Identification of Salt-Sensitive and Salt-Tolerant Genes through Weighted Gene Co-Expression Networks across Multiple Datasets: A Centralization and Differential Correlation Analysis
Abstract
1. Introduction
2. Results
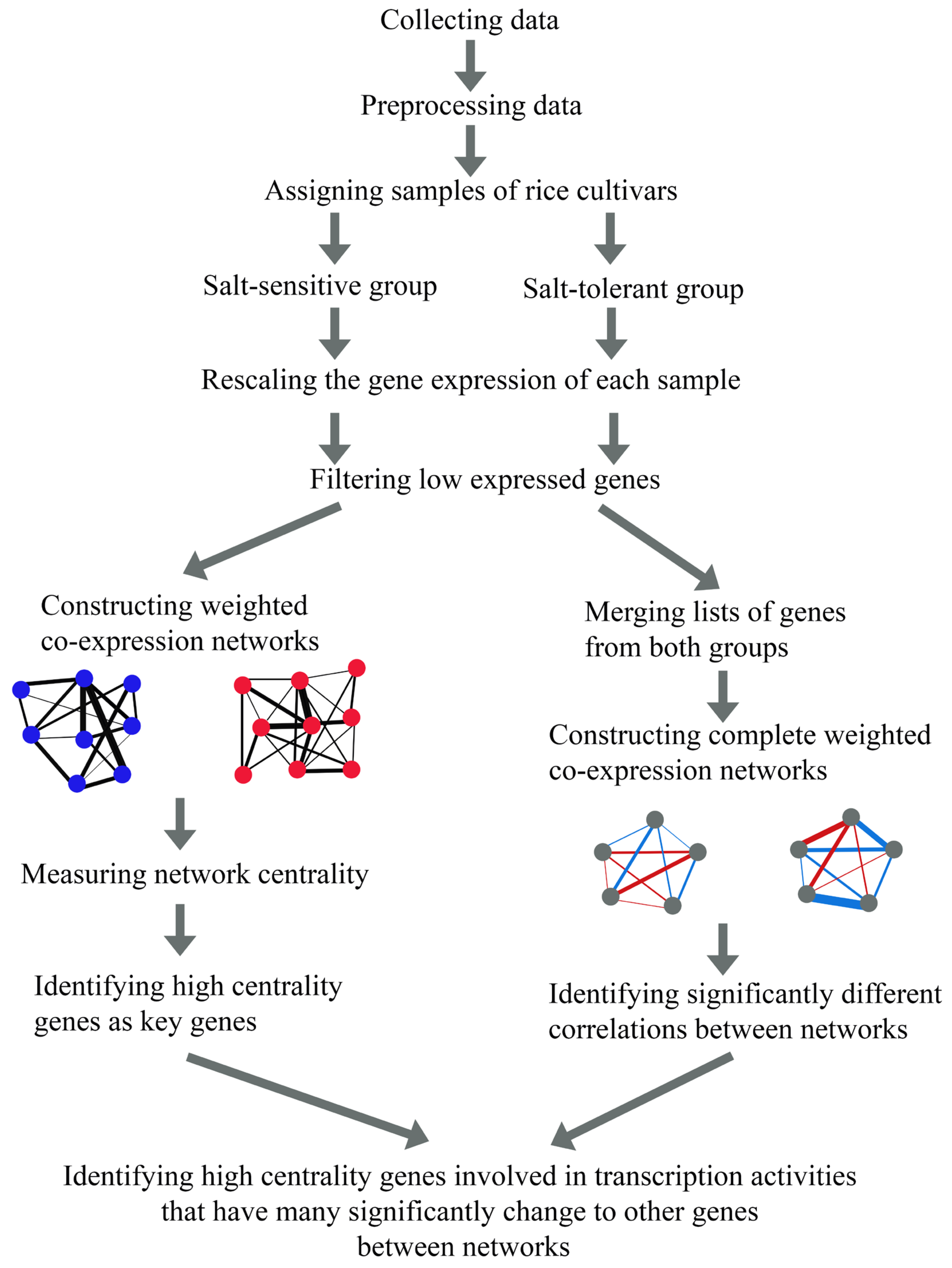
2.1. Characteristics of the Weighted Gene Co-Expression Networks
2.2. Network Parameters and Centralities
2.3. Genes with Highly Different Centrality Scores in Salt-Sensitive versus Salt-Tolerant Networks
2.3.1. Degree Centrality
2.3.2. GO Analysis of High-Degree Genes
2.3.3. Closeness Centrality
2.3.4. GO Analysis of High-Closeness Genes
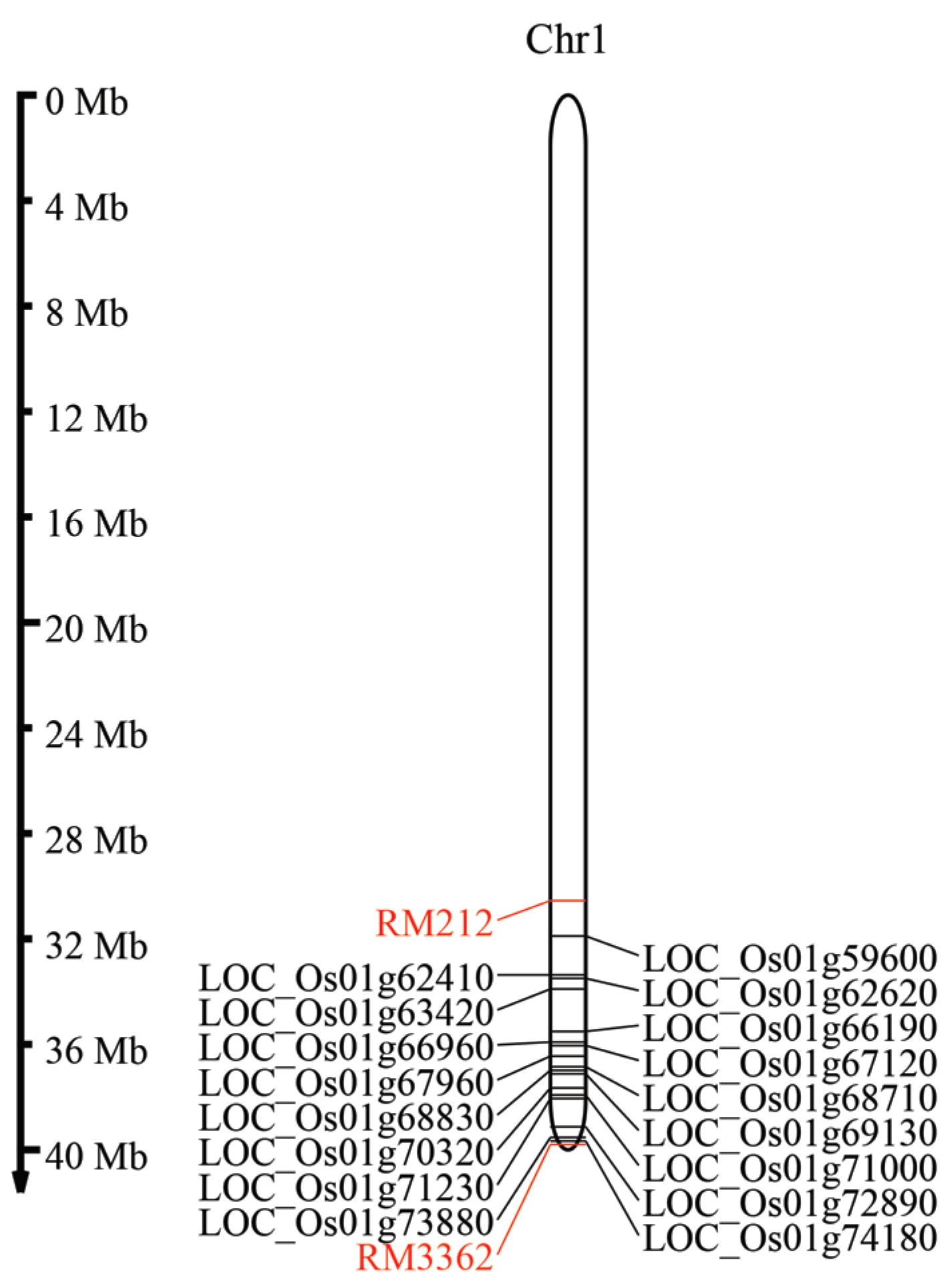
2.4. Identification of High-Centrality Genes Associated Meta-QTLs in Rice
2.5. Identification of Candidated Genes in Thai Rice
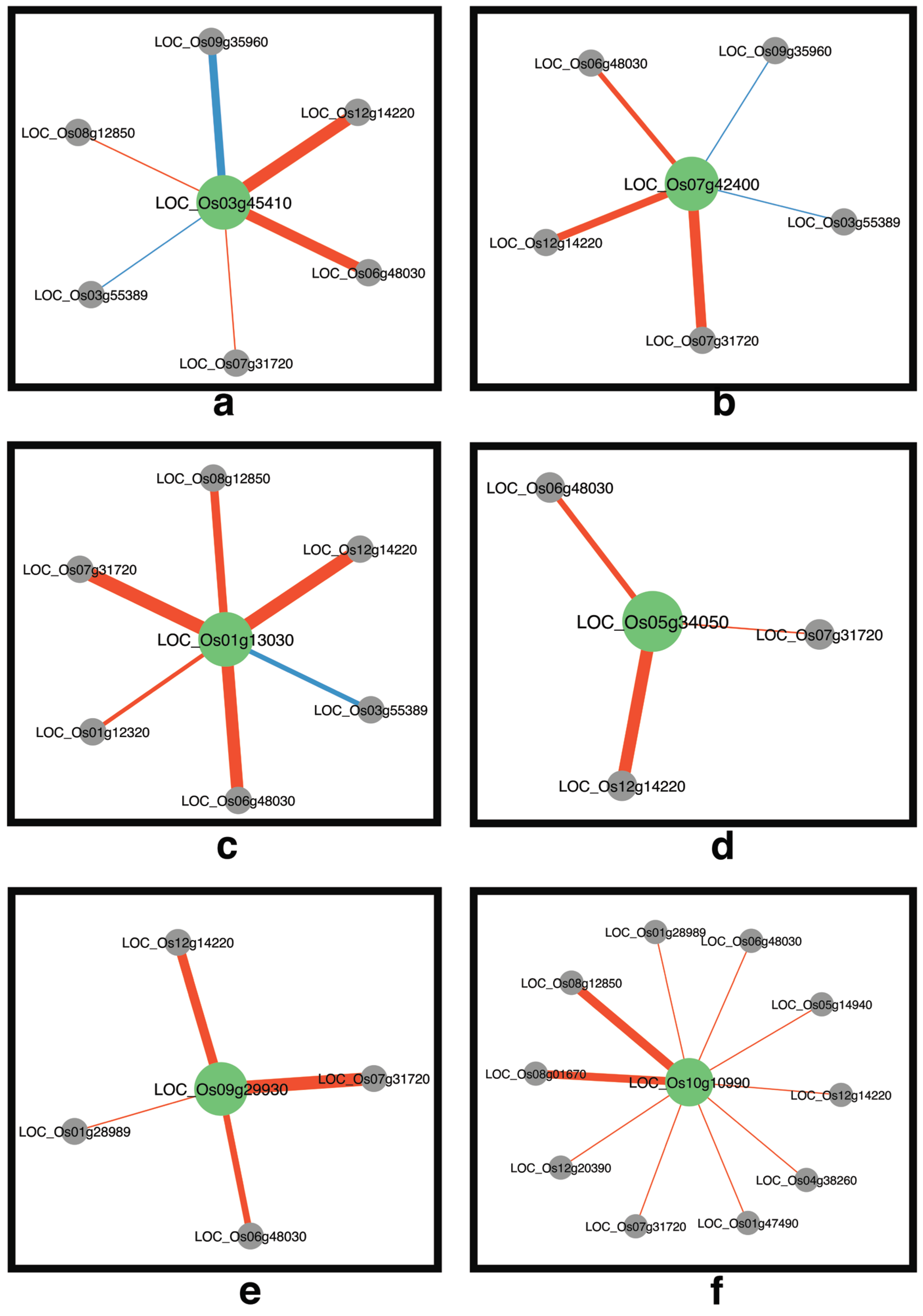
2.6. Identification of Transcription Factors Involving Various Significantly Different Correlations between the Networks
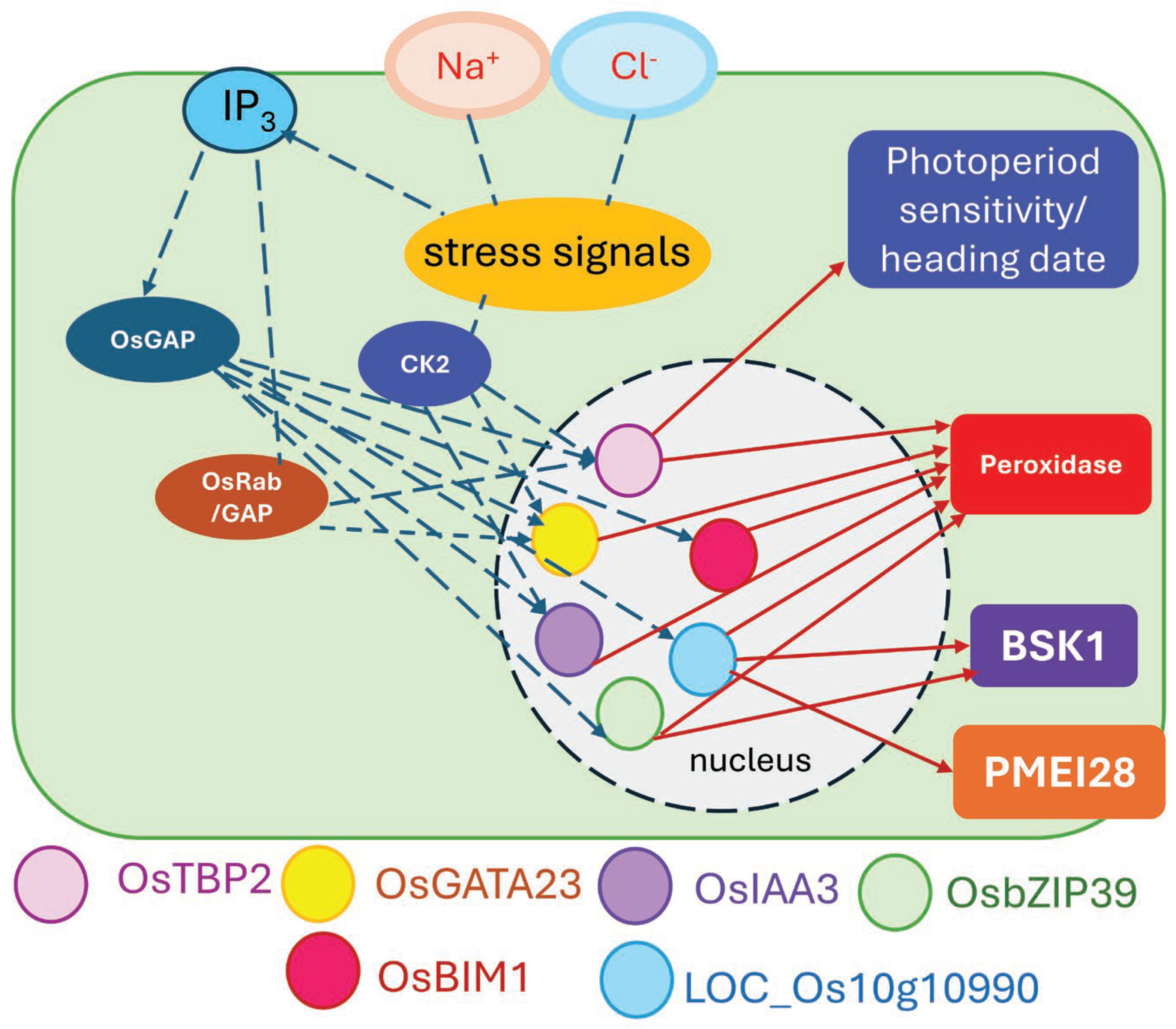
2.7. Salt-Stress Response Mechanisms
3. Discussion
4. Materials and Methods
4.1. Identification of Transcription Factors Involving Various Significantly Different Correlations between the Networks
4.2. Construction of Weighted Gene Co-Expression Network and Identification of High Centrality Genes
4.3. Identification of Genes with High Differential Correlations between Networks
4.4. Gene Ontology Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagarajan, S.; Varatharajan, N.; Gandhimeyyan, R.V. Understanding the Responses, Mechanism and Development of Salinity Stress Tolerant Cultivars in Rice. In Integrative Advances in Rice Research; Huang, M., Ed.; IntechOpen: London, UK, 2022; p. 91. [Google Scholar] [CrossRef]
- Rodríguez Coca, L.I.; García González, M.T.; Unday, Z.G.; Hernández, J.J.; Rodríguez Jáuregui, M.M.; Cancio, Y.F. Effects of Sodium Salinity on Rice (Oryza Sativa L.) Cultivation: A Review. Sustainability 2023, 15, 1804. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Hussain, S.; Yang, S.; Li, R.; Liu, S.; Chen, Y.; Wei, H.; Dai, Q.; Hou, H. Study on the Effect of Salt Stress on Yield and Grain Quality among Different Rice Varieties. Front. Plant Sci. 2022, 13, 918460. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef]
- Razzaq, A.; Ali, A.; Safdar, L.B.; Zafar, M.M.; Rui, Y.; Shakeel, A.; Shaukat, A.; Ashraf, M.; Gong, W.; Yuan, Y. Salt stress induces physiochemical alterations in rice grain composition and quality. J. Food Sci. 2020, 85, 14–20. [Google Scholar] [CrossRef]
- Haque, M.A.; Rafii, M.Y.; Yusoff, M.M.; Ali, N.S.; Yusuff, O.; Datta, D.R.; Anisuzzaman, M.; Ikbal, M.F. Advanced Breeding Strategies and Future Perspectives of Salinity Tolerance in Rice. Agronomy 2021, 11, 1631. [Google Scholar] [CrossRef]
- Qin, H.; Li, Y.; Huang, R. Advances and Challenges in the Breeding of Salt-Tolerant Rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef]
- Ammar, M.H.M.; Pandit, A.; Singh, R.K.; Sameena, S.; Chauhan, M.S.; Singh, A.K.; Sharma, P.C.; Gaikwad, K.; Sharma, T.R.; Mohapatra, T.; et al. Mapping of QTLs Controlling Na+, K+ and CI− Ion Concentrations in Salt Tolerant Indica Rice Variety CSR27. J. Plant Biochem. Biotechnol. 2009, 18, 139–150. [Google Scholar] [CrossRef]
- Thomson, M.J.; de Ocampo, M.; Egdane, J.; Rahman, M.A.; Sajise, A.G.; Adorada, D.L.; Tumimbang-Raiz, E.; Blumwald, E.; Seraj, Z.I.; Singh, R.K.; et al. Characterizing the Saltol Quantitative Trait Locus for Salinity Tolerance in Rice. Rice 2010, 3, 148–160. [Google Scholar] [CrossRef]
- Lekklar, C.; Pongpanich, M.; Suriya-Arunroj, D.; Chinpongpanich, A.; Tsai, H.; Comai, L.; Chadchawan, S.; Buaboocha, T. Genome-wide association study for salinity tolerance at the flowering stage in a panel of rice accessions from Thailand. BMC Genom. 2019, 20, 76. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, X.; Zhao, Y.; Khan, N.U.; Zhao, Z.; Zhang, Y.; Wen, X.; Tang, F.; Wang, F.; Li, Z. Genetic basis and identification of candidate genes for salt tolerance in rice by GWAS. Sci. Rep. 2020, 10, 9958. [Google Scholar] [CrossRef]
- Nayyeripasand, L.; Garoosi, G.A.; Ahmadikhah, A. Genome-Wide Association Study (GWAS) to Identify Salt-Tolerance QTLs Carrying Novel Candidate Genes in Rice during Early Vegetative Stage. Rice 2021, 14, 9. [Google Scholar] [CrossRef]
- Sonsungsan, P.; Chantanakool, P.; Suratanee, A.; Buaboocha, T.; Comai, L.; Chadchawan, S.; Plaimas, K. Identification of Key Genes in ‘Luang Pratahn’, Thai Salt-Tolerant Rice, Based on Time-Course Data and Weighted Co-expression Networks. Front. Plant Sci. 2021, 12, 744654. [Google Scholar] [CrossRef]
- Cartagena, J.A.; Yao, Y.; Mitsuya, S.; Tsuge, T. Comparative transcriptome analysis of root types in salt tolerant and sensitive rice varieties in response to salinity stress. Physiol. Plant 2021, 173, 1629–1642. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Ogata, Y.; Shibata, D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007, 48, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Charitou, T.; Bryan, K.; Lynn, D.J. Using biological networks to integrate, visualize and analyze genomics data. Genet. Sel. Evol. 2016, 48, 27. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A. DiffCorr: An R package to analyze and visualize differential correlations in biological networks. Gene 2013, 518, 209–214. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Mirdar Mansuri, R.; Shobbar, Z.S.; Babaeian Jelodar, N.; Ghaffari, M.; Mohammadi, S.M.; Daryani, P. Salt tolerance involved candidate genes in rice: An integrative meta-analysis approach. BMC Plant Biol. 2020, 20, 452. [Google Scholar] [CrossRef] [PubMed]
- Chutimanukul, P.; Kositsup, B.; Plaimas, K.; Buaboocha, T.; Siangliw, M.; Toojinda, T.; Comai, L.; Chadchawan, S. Photosynthetic responses and identification of salt tolerance genes in a chromosome segment substitution line of ‘Khao dawk Mali 105’ rice. Environ. Exp. Bot. 2018, 15, 497–508. [Google Scholar] [CrossRef]
- Chutimanukul, P.; Saputro, T.B.; Mahaprom, P.; Plaimas, K.; Comai, L.; Buaboocha, T.; Siangliw, M.; Toojinda, T.; Chadchawan, S. Combining Genome and Gene Co-expression Network Analyses for the Identification of Genes Potentially Regulating Salt Tolerance in Rice. Front. Plant Sci. 2021, 12, 704549. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.; Zhang, X.; et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Park, H.J.; You, Y.N.; Lee, A.; Jung, H.; Jo, S.H.; Oh, N.; Kim, H.S.; Lee, H.J.; Kim, J.K.; Kim, Y.S.; et al. OsFKBP20-1b interacts with the splicing factor OsSR45 and participates in the environmental stress response at the post-transcriptional level in rice. Plant J. 2020, 102, 992–1007. [Google Scholar] [CrossRef]
- Gu, X.; Gao, S.; Li, J.; Song, P.; Zhang, Q.; Guo, J.; Wang, X.; Han, X.; Wang, X.; Zhu, Y.; et al. The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice. Plant Physiol. Biochem. 2021, 169, 269–279. [Google Scholar] [CrossRef]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic Stresses Cause Differential Regulation of Alternative Splice Forms of GATA Transcription Factor in Rice. Front. Plant Sci. 2017, 8, 1944. [Google Scholar] [CrossRef]
- Huang, J.; Liu, F.; Chao, D.; Xin, B.; Liu, K.; Cao, S.; Chen, X.; Peng, L.; Zhang, B.; Fu, S.; et al. The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 11999. [Google Scholar] [CrossRef]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef]
- Teng, Y.; Lv, M.; Zhang, X.; Cai, M.; Chen, T. BEAR1, a bHLH Transcription Factor, Controls Salt Response Genes to Regulate Rice Salt Response. J. Plant Biol. 2022, 65, 217–230. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H.; et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 2021, 21, 546. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, D.B.; Chen, H.; Halay, E.D.; Usheva, A.A.; Hisatake, K.; Lee, D.K.; Roeder, R.G.; Burley, S.K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 1995, 377, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Orphanides, G.; Lagrange, T.; Reinberg, D. The general transcription factors of RNA polymerase II. Genes. Dev. 1996, 10, 2657–2683. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Ren, F.; Lu, Y.T. The Arabidopsis mutant stg1 identifies a function for TBP-associated factor 10 in plant osmotic stress adaptation. Plant Cell Physiol. 2006, 47, 1285–1294. [Google Scholar] [CrossRef]
- Grzesiak, M.T.; Hordynska, N.; Maksymowicz, A.; Grzesiak, S.; Szechynska-Hebda, M. Variation Among Spring Wheat (Triticum aestivum L.) Genotypes in Response to the Drought Stress. II-Root System Structure. Plants 2019, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Parvathi, M.S.; Nataraja, K.N. Discovery of Stress Responsive TATA-Box Binding Protein Associated Factor6 (TAF6) from Finger Millet (Eleusine coracana (L.) Gaertn). J. Plant Biol. 2017, 60, 335–342. [Google Scholar] [CrossRef]
- Zou, Y.; Huang, W.; Gu, Z.; Gu, X. Predominant gain of promoter TATA box after gene duplication associated with stress responses. Mol. Biol. Evol. 2011, 28, 2893–2904. [Google Scholar] [CrossRef]
- Zhang, Y.; Iqbal, M.F.; Wang, Y.; Qian, K.; Xiang, J.; Xu, G.; Fan, X. OsTBP2.1, a TATA-Binding Protein, Alters the Ratio of OsNRT2.3b to OsNRT2.3a and Improves Rice Grain Yield. Int. J. Mol. Sci. 2022, 23, 10795. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Xiao, H.; Chew, J.; Xiang, J.; Qian, K.; Fan, X. Knockdown of a Novel Gene OsTBP2.2 Increases Sensitivity to Drought Stress in Rice. Genes 2020, 11, 629. [Google Scholar] [CrossRef]
- Zhu, Q.; Ordiz, M.I.; Dabi, T.; Beachy, R.N.; Lamb, C. Rice TATA binding protein interacts functionally with transcription factor IIB and the RF2a bZIP transcriptional activator in an enhanced plant in vitro transcription system. Plant Cell 2002, 14, 795–803. [Google Scholar] [CrossRef]
- Jackson, T.R.; Kearns, B.G.; Theibert, A.B. Cytohesins and centaurins: Mediators of PI 3-kinase-regulated Arf signaling. Trends Biochem. Sci. 2000, 25, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Ganotra, J.; Sharma, B.; Biswal, B.; Bhardwaj, D.; Tuteja, N. Emerging role of small GTPases and their interactome in plants to combat abiotic and biotic stress. Protoplasma 2023, 260, 1007–1029. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Rho, H.S.; Kim, S.W.; Hwang, S.M.; Kwon, H.J.; Nahm, M.Y.; Bang, W.Y.; Bahk, J.D. OsGAP1 functions as a positive regulator of OsRab11-mediated TGN to PM or vacuole trafficking. Plant Cell Physiol. 2005, 46, 2005–2018. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, M.K.; Deswal, R.; Sopory, S.K. Plant RABs: Role in Development and in Abiotic and Biotic Stress Responses. Curr. Genom. 2021, 22, 26–40. [Google Scholar] [CrossRef]
- Huttner, S.; Veit, C.; Schoberer, J.; Grass, J.; Strasser, R. Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant Mol. Biol. 2012, 79, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cui, F.; Li, Q.; Yin, B.; Zhang, H.; Lin, B.; Wu, Y.; Xia, R.; Tang, S.; Xie, Q. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011, 21, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Gao, G.; Yang, X.; Yang, X.; Ma, P. Casein kinase CK2 structure and activities in plants. J. Plant Physiol. 2022, 276, 153767. [Google Scholar] [CrossRef]
- Hasan, M.M.; Rahman, M.A.; Corpas, F.J.; Rahman, M.M.; Jahan, M.S.; Liu, X.D.; Ghimire, S.; Alabdallah, N.M.; Wassem, M.; Alharbi, B.M.; et al. Salt stress tolerance in rice (Oryza sativa L.): A proteomic overview of recent advances and future prospects. Plant Stress 2024, 11, 100307. [Google Scholar] [CrossRef]
- Li, W.Q.; Zheng, W.J.; Peng, Y.; Shao, Y.; Liu, C.T.; Li, J.; Hu, Y.Y.; Zhao, B.R.; Mao, B.G. OsPMS1 Mutation Enhances Salt Tolerance by Suppressing ROS Accumulation, Maintaining Na+/K+ Homeostasis, and Promoting ABA Biosynthesis. Genes 2023, 14, 1621. [Google Scholar] [CrossRef] [PubMed]
- Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J. Exp. Bot. 2020, 71, 684–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, T.; Li, Z.; Huang, K.; Kim, N.E.; Ma, Z.; Kwon, S.W.; Jiang, W.; Du, X. OsGATA16, a GATA Transcription Factor, Confers Cold Tolerance by Repressing OsWRKY45-1 at the Seedling Stage in Rice. Rice 2021, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Umemura, I.; Gomi, K.; Hasegawa, Y.; Kitano, H.; Sazuka, T.; Matsuoka, M. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 2006, 46, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Li, G.; Jin, L.; Huang, Y.; Wang, Y.; Wei, C.; Xu, Z.; Yang, Z.; Wang, H.; Li, Y. Auxin response factors (ARFs) differentially regulate rice antiviral immune response against rice dwarf virus. PLoS Pathog. 2020, 16, e1009118. [Google Scholar] [CrossRef]
- Song, X.; Xiong, Y.; Kong, X.; Huang, G. Roles of auxin response factors in rice development and stress responses. Plant Cell Environ. 2023, 46, 1075–1086. [Google Scholar] [CrossRef]
- Carranco, R.; Espinosa, J.M.; Prieto-Dapena, P.; Almoguera, C.; Jordano, J. Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc. Natl. Acad. Sci. USA 2010, 107, 21908–21913. [Google Scholar] [CrossRef]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szecsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef]
- Aloryi, K.D.; Okpala, N.E.; Amo, A.; Bello, S.F.; Akaba, S.; Tian, X. A meta-quantitative trait loci analysis identified consensus genomic regions and candidate genes associated with grain yield in rice. Front. Plant Sci. 2022, 13, 1035851. [Google Scholar] [CrossRef]
- Gao, J.; Zhao, Y.; Zhao, Z.; Liu, W.; Jiang, C.; Li, J.; Zhang, Z.; Zhang, H.; Zhang, Y.; Wang, X.; et al. RRS1 shapes robust root system to enhance drought resistance in rice. New Phytol. 2023, 238, 1146–1162. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, Y.; Zou, X.; Li, F.; Zhang, J.; Kang, Z.; Li, X.; Yin, C.; Lin, Y. Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation. Cells 2020, 9, 916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Tang, Z.; Sun, X.; Zhang, H.; Yu, J.; Yao, G.; Li, G.; Guo, H.; Li, J.; et al. Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3-OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J. Exp. Bot. 2018, 69, 4723–4737. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Shao, J.; Wang, Q.; Chen, P.; Zhu, Y.; Yin, C. Supraoptimal Cytokinin Content Inhibits Rice Seminal Root Growth by Reducing Root Meristem Size and Cell Length via Increased Ethylene Content. Int. J. Mol. Sci. 2018, 19, 4051. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yang, X.; Xiao, C.; Li, J.; Chen, Y.; Li, R.; Li, S.; Lu, S.; Hu, H. GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 2020, 228, 1880–1896. [Google Scholar] [CrossRef]
- Lu, W.; Deng, M.; Guo, F.; Wang, M.; Zeng, Z.; Han, N.; Yang, Y.; Zhu, M.; Bian, H. Suppression of OsVPE3 Enhances Salt Tolerance by Attenuating Vacuole Rupture during Programmed Cell Death and Affects Stomata Development in Rice. Rice 2016, 9, 65. [Google Scholar] [CrossRef]
- Takahashi, H.; Kawakatsu, T.; Wakasa, Y.; Hayashi, S.; Takaiwa, F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 2012, 53, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, W.; Zhou, J.; He, H.; Chen, L.; Chen, H.; Deng, X.W. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci. 2012, 193–194, 8–17. [Google Scholar] [CrossRef]
- Gai, W.X.; Ma, X.; Qiao, Y.M.; Shi, B.H.; Ul Haq, S.; Li, Q.H.; Wei, A.M.; Liu, K.K.; Gong, Z.H. Characterization of the bZIP Transcription Factor Family in Pepper (Capsicum annuum L.): CabZIP25 Positively Modulates the Salt Tolerance. Front. Plant Sci. 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Zhou, R.; Dossa, K.; Yu, J.; Li, D.; Liu, A.; Mmadi, M.A.; Zhang, X.; You, J. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS ONE 2018, 13, e0200850. [Google Scholar] [CrossRef]
- Amir Hossain, M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef]
- Das, P.; Lakra, N.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice 2019, 12, 58. [Google Scholar] [CrossRef]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Liu, C.; Wu, Y.; Wang, X. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice. Planta 2012, 235, 1157–1169. [Google Scholar] [CrossRef]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 260. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Luan, J.; Guo, C.; Shi, X.; Deng, P.; Zhou, Z.; Zhang, W.; Shen, L. A bHLH protein, OsBIM1, positively regulates rice leaf angle by promoting brassinosteroid signaling. Biochem. Biophys. Res. Commun. 2021, 578, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Ehleringer, J.R.; Comstock, J. Leaf absorptance and leaf angle: Mechanisms for stress avoidance. In Plant Response to Stress; NATO ASI Series; Springer: Berlin/Heidelberg, Germany, 1987. [Google Scholar]
- Geldhof, B.; Pattyn, J.; Eyland, D.; Carpentier, S.; Van de Poel, B. A digital sensor to measure real-time leaf movements and detect abiotic stress in plants. Plant Physiol. 2021, 187, 1131–1148. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Sun, B.; He, H.; Zhang, Y.; Tian, H.; Wang, B. Current Understanding of bHLH Transcription Factors in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Goswami, K.; Tripathi, A.; Sanan-Mishra, N. Comparative miRomics of Salt-Tolerant and Salt-Sensitive Rice. J. Integr. Bioinform. 2017, 14, 20170002. [Google Scholar] [CrossRef] [PubMed]
- Vitoriano, C.B.; Calixto, C.P.G. Reading between the Lines: RNA-seq Data Mining Reveals the Alternative Message of the Rice Leaf Transcriptome in Response to Heat Stress. Plants 2021, 10, 1647. [Google Scholar] [CrossRef]
- Yan, J.; He, H.; Fang, L.; Zhang, A. Pectin methylesterase31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 496, 497–501. [Google Scholar] [CrossRef]
- Jithesh, M.N.; Wally, O.S.D.; Manfield, I.; Critchley, A.T.; Hiltz, D.; Prithiviraj, B. Analysis of seaweed extract-induced transcriptome leads to identification of a negative regulator of salt tolerance in Arabidopsis. HortScience 2012, 46, 704–709. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Zhang, Q.; Zhang, Y.; Ou, X.; An, L.; Feng, H.; Zhao, Z. A cold-induced pectin methyl-esterase inhibitor gene contributes negatively to freezing tolerance but positively to salt tolerance in Arabidopsis. J. Plant Physiol. 2018, 222, 67–78. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.; Yoon, I.; Kim, K.; Kwon, T. Salt Tolerance in Rice: Focus on Mechanisms and Approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Hamada, K.; Hongo, K.; Suwabe, K.; Shimizu, A.; Nagayama, T.; Abe, R.; Kikuchi, S.; Yamamoto, N.; Fujii, T.; Yokoyama, K.; et al. OryzaExpress: An integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011, 52, 220–229. [Google Scholar] [CrossRef]
- Sha, Y.; Phan, J.H.; Wang, M.D. Effect of low-expression gene filtering on detection of differentially expressed genes in RNA-seq data. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 6461–6464. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Buell, C.R. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004, 32, D360–D363. [Google Scholar] [CrossRef] [PubMed]
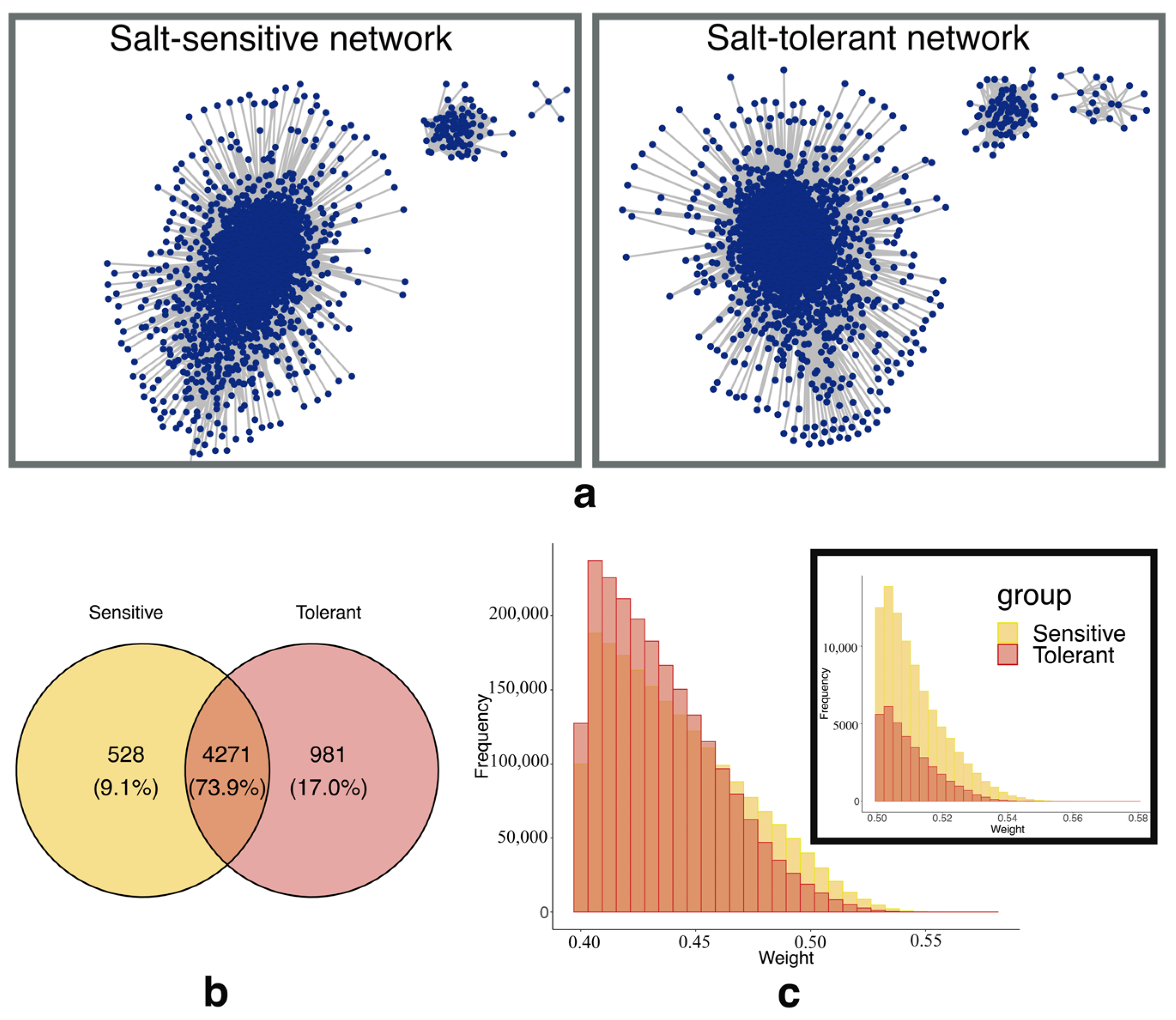



| Properties | Salt-Sensitive Network | Salt-Tolerant Network |
|---|---|---|
| The network diameter | 7 | 4 |
| The network radius | 4 | 2 |
| The average number of neighbors | 861.908 | 833.153 |
| The average shortest path length | 1.966 | 1.902 |
| The network density | 0.183 | 0.162 |
| The network heterogeneity | 0.999 | 1.098 |
| The network clustering coefficient | 0.824 | 0.851 |
| Genes | Function | DG in the Salt-Sensitive Network | DG in the Salt-Tolerant Network |
|---|---|---|---|
| LOC_Os09g32430 | NADH: ubiquinone oxidoreductase | 3401 | 0 |
| LOC_Os06g05150 | expressed protein | 0 | 4320 |
| LOC_Os02g42550 | expressed protein | 0 | 4050 |
| LOC_Os06g42430 | DUF1950 domain containing protein | 0 | 3956 |
| LOC_Os01g17260 | TGA transcription factor, bZIP transcription factor, de-fense response, multiple abiotic stress tolerance | 0 | 3882 |
| LOC_Os06g21580 | HMG-I and HMG-Y, DNA-binding domain containing protein | 0 | 3708 |
| LOC_Os07g42395 | DNA-directed RNA polymerase II subunit RPB9 | 0 | 3544 |
| LOC_Os03g60360 | protein kinase PKN/PRK1, effector | 0 | 3384 |
| LOC_Os03g10200 | ethylene-responsive element-binding protein | 0 | 3380 |
| LOC_Os02g45920 | expressed protein | 0 | 3336 |
| LOC_Os02g57370 | DHHC zinc finger domain containing protein | 0 | 3325 |
| LOC_Os08g01660 | mago nashi | 0 | 3162 |
| LOC_Os10g37690 | inner membrane protein | 0 | 2962 |
| LOC_Os09g28590 | AGAP010609-PA | 0 | 2898 |
| LOC_Os03g37330 | KIN, antigenic determinant of recA protein | 0 | 2892 |
| LOC_Os02g48590 | expressed protein | 0 | 2887 |
| LOC_Os07g27760 | expressed protein | 0 | 2884 |
| LOC_Os03g26090 | GPI-anchored wall transfer protein 1 | 0 | 2824 |
| LOC_Os03g55784 | tumor necrosis factor superfamily, member 5-induced protein 1 | 0 | 2779 |
| LOC_Os10g32870 | L11 domain containing ribosomal protein | 0 | 2749 |
| LOC_Os01g46580 | actin-related protein 2/3 complex subunit 2 | 0 | 2739 |
| LOC_Os03g63140 | L11 domain containing ribosomal protein | 0 | 2735 |
| LOC_Os03g46610 | DEAD-box ATP-dependent RNA helicase | 0 | 2733 |
| LOC_Os06g05250 | GTP-binding protein GUF1 | 0 | 2721 |
| LOC_Os02g10050 | expressed protein | 0 | 2698 |
| LOC_Os05g23530 | expressed protein | 0 | 2684 |
| Genes | Function | CN in the Salt-Sensitive Network | CN in the Salt-Tolerant Network |
|---|---|---|---|
| LOC_Os04g52450 | γ-Aminobutyric acid transaminase | 1 | 0 |
| LOC_Os03g23980 | NAD dependent epimerase/dehydratase family protein | 0.8257 | 0 |
| LOC_Os09g32430 | NADH: ubiquinone oxidoreductase | 0.7693 | 0 |
| LOC_Os06g05150 | expressed protein | 0 | 0.8577 |
| LOC_Os02g42550 | expressed protein | 0 | 0.8181 |
| LOC_Os06g42430 | DUF1950 domain containing protein | 0 | 0.8087 |
| LOC_Os01g17260 | TGA transcription factor, bZIP transcription factor, defense response, multiple abiotic stress tolerance | 0 | 0.7966 |
| LOC_Os06g21580 | HMG-I and HMG-Y, DNA-binding domain containing protein | 0 | 0.7783 |
| LOC_Os07g42395 | DNA-directed RNA polymerase II subunit RPB9 | 0 | 0.7570 |
| LOC_Os10g42500 | PAP fibrillin family domain containing protein | 0 | 0.7565 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonsungsan, P.; Suratanee, A.; Buaboocha, T.; Chadchawan, S.; Plaimas, K. Identification of Salt-Sensitive and Salt-Tolerant Genes through Weighted Gene Co-Expression Networks across Multiple Datasets: A Centralization and Differential Correlation Analysis. Genes 2024, 15, 316. https://doi.org/10.3390/genes15030316
Sonsungsan P, Suratanee A, Buaboocha T, Chadchawan S, Plaimas K. Identification of Salt-Sensitive and Salt-Tolerant Genes through Weighted Gene Co-Expression Networks across Multiple Datasets: A Centralization and Differential Correlation Analysis. Genes. 2024; 15(3):316. https://doi.org/10.3390/genes15030316
Chicago/Turabian StyleSonsungsan, Pajaree, Apichat Suratanee, Teerapong Buaboocha, Supachitra Chadchawan, and Kitiporn Plaimas. 2024. "Identification of Salt-Sensitive and Salt-Tolerant Genes through Weighted Gene Co-Expression Networks across Multiple Datasets: A Centralization and Differential Correlation Analysis" Genes 15, no. 3: 316. https://doi.org/10.3390/genes15030316
APA StyleSonsungsan, P., Suratanee, A., Buaboocha, T., Chadchawan, S., & Plaimas, K. (2024). Identification of Salt-Sensitive and Salt-Tolerant Genes through Weighted Gene Co-Expression Networks across Multiple Datasets: A Centralization and Differential Correlation Analysis. Genes, 15(3), 316. https://doi.org/10.3390/genes15030316






