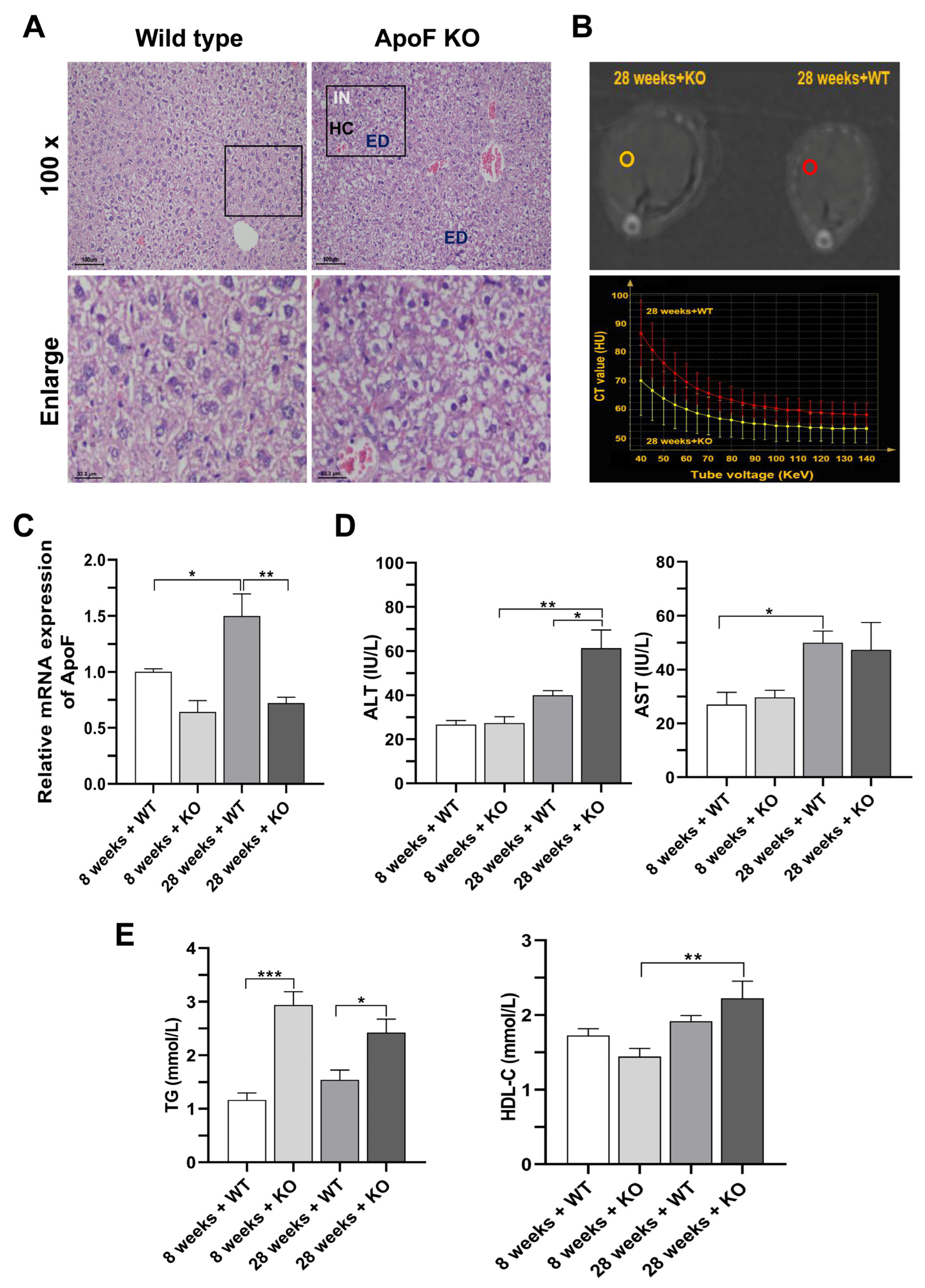
3.2. Histopathological Observation and Detection of Enzyme Activity and Dyslipidemia in ApoF Knockout Mice
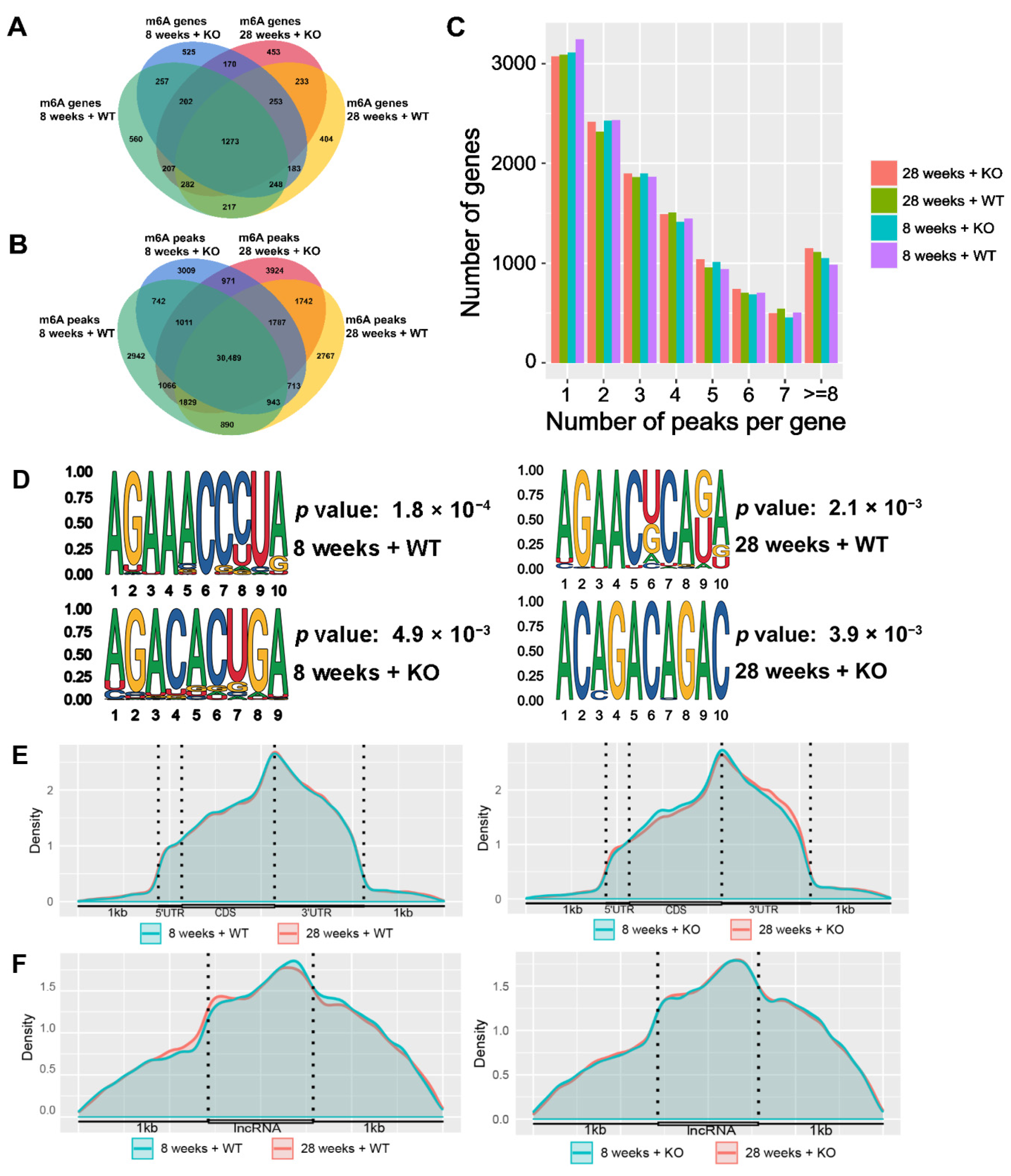
3.3. Description of the m6A-Modified Genes and Peaks
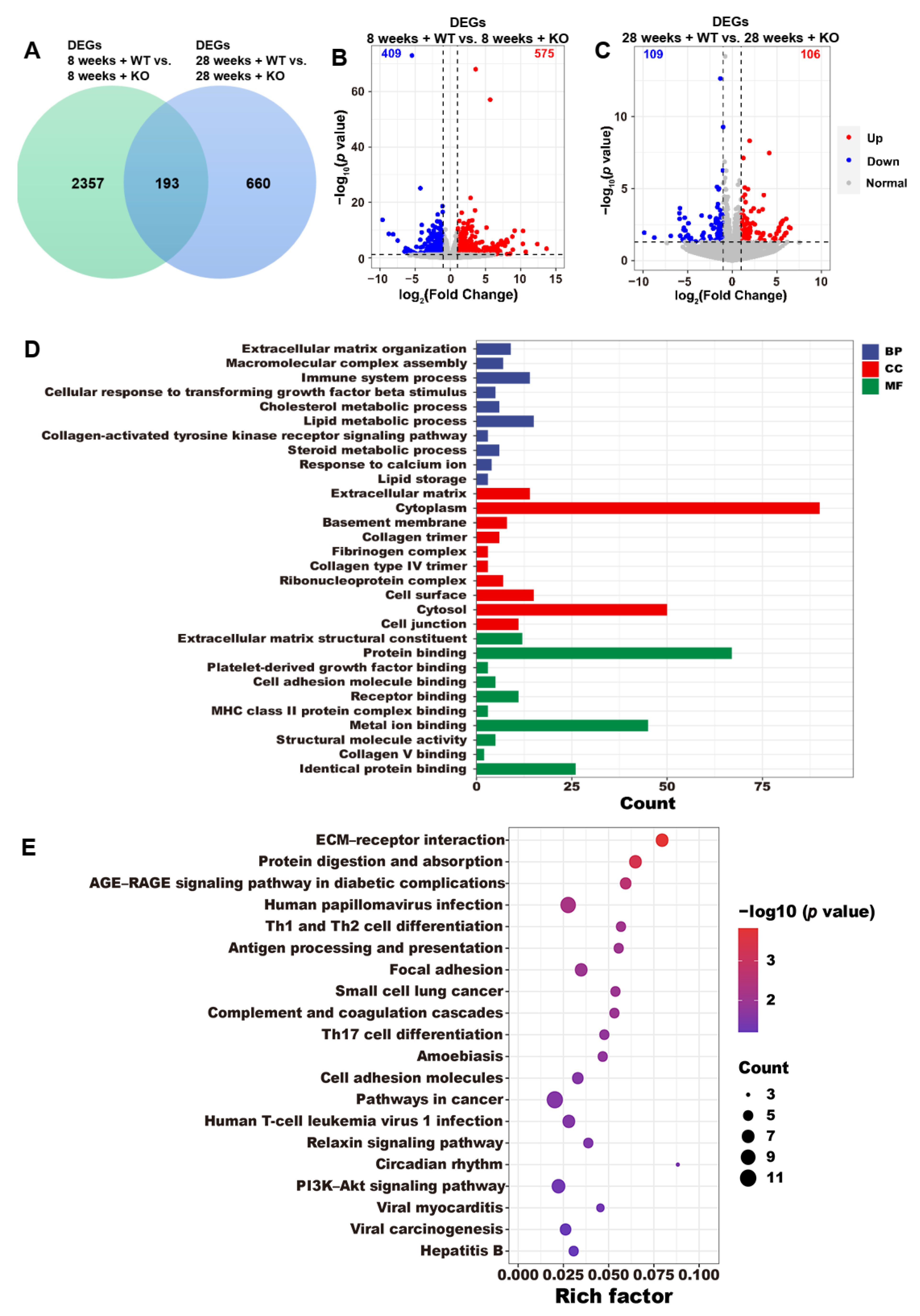
3.4. DEG Annotations
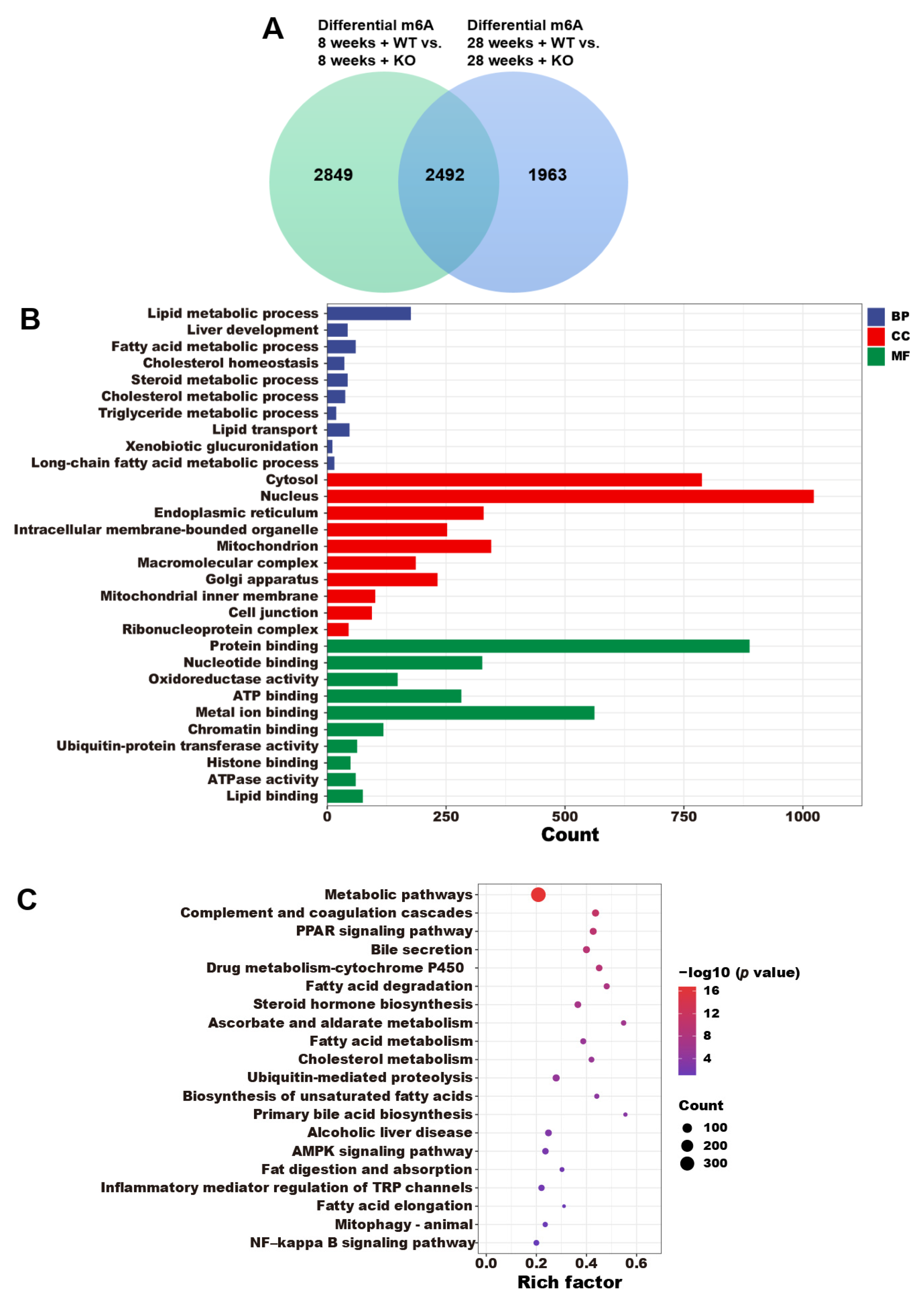
3.5. The Annotations of Differential m6A Genes
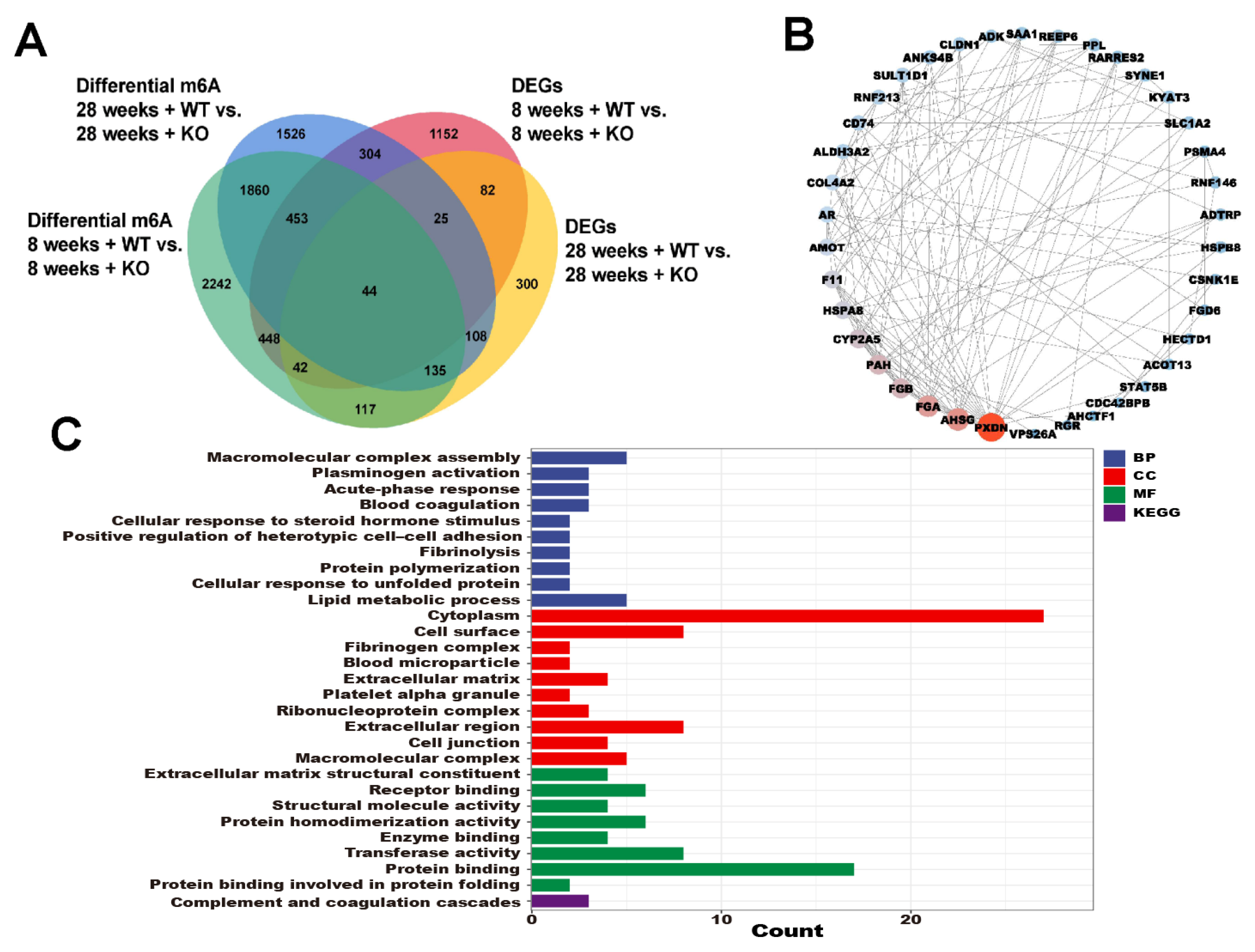
3.6. The Annotations for Overlapping Genes between the DEGs and Differential m6A Genes
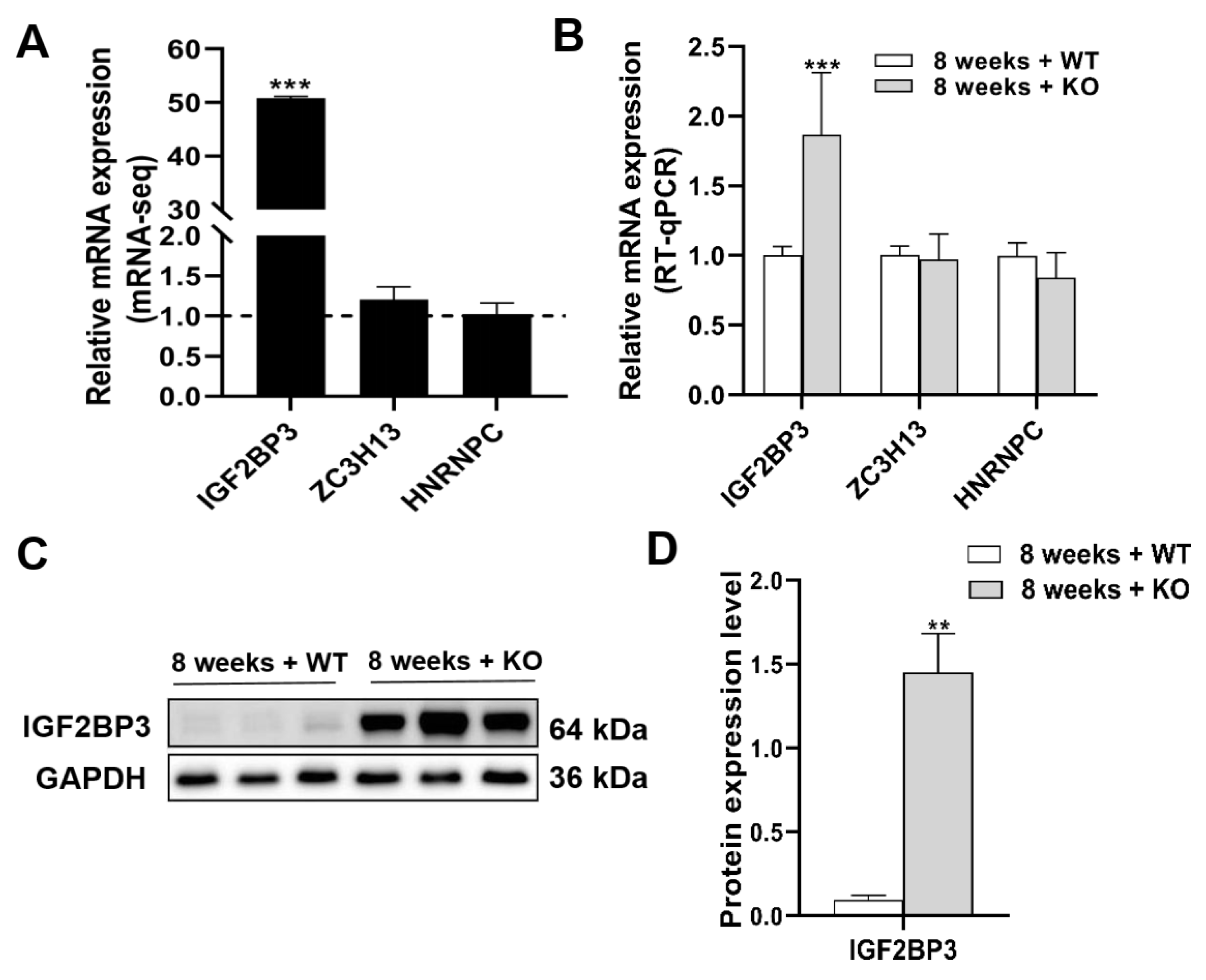
3.7. The Expression Levels of mRNA and Protein of m6A Regulators
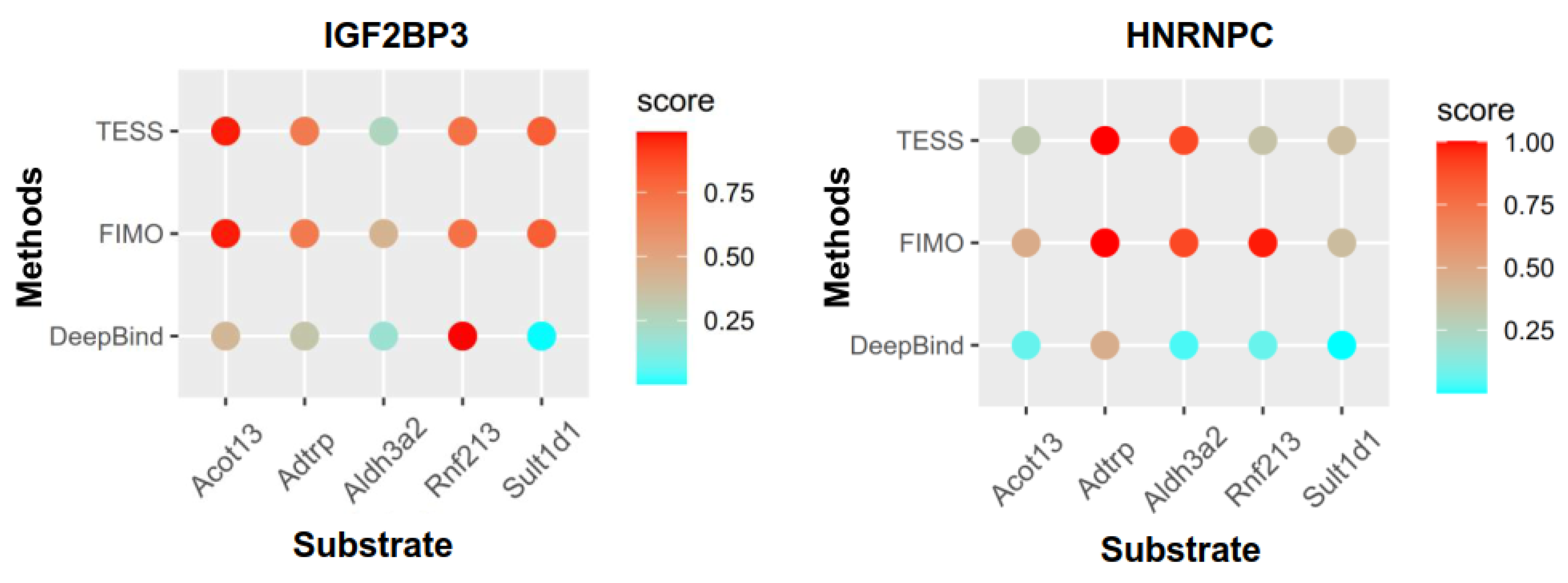
3.8. The Role of m6A Regulators in Lipid Metabolism
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morton, R.E. Cholesteryl ester transfer protein and its plasma regulator: Lipid transfer inhibitor protein. Curr. Opin. Lipidol. 1999, 10, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Morton, R.E.; Nunes, V.; Izem, L.; Quintão, E. Markedly elevated lipid transfer inhibitor protein in hypercholesterolemic subjects is mitigated by plasma triglyceride levels. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1642–1649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deprince, A.; Hennuyer, N.; Kooijman, S.; Pronk, A.C.M.; Baugé, E.; Lienard, V.; Verrijken, A.; Dirinck, E.; Vonghia, L.; Woitrain, E.; et al. Apolipoprotein F is reduced in humans with steatosis and controls plasma triglyceride-rich lipoprotein metabolism. Hepatology 2023, 77, 1287–1302. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Innerarity, T.L.; Rall, S.C., Jr.; Weisgraber, K.H. Plasma lipoproteins: Apolipoprotein structure and function. J. Lipid Res. 1984, 25, 1277–1294. [Google Scholar] [CrossRef]
- Duffy, D.; Rader, D.J. Emerging therapies targeting high-density lipoprotein metabolism and reverse cholesterol transport. Circulation 2006, 113, 1140–1150. [Google Scholar] [CrossRef]
- Singh, I.M.; Shishehbor, M.H.; Ansell, B.J. High-density lipoprotein as a therapeutic target: A systematic review. JAMA 2007, 298, 786–798. [Google Scholar] [CrossRef]
- Morton, R.E.; Greene, D.J. CETP and lipid transfer inhibitor protein are uniquely affected by the negative charge density of the lipid and protein domains of LDL. J. Lipid Res. 2003, 44, 2287–2296. [Google Scholar] [CrossRef]
- Lagor, W.R.; Fields, D.W.; Khetarpal, S.A.; Kumaravel, A.; Lin, W.; Weintraub, N.; Wu, K.; Hamm-Alvarez, S.F.; Drazul-Schrader, D.; de la Llera-Moya, M.; et al. The effects of apolipoprotein F deficiency on high density lipoprotein cholesterol metabolism in mice. PLoS ONE 2012, 7, e31616. [Google Scholar] [CrossRef]
- Li, L.; Liu, H.; Peng, J.; Wang, Y.; Zhang, Y.; Dong, J.; Liu, X.; Guo, D.; Jiang, Y. Farnesoid X receptor up-regulates expression of lipid transfer inhibitor protein in liver cells and mice. Biochem. Biophys. Res. Commun. 2013, 441, 880–885. [Google Scholar] [CrossRef][Green Version]
- Shen, X.-B.; Huang, L.; Zhang, S.-H.; Wang, D.-P.; Wu, Y.-L.; Chen, W.-N.; Xu, S.-H.; Lin, X. Transcriptional regulation of the apolipoprotein F (ApoF) gene by ETS and C/EBPα in hepatoma cells. Biochimie 2015, 112, 1–9. [Google Scholar] [CrossRef]
- Morton, R.E.; Gnizak, H.M.; Greene, D.J.; Cho, K.-H.; Paromov, V.M. Lipid transfer inhibitor protein (apolipoprotein F) concentration in normolipidemic and hyperlipidemic subjects. J. Lipid Res. 2008, 49, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Peloso, G.M.; Demissie, S.; Collins, D.; Mirel, D.B.; Gabriel, S.B.; Cupples, L.A.; Robins, S.J.; Schaefer, E.J.; Brousseau, M.E. Common genetic variation in multiple metabolic pathways influences susceptibility to low HDL-cholesterol and coronary heart disease. J. Lipid Res. 2010, 51, 3524–3532. [Google Scholar] [CrossRef] [PubMed]
- Izem, L.; Morton, R.E. Molecular cloning of hamster lipid transfer inhibitor protein (apolipoprotein F) and regulation of its expression by hyperlipidemia. J. Lipid Res. 2009, 50, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.G.; Sacco, M.T.; Horner, S.M. How RNA modifications regulate the antiviral response. Immunol. Rev. 2021, 304, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Li, L.; Wang, Y.; Wu, P.; Hou, X.; Ouyang, J.; Fan, C.; Li, Z.; Wang, F.; Guo, C.; et al. RNA modifications in cancer. Br. J. Cancer 2023, 129, 204–221. [Google Scholar] [CrossRef] [PubMed]
- He, P.C.; He, C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Zheng, X.; Guo, Y.; Cao, J.; Zhang, Y.; Wen, S.; Gao, W.; Wu, Y. Regulatory role and mechanism of m6A RNA modification in human metabolic diseases. Mol. Ther. Oncolytics 2021, 22, 52–63. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Gu, J.; Su, T.; Gu, X.; Feng, Y. The role of RNA m6A methylation in lipid metabolism. Front. Endocrinol. 2022, 13, 866116. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, G.-L.; Zhu, X.; Peng, T.-H.; Lv, Y.-C. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2021, 9, 51–61. [Google Scholar] [CrossRef]
- Zeng, B.; Wu, R.; Chen, Y.; Chen, W.; Liu, Y.; Liao, X.; Guo, G.; Wang, X. FTO knockout in adipose tissue effectively alleviates hepatic steatosis partially via increasing the secretion of adipocyte-derived IL-6. Gene 2022, 818, 146224. [Google Scholar] [CrossRef]
- Huang, J.; Sun, W.; Wang, Z.; Lv, C.; Zhang, T.; Zhang, D.; Dong, W.; Shao, L.; He, L.; Ji, X.; et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J. Exp. Clin. Cancer Res. 2022, 41, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Cui, G.; Zhao, F.; Tian, X.; Sun, B.-F.; Yang, Y.; Li, W. m6A Regulates Liver Metabolic Disorders and Hepatogenous Diabetes. Genom. Proteom. Bioinform. 2020, 18, 371–383. [Google Scholar] [CrossRef]
- Fong, A.S.; Pan, Y.; Yu, J.; Wong, C.C. Interplay between the m6A Epitranscriptome and Tumor Metabolism: Mechanisms and Therapeutic Implications. Biomedicines 2022, 10, 2589. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Chang, X.; Sun, Y.; Zou, L.; Su, L.; Sun, Y.; Li, S.; Liu, S.; Sun, Y.; Zhou, H.; et al. Role of Oxidative Stress and Inflammatory Response in Subchronic Pulmonary Toxicity Induced by Nano Nickel Oxide in Rats. J. Nanosci. Nanotechnol. 2017, 17, 1753–1761. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Meng, J.; Cui, X.; Rao, M.K.; Chen, Y.; Huang, Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics 2013, 29, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Duan, D.; Tang, W.; Wang, R.; Guo, Z.; Feng, H. Evaluation of epitranscriptome-wide N6-methyladenosine differential analysis methods. Brief Bioinform. 2023, 24, bbad139. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, K.; Wei, Z.; Coenen, F.; Su, J.; Meng, J. MetaTX: Deciphering the distribution of mRNA-related features in the presence of isoform ambiguity, with applications in epitranscriptome analysis. Bioinformatics 2021, 37, 1285–1291. [Google Scholar] [CrossRef]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, S.; Zhu, Y.; Xi, X.; Bao, P.; Ma, Z.; Kapral, T.H.; Chen, S.; Zagrovic, B.; Yang, Y.T.; et al. POSTAR3: An updated platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2022, 50, D287–D294. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wong, V.W.-S.; Dufour, J.-F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Sanyal, A.J.; George, J.; on behalf of the International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014. [Google Scholar] [CrossRef] [PubMed]
- Baffy, G.; Brunt, E.M.; Caldwell, S.H. Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J. Hepatol. 2012, 56, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-M.; Pu, C.-W.; Hou, Y.-H.; Chen, Z.; Alanazy, M.; Hebbard, L. Non alcoholic steatohepatitis a precursor for hepatocellular carcinoma development. World J. Gastroenterol. 2014, 20, 16464–16473. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J.; Brunt, E.M.; Kleiner, D.E.; Kowdley, K.V.; Chalasani, N.; LaVine, J.E.; Ratziu, V.; McCullough, A. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology 2011, 54, 344–353. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, A.; Wu, Y.; Zhao, Y.; Yang, X. Soybean soluble polysaccharides enhance bioavailability of genistein and its prevention against obesity and metabolic syndrome of mice with chronic high fat consumption. Food Funct. 2019, 10, 4153–4165. [Google Scholar] [CrossRef]
- Kuhnast, S.; Van Der Tuin, S.J.L.; Van Der Hoorn, J.W.A.; Van Klinken, J.B.; Simic, B.; Pieterman, E.; Havekes, L.M.; Landmesser, U.; Luscher, T.F.; Van Dijk, K.W.; et al. Anacetrapib reduces progression of atherosclerosis, mainly by reducing non-HDL-cholesterol, improves lesion stability and adds to the beneficial effects of atorvastatin. Eur. Heart J. 2015, 36, 39–48. [Google Scholar] [CrossRef]
- Schaefer, M.R. The Regulation of RNA Modification Systems: The Next Frontier in Epitranscriptomics? Genes 2021, 12, 345. [Google Scholar] [CrossRef]
- Xiong, X.; Yi, C.; Peng, J. Epitranscriptomics: Toward A Better Understanding of RNA Modifications. Genom. Proteom. Bioinform. 2017, 15, 147–153. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Liu, B.; Xu, J.; Cai, B.; Yang, H.; Straube, J.; Yu, X.; Ma, T. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. Biomark. Res. 2022, 10, 15. [Google Scholar] [CrossRef]
- Malovic, E.; Ealy, A.; Kanthasamy, A.; Kanthasamy, A.G. Emerging Roles of N6-Methyladenosine (m6A) Epitranscriptomics in Toxicology. Toxicol. Sci. 2021, 181, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, X.; Bao, W.; Hong, X.; Li, C.; Lu, J.; Zhang, D.; Zhu, A. Effect of Humantenine on mRNA m6A Modification and Expression in Human Colon Cancer Cell Line HCT116. Genes 2022, 13, 781. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.M.; Huo, F.C.; Zhang, J.; Shan, H.J.; Pei, D.S. Crosstalk between m6A modification and alternative splicing during cancer progression. Clin Transl Med 2023, 13, e1460. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, J.; Ma, J.; Wei, Z.; Wang, Y.; Song, B.; Meng, J.; Jia, G.; de Magalhães, J.P.; Rigden, D.J.; et al. DirectRMDB: A database of post-transcriptional RNA modifications unveiled from direct RNA sequencing technology. Nucleic Acids Res. 2023, 51, D106–D116. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Meng, J.; Su, R.; Zhang, J.; Chen, J.; Ma, X.; Xia, Q. Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J. Hepatol. 2020, 73, 664–679. [Google Scholar] [CrossRef]
- Cheng, C.; Yu, F.; Yuan, G.; Jia, J. Update on N6-methyladenosine methylation in obesity-related diseases. Obesity 2024, 32, 240–251. [Google Scholar] [CrossRef]
- Tang, Z.; Sun, C.; Yan, Y.; Niu, Z.; Li, Y.; Xu, X.; Zhang, J.; Wu, Y.; Li, Y.; Wang, L.; et al. Aberrant elevation of FTO levels promotes liver steatosis by decreasing the m6A methylation and increasing the stability of SREBF1 and ChREBP mRNAs. J. Mol. Cell Biol. 2023, 14, mjac061. [Google Scholar] [CrossRef] [PubMed]
- Karthiya, R.; Khandelia, P. m6A RNA Methylation: Ramifications for Gene Expression and Human Health. Mol. Biotechnol. 2020, 62, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Hu, C.; Liu, T.; Sun, Y.; Hu, F.; He, Y.; Zhang, J.; Chen, J.; Ding, J.; Fan, J.; et al. IGF2BP3 enhances lipid metabolism in cervical cancer by upregulating the expression of SCD. Cell Death Dis. 2024, 15, 138. [Google Scholar] [CrossRef]
- Wu, Z.; Zuo, X.; Zhang, W.; Li, Y.; Gui, R.; Leng, J.; Shen, H.; Pan, B.; Fan, L.; Li, J.; et al. m6A-Modified circTET2 Interacting with HNRNPC Regulates Fatty Acid Oxidation to Promote the Proliferation of Chronic Lymphocytic Leukemia. Adv. Sci. 2023, 10, e2304895. [Google Scholar] [CrossRef]
- Parsons, W.H.; Kolar, M.J.; Kamat, S.S.; Cognetta, A.B., III; Hulce, J.J.; Saez, E.; Kahn, B.B.; Saghatelian, A.; Cravatt, B.F. AIG1 and ADTRP are atypical integral membrane hydrolases that degrade bioactive FAHFAs. Nat. Chem. Biol. 2016, 12, 367–372. [Google Scholar] [CrossRef]
| Gene | Accession Number | Primer Sequence (5′ → 3′) | |
|---|---|---|---|
| ApoF | NM_133997 | Forward | CACTTCACACGGAGAGGCAACC |
| Reverse | GAGCAGCATCTGGCAGGACAAG | ||
| IGF2BP3 | NM_023670 | Forward | CATCTGTTTATTCCCGCCCTGTC |
| Reverse | AGCCTTGAACTGAGCCTCTGG | ||
| ZC3H13 | NM_026083 | Forward | GGCAAAGAGAGTGGGAAGATAA |
| Reverse | CCTGTCCTCTCGAACATGAATA | ||
| HNRNPC | NM_001170981 | Forward | TTAATGAAAGAAATGCCCGAGC |
| Reverse | CTCTGCAGCCAGGTTAATATCT | ||
| GAPDH | NM_001289726 | Forward | ACTCCACTCACGGCAAATTCAAC |
| Reverse | ACACCAGTAGACTCCACGACATAC | ||
| Genes | Regulators | 8-Week-Old WT vs. 8-Week-Old KO | 28-Week-Old WT vs. 28-Week-Old KO | ||
|---|---|---|---|---|---|
| log2(Fold Change) | p Value | log2(Fold Change) | p Value | ||
| IGF2BP3 | reader | 5.6676 | 8.96 × 10−58 | 0.3463 | 5.66 × 10−1 |
| METTL5 | writer | 0.3457 | 6.68 × 10−2 | 0.0420 | 7.95 × 10−1 |
| ZC3H13 | writer | 0.2689 | 8.20 × 10−2 | −0.0542 | 7.69 × 10−1 |
| ALKBH5 | eraser | −0.2811 | 1.03 × 10−1 | −0.0004 | 9.98 × 10−1 |
| HNRNPA2B1 | reader | 0.2657 | 1.15 × 10−1 | −0.0270 | 8.63 × 10−1 |
| VIRMA | writer | 0.2376 | 1.84 × 10−1 | −0.1127 | 4.81 × 10−1 |
| METTL14 | writer | 0.2085 | 2.03 × 10−1 | −0.0975 | 6.00 × 10−1 |
| IGF2BP2 | reader | 0.5964 | 2.35 × 10−1 | −0.4649 | 2.78 × 10−1 |
| FTO | eraser | −0.1363 | 3.62 × 10−1 | −0.1105 | 4.17 × 10−1 |
| WTAP | writer | 0.1078 | 4.37 × 10−1 | 0.1507 | 3.54 × 10−1 |
| METTL3 | writer | −0.1495 | 4.54 × 10−1 | −0.0561 | 7.57 × 10−1 |
| YTHDF1 | reader | 0.1498 | 4.73 × 10−1 | −0.0304 | 8.99 × 10−1 |
| YTHDF2 | reader | 0.1664 | 5.16 × 10−1 | 0.0396 | 8.67 × 10−1 |
| RBM15B | writer | −0.1015 | 6.50 × 10−1 | 0.1865 | 5.26 × 10−1 |
| FMR1 | reader | −0.0769 | 6.66 × 10−1 | −0.0056 | 9.76 × 10−1 |
| YTHDC2 | reader | −0.0711 | 7.36 × 10−1 | −0.2400 | 2.33 × 10−1 |
| RBM15 | writer | −0.0473 | 7.91 × 10−1 | 0.1011 | 5.50 × 10−1 |
| HNRNPC | reader | 0.0240 | 8.64 × 10−1 | 0.4048 | 6.86 × 10−4 |
| CBLL1 | writer | −0.0065 | 9.77 × 10−1 | 0.1255 | 5.85 × 10−1 |
| IGF2BP1 | reader | Not detected | Not detected | −2.1825 | 5.87 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Chen, M.; Zhang, J.; Lin, Y.; Gao, X.; Tu, J.; Chen, K.; Zhu, A.; Xu, S. Unveiling the Impact of ApoF Deficiency on Liver and Lipid Metabolism: Insights from Transcriptome-Wide m6A Methylome Analysis in Mice. Genes 2024, 15, 347. https://doi.org/10.3390/genes15030347
Shen X, Chen M, Zhang J, Lin Y, Gao X, Tu J, Chen K, Zhu A, Xu S. Unveiling the Impact of ApoF Deficiency on Liver and Lipid Metabolism: Insights from Transcriptome-Wide m6A Methylome Analysis in Mice. Genes. 2024; 15(3):347. https://doi.org/10.3390/genes15030347
Chicago/Turabian StyleShen, Xuebin, Mengting Chen, Jian Zhang, Yifan Lin, Xinyue Gao, Jionghong Tu, Kunqi Chen, An Zhu, and Shanghua Xu. 2024. "Unveiling the Impact of ApoF Deficiency on Liver and Lipid Metabolism: Insights from Transcriptome-Wide m6A Methylome Analysis in Mice" Genes 15, no. 3: 347. https://doi.org/10.3390/genes15030347
APA StyleShen, X., Chen, M., Zhang, J., Lin, Y., Gao, X., Tu, J., Chen, K., Zhu, A., & Xu, S. (2024). Unveiling the Impact of ApoF Deficiency on Liver and Lipid Metabolism: Insights from Transcriptome-Wide m6A Methylome Analysis in Mice. Genes, 15(3), 347. https://doi.org/10.3390/genes15030347








