Genomic and Expression Analysis of Cassava (Manihot esculenta Crantz) Chalcone Synthase Genes in Defense against Tetranychus cinnabarinus Infestation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of CHS Genes in Cassava
2.2. Characteristics of CHS Genes
2.3. Phylogenetic, Conserved Motifs and Gene Structure Analysis
2.4. Chromosomal Localization and Gene Duplication Analysis
2.5. Cis-Elements in the Promoter Regions Analysis
2.6. Protein-Protein Interaction (PPI) Network Analysis
2.7. Cultivation of Cassava and Rearing of T. cinnabarinus
2.8. RT-PCR and Proteomic Analysis
3. Results
3.1. Identification of CHS Genes Family from Cassava Genome Resources
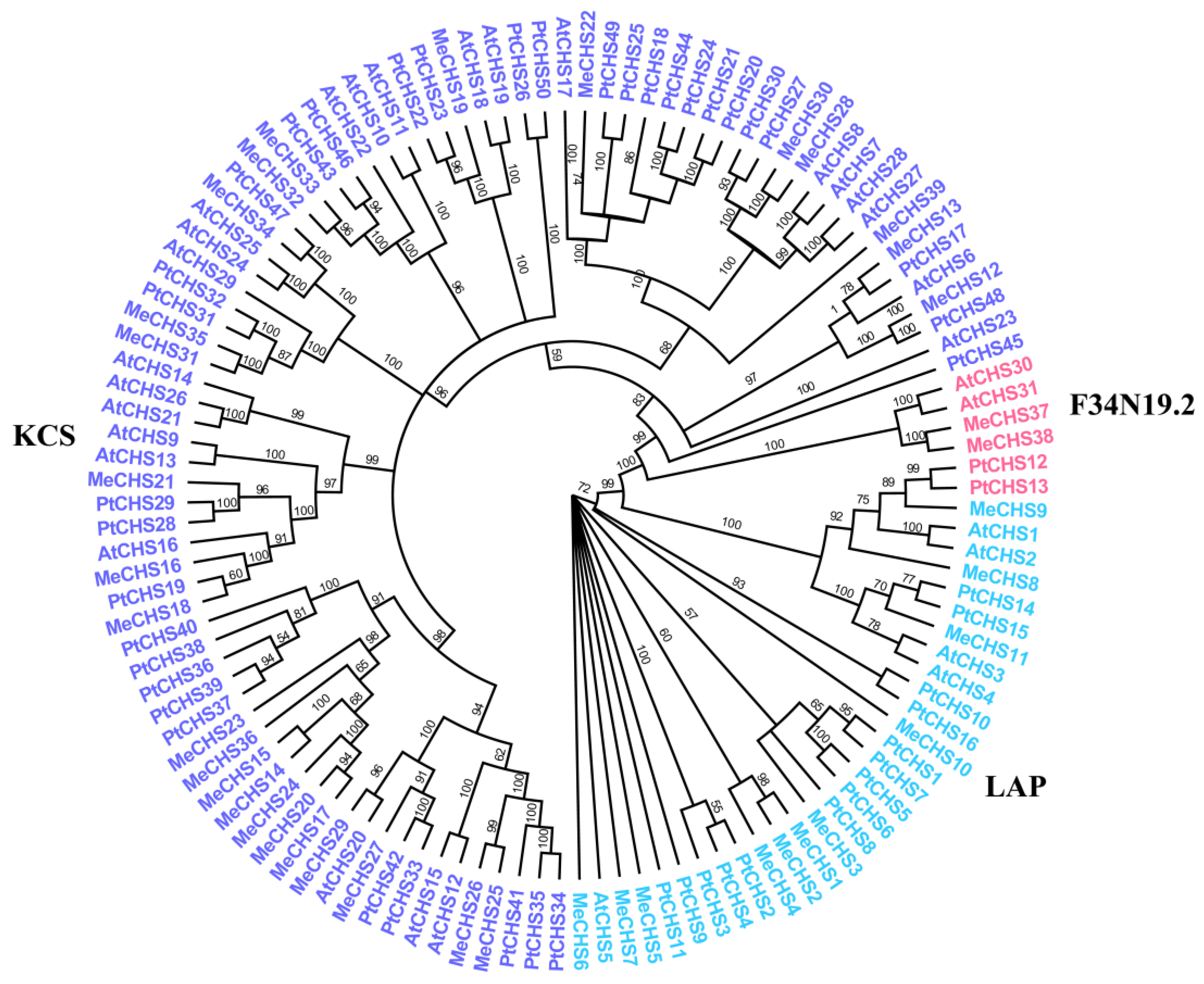
3.2. Phylogenetic Relationship of MeCHS Family
3.3. Conserved Motifs and Gene Structure Analysis

3.4. Chromosomal Localization Analysis
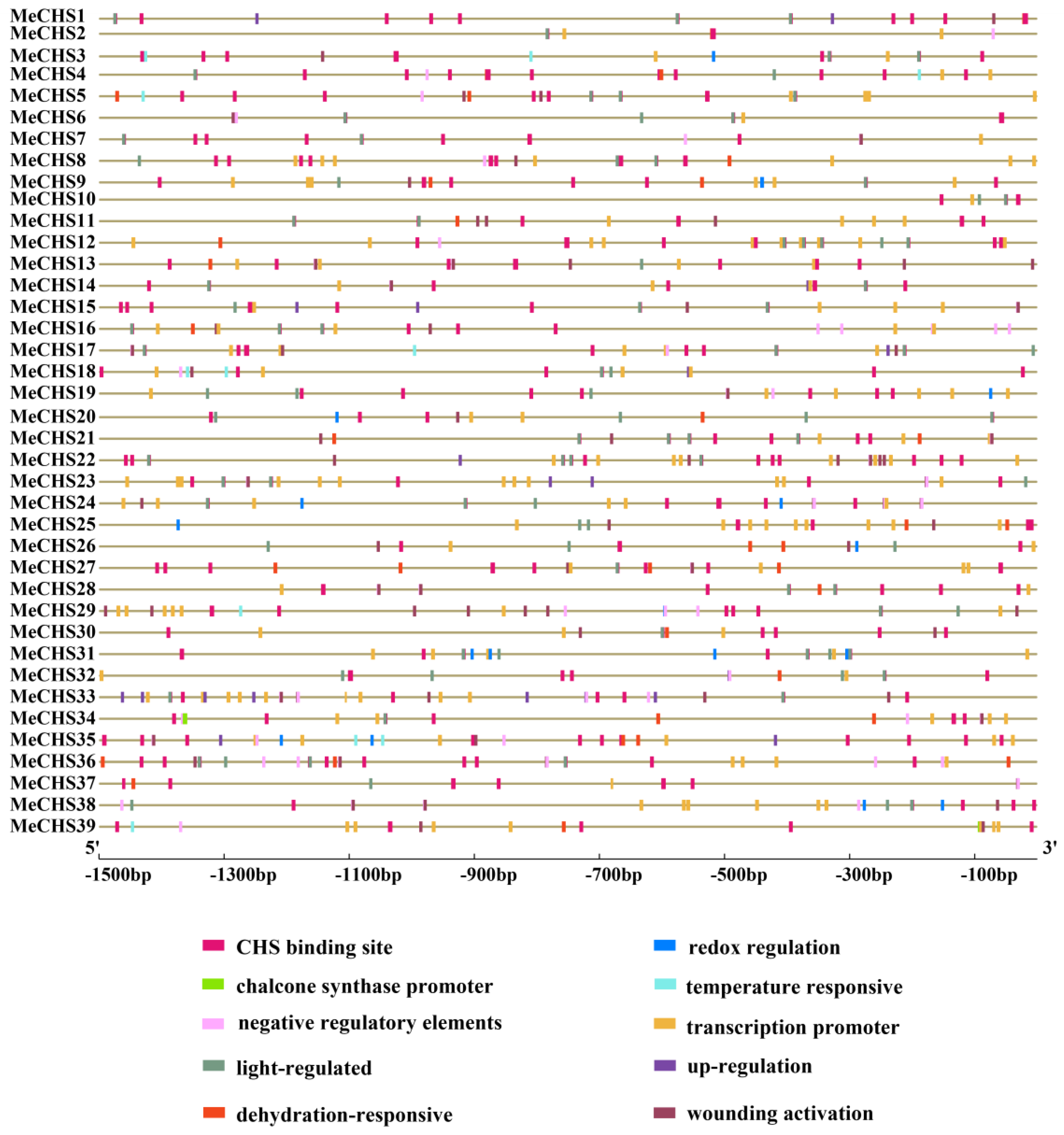
3.5. Promoters Cis-Elements Analysis
3.6. Protein-Protein Interaction (PPI) Network Analysis
3.7. Analysis of MeCHS Gene Expression in Different Tissues
3.8. Verification of MeCHS Gene Expression
3.9. Gene Ontology (GO) Enrichment Analysis of MeCHS Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Schröder, J. A family of plant-specific polyketide synthases: Facts and predictions. Trends Plant Sci. 1997, 2, 373–378. [Google Scholar] [CrossRef]
- Austin, M.B. The chalcone synthase superfamily of type III polyketide synthases. Cheminform 2003, 20, 79–110. [Google Scholar]
- Martin, C.R. Structure, function, and regulation of the chalcone synthase. Int. Rev. Cytol.—Surv. Cell Biol. 1993, 147, 233–284. [Google Scholar]
- Sanjari, S.; Shobbar, Z.S.; Ebrahimi, M.; Hasanloo, T.; Tirnaz, S. Chalcone synthase genes from milk thistle (Silybum marianum): Isolation and expression analysis. J. Genet. 2015, 94, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, Y.; Zhan, Y.; Li, Y.; Kawabata, S. Chalcone synthase family genes have redundant roles in anthocyanin biosynthesis and in response to blue/UV-A light in turnip (Brassica rapa; Brassicaceae). Am. J. Bot. 2013, 100, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New Roles for Old Molecules. J. Integr. Plant Biol. 2010, 52, 98–111. [Google Scholar] [CrossRef]
- Ferreyra, M.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222–236. [Google Scholar]
- Dela, G.; Or, E.; Ovadia, R.; Nissim-Levi, A.; Oren-Shamir, M. Changes in anthocyanin concentration and composition in ‘Jaguar’ rose flowers due to transient high-temperature conditions. Plant Sci. 2003, 164, 333–340. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagnè, D.; Rowan, D.D.; Troggio, M. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Lv, L.L.; Feng, X.F.; Li, W.; Ke, L. High temperature reduces peel color in eggplant (Solanum melongena) as revealed by RNA-seq analysis. Genome 2019, 62, 1139–1156. [Google Scholar] [CrossRef]
- Chennupati, P.; Seguin, P.; Chamoun, R.; Jabaji, S. Effects of High-Temperature Stress on Soybean Isoflavone Concentration and Expression of Key Genes Involved in Isoflavone Synthesis. J. Agric. Food Chem. 2012, 60, 12421–12427. [Google Scholar] [CrossRef] [PubMed]
- Correia, B.; Rodriguez, J.L.; Valledor, L.; Almeida, T.; Pinto, G. Analysis of the expression of putative heat-stress related genes in relation to thermotolerance of cork oak. J. Plant Physiol. 2014, 171, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Schijlen, E.G. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 2007, 144, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Pandith, S.A.; Ramazan, S.; Khan, M.I.; Reshi, Z.A.; Shah, M.A. Chalcone synthases (CHSs): The symbolic type III polyketide synthases. Planta 2020, 251, 106–113. [Google Scholar] [CrossRef]
- Reimold, U.; Kröger, M.; Kreuzaler, F.; Hahlbrock, K. Coding and 3’ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J. 1983, 2, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 775–784. [Google Scholar]
- Dao, T.; Linthorst, H.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Yazaki, K.; Sugiyama, A.; Morita, M.; Shitan, N. Secondary transport as an efficient membrane transport mechanism for plant secondary metabolites. Phytochem. Rev. 2008, 7, 513–524. [Google Scholar] [CrossRef]
- Goławska, S.; Sprawka, I.; Łukasik, I.; Goławski, A. Are naringenin and quercetin useful chemicals in pest-management strategies? J. Pest Sci. 2014, 87, 173–180. [Google Scholar] [CrossRef]
- Gabriele, M.; Frassinetti, S.; Caltavuturo, L.; Montero, L.; Dinelli, G.; Longo, V.; Gioia, D.D.; Pucci, L. Citrus bergamia powder: Antioxidant, antimicrobial and anti-inflammatory properties. J. Funct. Foods 2017, 31, 255–265. [Google Scholar] [CrossRef]
- Lawton, M.A.; Dixon, R.A.; Hahlbrock, K.; Lamb, C.J. Elicitor induction of mRNA activity. Rapid effects of elicitor on phenylalanine ammonia-lyase and chalcone synthase mRNA activities in bean cells. Eur. J. Biochem. 2010, 130, 131–139. [Google Scholar] [CrossRef]
- Ran, L.; Xu, C.; Zhang, Q.; Wang, S.; Fang, W. Evolution of the chitin synthase gene family correlates with fungal morphogenesis and adaption to ecological niches. Sci. Rep. 2017, 7, 44527. [Google Scholar]
- Castelblanco, W.; Fregene, M. SSCP-SNP-based conserved ortholog set (COS) markers for comparative genomics in cassava (Manihot esculenta crantz). Plant Mol. Biol. Rep. 2006, 24, 229–236. [Google Scholar] [CrossRef]
- Prochnik, S.; Marri, P.R.; Desany, B.; Rabinowicz, P.D.; Kodira, C.; Mohiuddin, M.; Rodriguez, F.; Fauquet, C.; Tohme, J.; Harkins, T. The Cassava Genome: Current Progress, Future Directions. Trop. Plant Biol. 2012, 5, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Tian, Y.; Jiang, C.; Yang, Q.; Wang, H.; Li, Q. Laboratory assays on the effects of a novel acaricide, SYP-9625 on Tetranychus cinnabarinus (Boisduval) and its natural enemy, Neoseiulus californicus (McGregor). PLoS ONE 2018, 13, e0199269. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Luo, X.; Wei, W.; Fan, Z.; Pan, X. Analysis of leaf morphology, secondary metabolites and proteins related to the resistance to Tetranychus cinnabarinus in cassava (Manihot esculenta Crantz). Sci. Rep. 2020, 10, 14197. [Google Scholar] [CrossRef]
- Park, H.L.; Yoo, Y.; Bhoo, S.H.; Lee, T.H.; Cho, M.H. Two Chalcone Synthase Isozymes Participate Redundantly in UV-Induced Sakuranetin Synthesis in Rice. Int. J. Mol. Sci. 2020, 21, 3777. [Google Scholar] [CrossRef]
- Han, Y.; Cao, Y.; Jiang, H.; Ding, T. Genome-wide dissection of the chalcone synthase gene family in Oryza sativa. Mol. Breed. 2017, 37, 119. [Google Scholar] [CrossRef]
- Hu, L.; He, H.; Zhu, C.; Peng, X.; Liu, S. Genome-wide identification and phylogenetic analysis of the chalcone synthase gene family in rice. J. Plant Res. 2016, 130, 1–11. [Google Scholar] [CrossRef]
- Kong, X.; Khan, A.; Li, Z.; You, J.; Zhou, R. Identification of chalcone synthase genes and their expression patterns reveal pollen abortion in cotton. Saudi J. Biol. Sci. 2020, 27, 3691–3699. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-Wide Identification, Characterization and Expression Analysis of the Chalcone Synthase Family in Maize. Int. J. Mol. Sci. 2016, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.E.; Spelt, C.E.; Van, D.; Mol, J. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 1989, 81, 245–257. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Ichinose, Y.; Yamada, T.; Tanaka, Y.; Oku, H. Organization of the genes encoding chalcone synthase in Pisum sativum. Plant Mol. Biol. 1993, 21, 789. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, S.; Liu, X.; Shang, J.; Zha, D. Chalcone synthase (CHS) family members analysis from eggplant (Solanum melongena L.) in the flavonoid biosynthetic pathway and expression patterns in response to heat stress. PLoS ONE 2020, 15, e0226537. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Wang, T.; Wang, H.; Liu, H.; Liu, S.; Zhao, Q.; Chen, K.; Zhang, P. Identification, characterization and expression analysis of the chalcone synthase family in the Antarctic moss Pohlia nutans. Antarct. Sci. 2019, 31, 23–33. [Google Scholar] [CrossRef]
- Clegg, M.T.; Durbin, M.L. Flower color variation: A model for the experimental study of evolution. Proc. Natl. Acad. Sci. USA 2000, 97, 7016–7023. [Google Scholar] [CrossRef]
- Chen, S.; Pan, X.; Li, Y.; Cui, L.; Zhang, Y.; Zhang, Z.; Pan, G.; Yang, J.; Cao, P.; Yang, A. Identification and Characterization of Chalcone Synthase Gene Family Members in Nicotiana tabacum. J. Plant Growth Regul. 2017, 36, 374–384. [Google Scholar] [CrossRef]
- Samappito, S. Molecular characterization of root-specific chalcone synthases from Cassia alata. Planta 2002, 216, 64–71. [Google Scholar] [CrossRef]
- Shan, L.W.; Wang, Y.; Wang, M.L.; Wang, Z.H. Cloning and Expression Analysis of CHS Genes Involved in the Biosynthesis of Flavonoids in Soybean. Acta Bot. Boreali—Occident. Sin. 2012, 56, 458–464. [Google Scholar]
- Wannapinpong, S.; Srikulnath, K.; Thongpan, A.; Choowongkomon, K.; Peyachoknagul, S. Molecular cloning and characterization of the CHS gene family in turmeric (Curcuma longa Linn.). J. Plant Biochem. Biotechnol. 2015, 24, 25–33. [Google Scholar]
- Wang, Z.; Yu, Q.; Shen, W.; El, M.; Zhao, X.; Gmitter, F.G. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Yahyaa, M.; Ali, S.; Davidovich-Rikanati, R.; Ibdah, M.; Ibdah, M. Characterization of three chalcone synthase-like genes from apple (Malus × domestica Borkh.). Phytochemistry 2017, 140, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Derynck, M.R.; Chen, L.; Dhaubhadel, S. Differential expression of CHS7 and CHS8 genes in soybean. Planta 2010, 231, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, J.H. Tissue-Specific Gene Silencing Mediated by a Naturally Occurring Chalcone Synthase Gene Cluster in Glycine max. Plant Cell 2004, 16, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhi, S.; Liu, C.; Xu, F.; Zhao, A.; Wang, X.; Tang, X.; Li, Z.; Huang, P.; Yu, M. Isolation and characterization of a novel chalcone synthase gene family from mulberry. Plant Physiol. Biochem. 2017, 115, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Farzad, M.; Soria-Hernanz, D.F.; Altura, M.; Hamilton, M.B.; Weiss, M.R.; Elmendorf, H.G. Molecular evolution of the chalcone synthase gene family and identification of the expressed copy in flower petal tissue of Viola cornuta. Plant Sci. 2005, 168, 1127–1134. [Google Scholar] [CrossRef]
- Finn, R.D.; Alex, B.; Jody, C.; Penelope, C.; Eberhardt, R.Y.; Eddy, S.R.; Andreas, H.; Kirstie, H.; Liisa, H.; Jaina, M. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Chakrabarty, B.; Parekh, N. Identifying tandem Ankyrin repeats in protein structures. BMC Bioinform. 2014, 15, 6599. [Google Scholar] [CrossRef]
- Gu, Z.; Andre, C.; Chen, F.; Peter, B.; Li, W. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Andrea, F.; Damian, S.; Sune, F.; Michael, K.; Milan, S.; Alexander, R.; Lin, J.; Pablo, M.; Peer, B.; Christian, V.M. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar]
- Eom, H.; Su, J.P.; Min, K.K.; Kim, H.; Lee, I. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J. Cell Mol. Biol. 2018, 93, 79–91. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mao, X.; Huang, J.; Yang, D.; Wu, J.; Dong, S.; Lei, K.; Ge, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar]
- Cen, W.; Liu, J.; Lu, S.; Jia, P.; Kai, Y.; Yue, H.; Li, R.; Luo, J. Comparative proteomic analysis of QTL CTS-12 derived from wild rice (Oryza rufipogon Griff.), in the regulation of cold acclimation and de-acclimation of rice (Oryza sativa L.) in response to severe chilling stress. BMC Plant Biol. 2018, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Sheng, F.; Dong, Z.; Zhang, L.; Cai, G.; Xin, M.; Mobeen, T.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genomics 2017, 18, 827. [Google Scholar]
- Long, M. Gene Duplication and Evolution. Science 2001, 293, 1551. [Google Scholar] [CrossRef]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Depamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2010, 50, 873–885. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Report. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Bartling, D.; Nosek, J. Molecular and immunological characterization of leucine aminopeptidase in Arabidopsis thaliana: A new antibody suggests a semi-constitutive regulation of a phylogenetically old enzyme. Plant Sci. 1994, 99, 199–209. [Google Scholar] [CrossRef]
- Jerome, G.; Patricia, A.R.; Teixeira, R.T.; Martinez-Zapater, J.M.; Fortes, A.M. Structural and Functional Analysis of the GRAS Gene Family in Grapevine Indicates a Role of GRAS Proteins in the Control of Development and Stress Responses. Front. Plant 2016, 7, 353. [Google Scholar]
- Sommer, H.; Saedler, H. Structure of the chalcone synthase gene of Antirrhinum majus. Mol. Gen. Genet. MGG 1986, 202, 429–434. [Google Scholar] [CrossRef]
- Radhakrishnan, E.K.; Varghese, R.T.; Vasudevan, S.E. Unusual intron in the second exon of a Type III polyketide synthase gene of Alpinia calcarata Rosc. Genet. Mol. Biol. 2010, 33, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Schommer, C.K.; Sun, Y.K.; Suh, D.Y. Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 2006, 67, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Pandith, S.A.; Dhar, N.; Rana, S.; Bhat, W.W.; Lattoo, S.K. Functional Promiscuity of Two Divergent Paralogs of Type III Plant Polyketide Synthases. Plant Physiol. 2016, 171, 2599–2619. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.Q.; Pang, X.B.; Shen, H.Y.; Pu, G.B.; Wang, H.H.; Lei, C.Y.; Hong, W.; Li, G.F.; Liu, B.Y.; Ye, H.C. A novel type III polyketide synthase encoded by a three-intron gene from Polygonum cuspidatum. Planta 2008, 229, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchishinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Michael, D.M.; Thomashow, M.F. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2010, 60, 328–339. [Google Scholar]
- Mundy, J. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. USA 1990, 87, 1406–1410. [Google Scholar] [CrossRef]
- Chaudhary, P.R.; Bang, H.; Jayaprakasha, G.K.; Patil, B.S. Variation in key flavonoid biosynthetic enzymes and phytochemicals in ‘Rio Red’ grapefruit (Citrus paradisi Macf) during fruit development. J. Agric. Food Chem. 2016, 64, 2975–3003. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Lewinsohn, E.; Britsch, L.; Mazur, Y.; Gressel, J. Flavanone Glycoside Biosynthesis in Citrus. Plant Physiol. 1989, 91, 1323–1328. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Liu, M.; Huang, Z. Genomic and Expression Analysis of Cassava (Manihot esculenta Crantz) Chalcone Synthase Genes in Defense against Tetranychus cinnabarinus Infestation. Genes 2024, 15, 336. https://doi.org/10.3390/genes15030336
Yang Y, Liu M, Huang Z. Genomic and Expression Analysis of Cassava (Manihot esculenta Crantz) Chalcone Synthase Genes in Defense against Tetranychus cinnabarinus Infestation. Genes. 2024; 15(3):336. https://doi.org/10.3390/genes15030336
Chicago/Turabian StyleYang, Yanni, Ming Liu, and Zenghui Huang. 2024. "Genomic and Expression Analysis of Cassava (Manihot esculenta Crantz) Chalcone Synthase Genes in Defense against Tetranychus cinnabarinus Infestation" Genes 15, no. 3: 336. https://doi.org/10.3390/genes15030336
APA StyleYang, Y., Liu, M., & Huang, Z. (2024). Genomic and Expression Analysis of Cassava (Manihot esculenta Crantz) Chalcone Synthase Genes in Defense against Tetranychus cinnabarinus Infestation. Genes, 15(3), 336. https://doi.org/10.3390/genes15030336




