Author Contributions
Conceptualization, L.A. (Lucía Almorox), L.A. (Laura Antequera), I.R., L.J.H. and F.M.O.; methodology, L.A. (Lucía Almorox), L.A. (Laura Antequera), L.J.H. and F.M.O.; software, L.A. (Lucía Almorox) and L.A. (Laura Antequera); validation, L.A. (Lucía Almorox) and L.A. (Laura Antequera); formal analysis, L.A. (Lucía Almorox), L.A. (Laura Antequera), L.J.H. and F.M.O.; investigation, L.A. (Lucía Almorox), L.A. (Laura Antequera), I.R., L.J.H. and F.M.O.; resources, L.A. (Lucía Almorox), L.A. (Laura Antequera), L.J.H. and F.M.O.; data curation, L.A. (Lucía Almorox), L.A. (Laura Antequera) and F.M.O.; writing—original draft preparation, L.A. (Lucía Almorox), L.A. (Laura Antequera), L.J.H. and F.M.O.; writing—review and editing, I.R., L.J.H. and F.M.O.; visualization, L.A. (Lucía Almorox) and L.A. (Laura Antequera); supervision, I.R., L.J.H. and F.M.O.; project administration, I.R., L.J.H. and F.M.O.; funding acquisition, I.R., L.J.H. and F.M.O. All authors have read and agreed to the published version of the manuscript.
Figure 1.
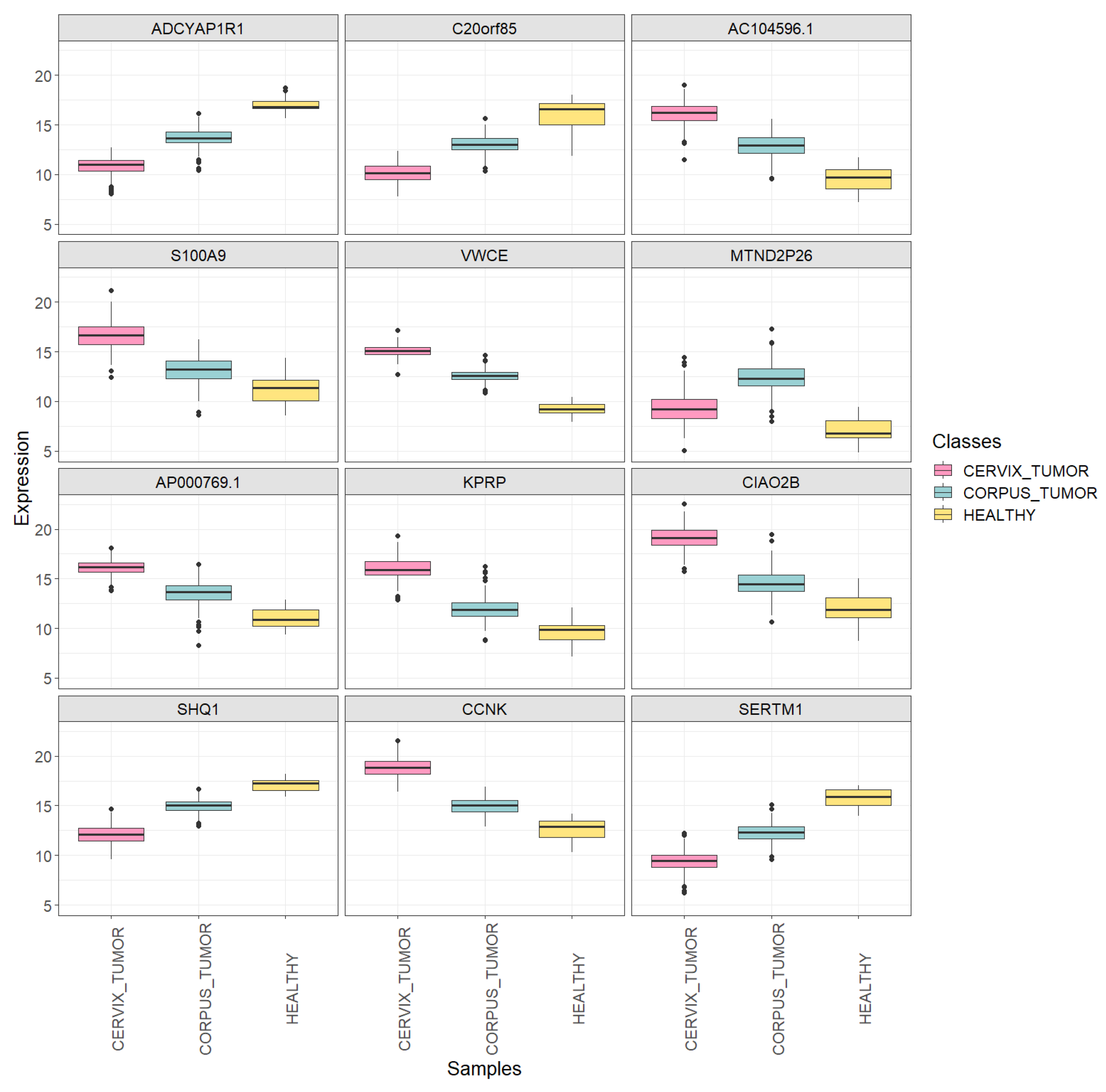
Boxplots showing the expression of the first 12 extracted DEGs in each uterine sample class (CORPUS_TUMOR, CERVIX_TUMOR and HEALTHY) using the training set.
Figure 1.
Boxplots showing the expression of the first 12 extracted DEGs in each uterine sample class (CORPUS_TUMOR, CERVIX_TUMOR and HEALTHY) using the training set.
Figure 2.
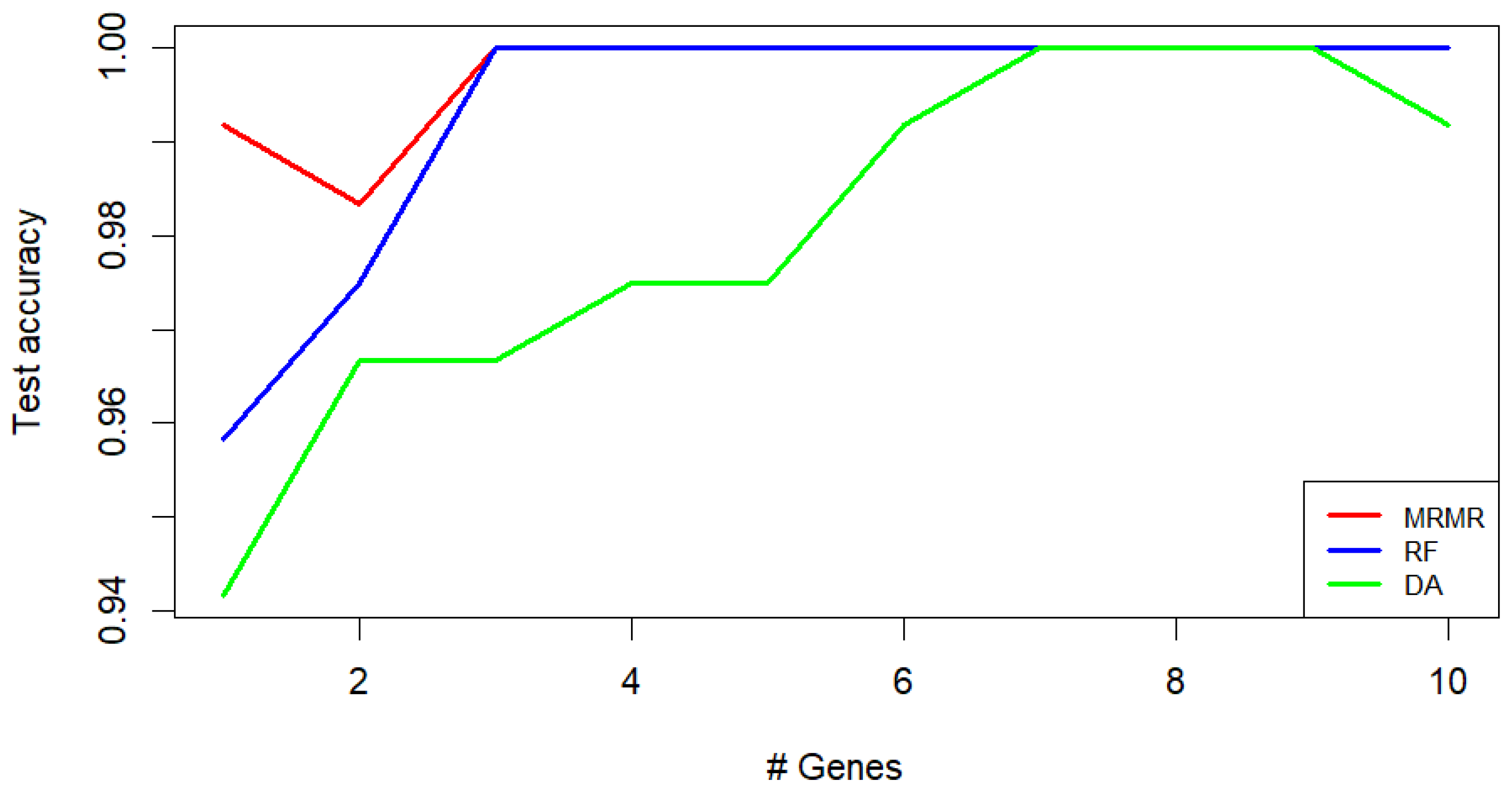
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: kNN test accuracy obtained using different feature selection methods (MRMR, RF, and DA). The values are presented as a function of the number of genes used.
Figure 2.
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: kNN test accuracy obtained using different feature selection methods (MRMR, RF, and DA). The values are presented as a function of the number of genes used.
Figure 3.
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: kNN mean training and testing accuracy of the 5-fold cross-validation using MRMR. The values are presented as a function of the number of genes used.
Figure 3.
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: kNN mean training and testing accuracy of the 5-fold cross-validation using MRMR. The values are presented as a function of the number of genes used.
Figure 4.
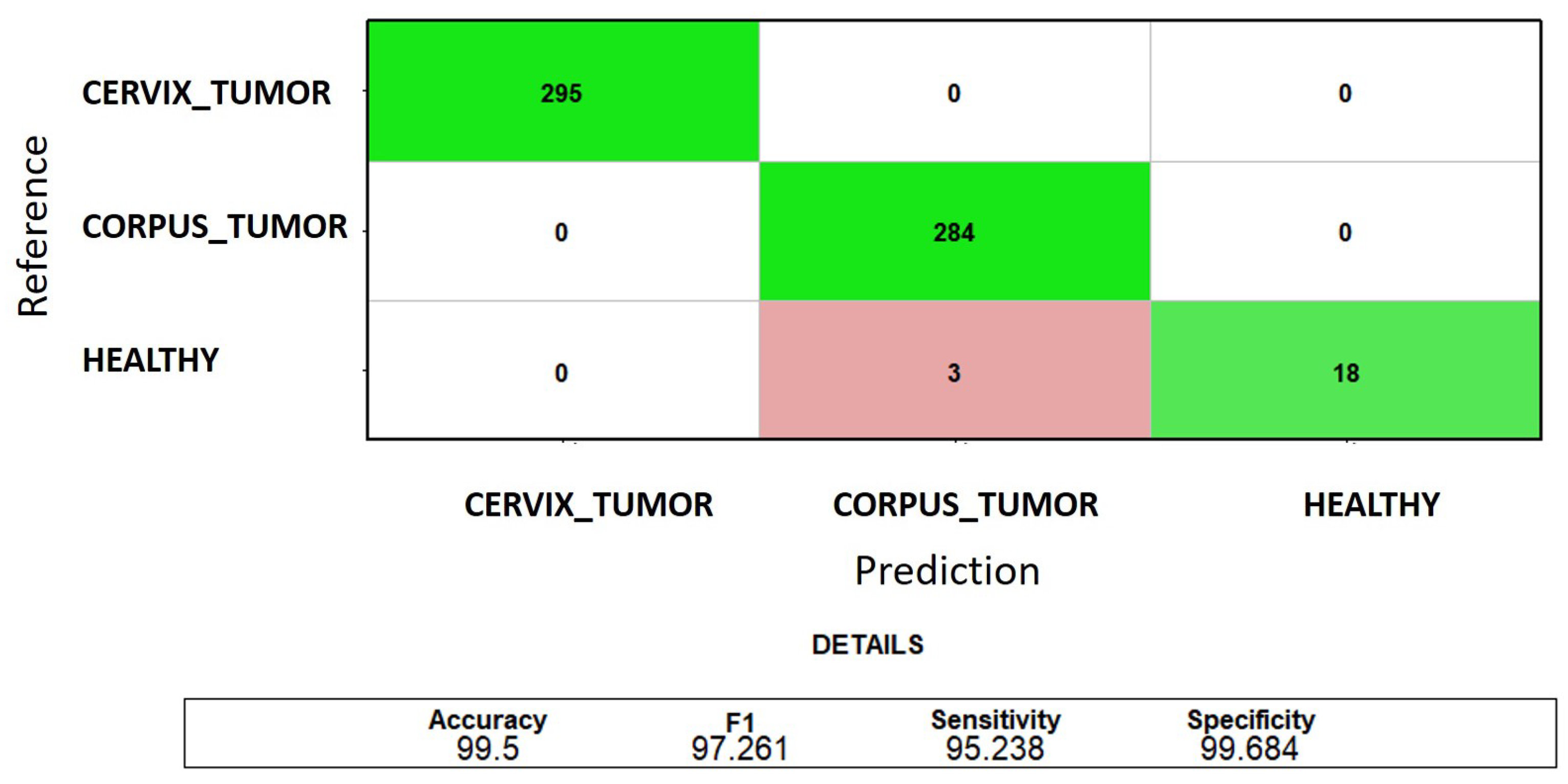
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: the sum of the test confusion matrices of each fold of the 5-fold cross-validation when using the first three MRMR-selected genes. The green color indicates correct predictions, and the red color indicates incorrect predictions.
Figure 4.
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: the sum of the test confusion matrices of each fold of the 5-fold cross-validation when using the first three MRMR-selected genes. The green color indicates correct predictions, and the red color indicates incorrect predictions.
Figure 5.
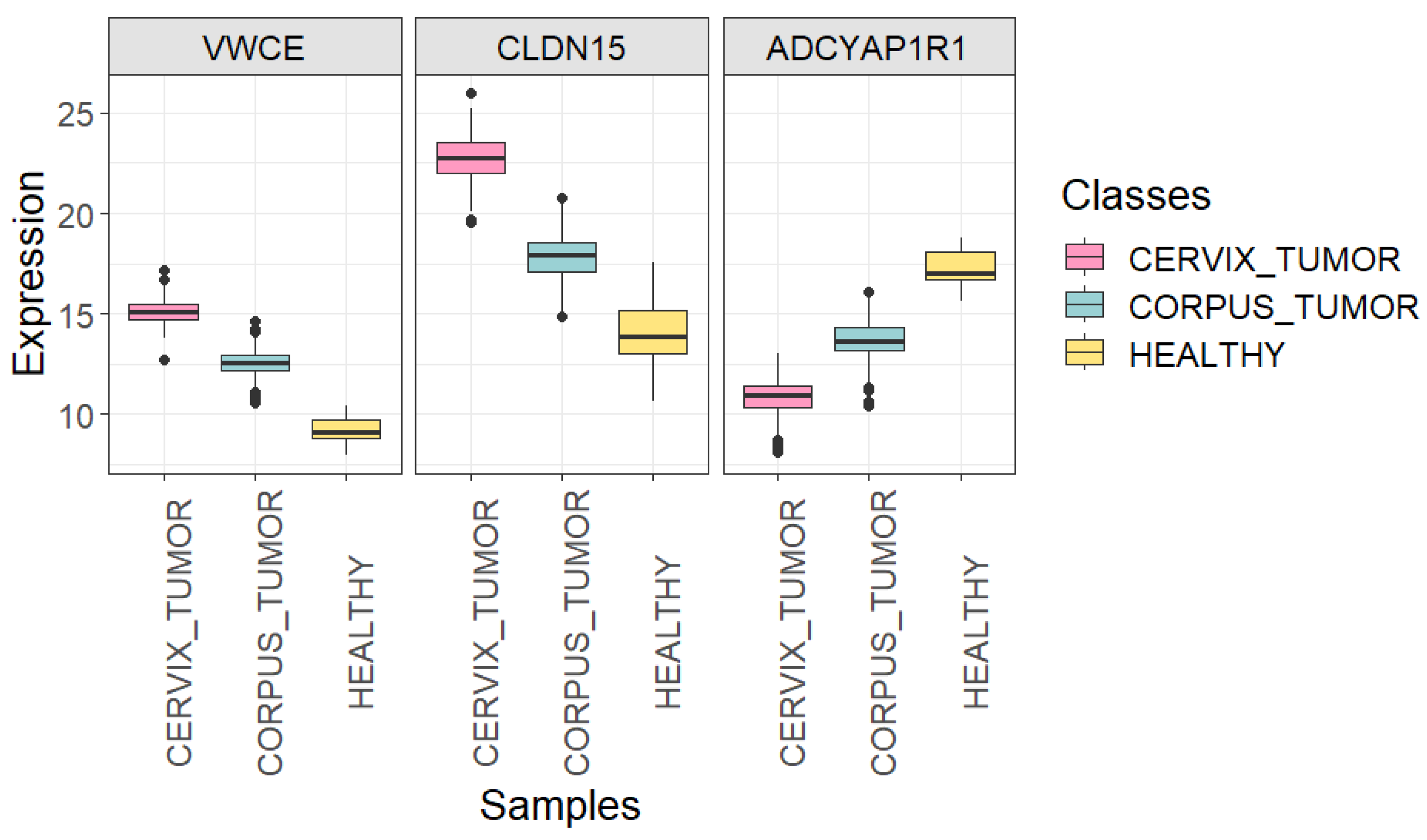
Boxplots showing the expression of VWCE, CLDN15, and ADCYAP1R1 genes in each uterine sample class (CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY) using all quality samples.
Figure 5.
Boxplots showing the expression of VWCE, CLDN15, and ADCYAP1R1 genes in each uterine sample class (CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY) using all quality samples.
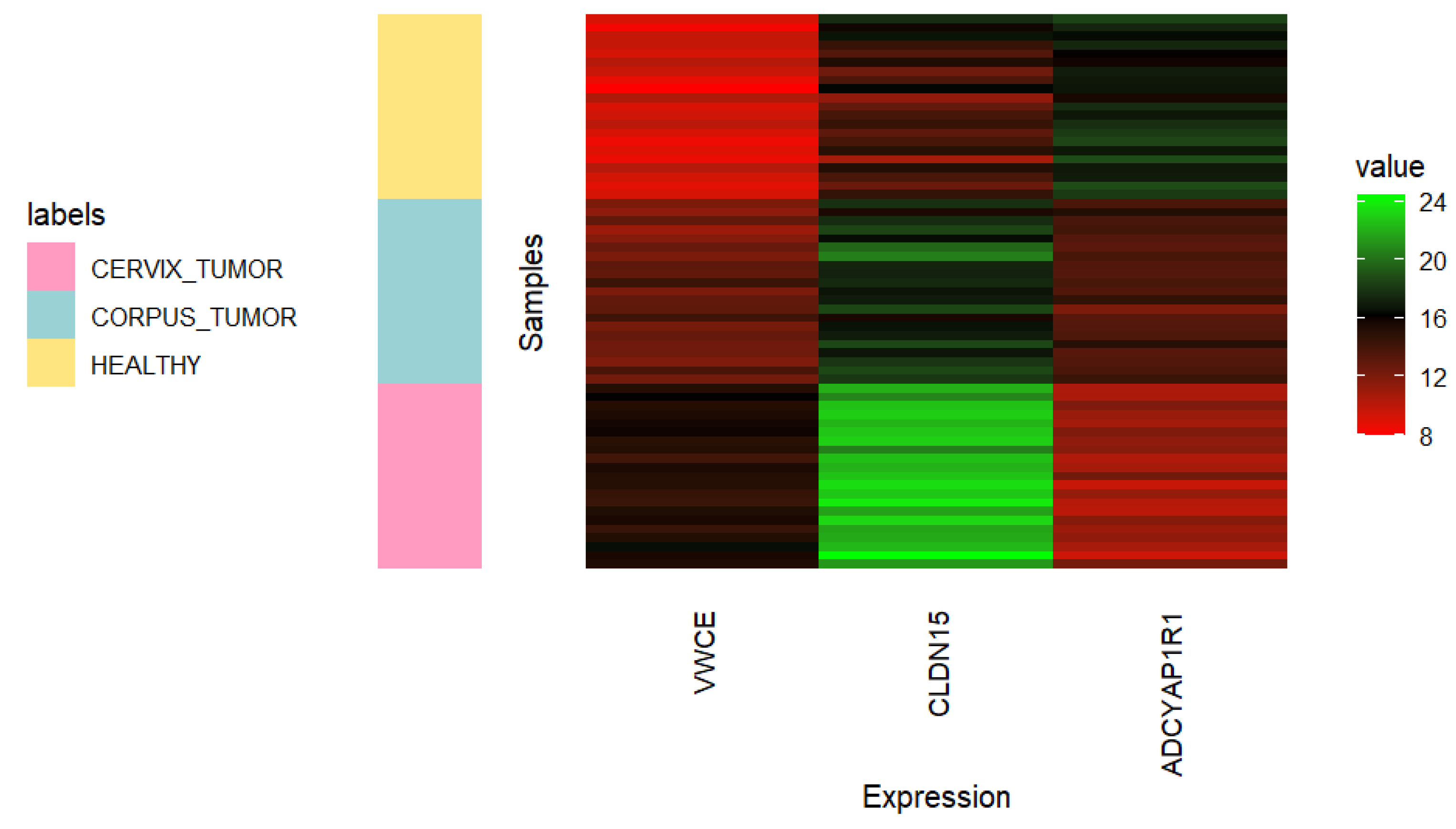
Figure 6.
Heatmap showing the expression of VWCE, CLDN15, and ADCYAP1R1 genes in 21 randomly selected samples of each uterine class (CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY). Undersampling of cancerous classes was carried out using all quality samples.
Figure 6.
Heatmap showing the expression of VWCE, CLDN15, and ADCYAP1R1 genes in 21 randomly selected samples of each uterine class (CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY). Undersampling of cancerous classes was carried out using all quality samples.
Figure 7.
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: sum of the test confusion matrices of each fold of the 5-fold cross-validation using the complete gene signature (VWCE, CLDN15, and ADCYAP1R1) for feature selection. The green color indicates correct predictions.
Figure 7.
Classification of CORPUS_TUMOR, CERVIX_TUMOR, and HEALTHY uterine samples: sum of the test confusion matrices of each fold of the 5-fold cross-validation using the complete gene signature (VWCE, CLDN15, and ADCYAP1R1) for feature selection. The green color indicates correct predictions.
Figure 8.
Boxplots showing the expression of the ICA1L gene in each cervix cancer class (SQUAMOUS and ADENO) using all TCGA-CESC quality samples.
Figure 8.
Boxplots showing the expression of the ICA1L gene in each cervix cancer class (SQUAMOUS and ADENO) using all TCGA-CESC quality samples.
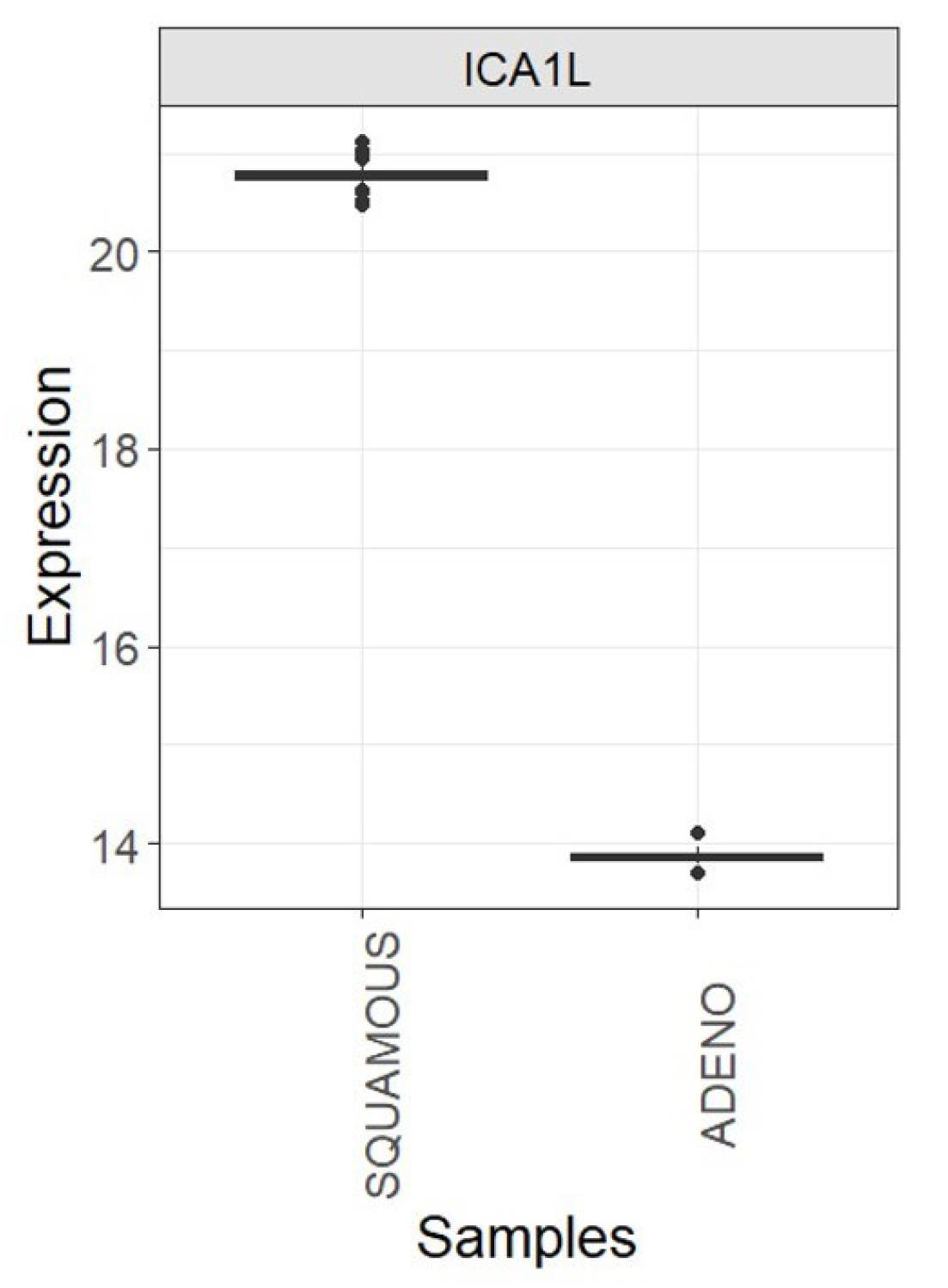
Figure 9.
Boxplots showing the expression of the ICA1L gene in each cervix cancer class (SQUAMOUS and ADENO): TCGA-CESC vs. CGCI-HTMCP-CC samples after joint normalization of both datasets.
Figure 9.
Boxplots showing the expression of the ICA1L gene in each cervix cancer class (SQUAMOUS and ADENO): TCGA-CESC vs. CGCI-HTMCP-CC samples after joint normalization of both datasets.
Figure 10.
Top 12 DEGs between the SQUAMOUS and ADENO cervix cancer classes: boxplots showing their expression in the SQUAMOUS, ADENO, and ADENO_SQUAMOUS classes, using the combined dataset encompassing TCGA-CESC and CGCI-HTMCP-CC samples.
Figure 10.
Top 12 DEGs between the SQUAMOUS and ADENO cervix cancer classes: boxplots showing their expression in the SQUAMOUS, ADENO, and ADENO_SQUAMOUS classes, using the combined dataset encompassing TCGA-CESC and CGCI-HTMCP-CC samples.
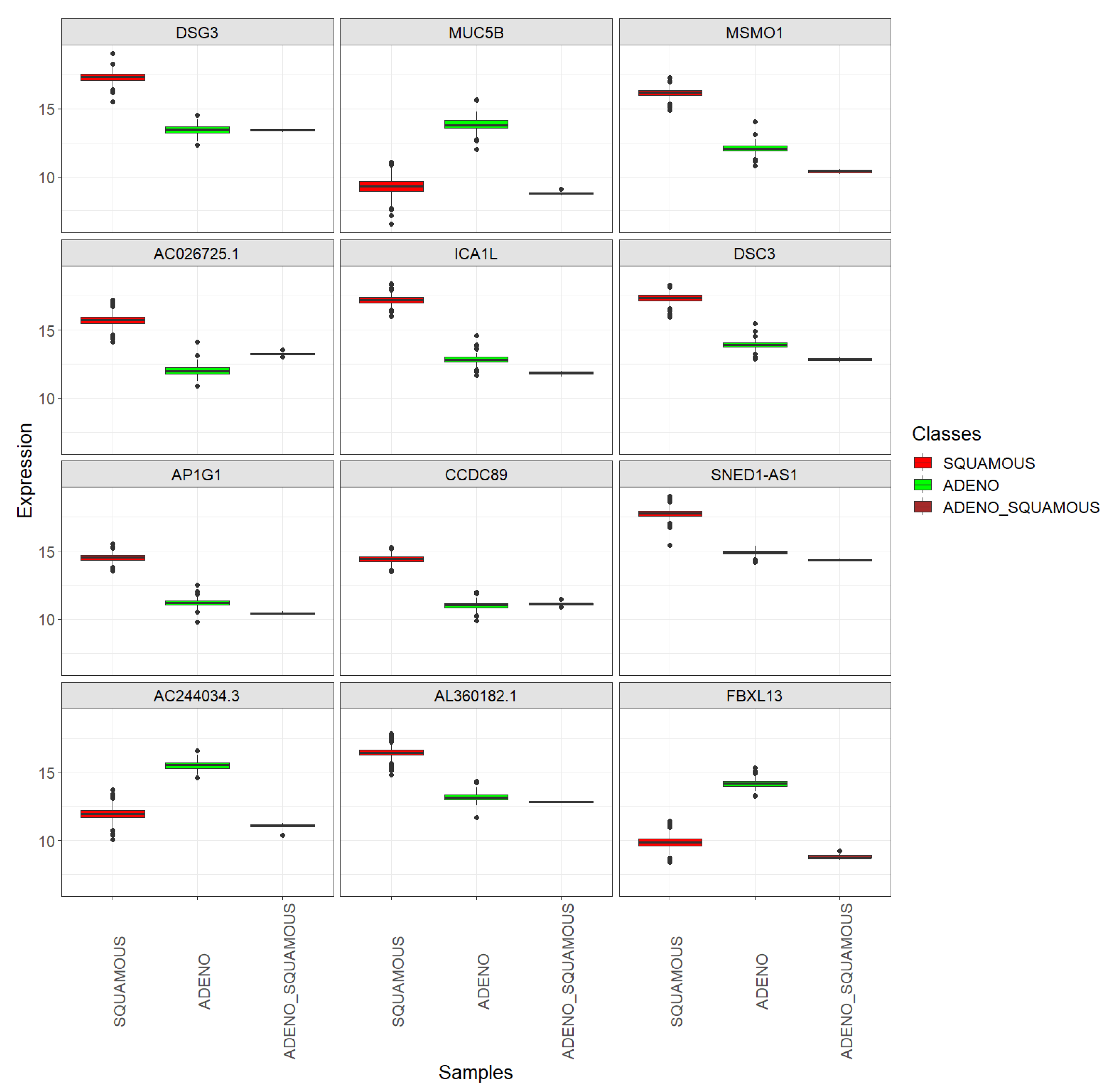
Figure 11.
Top 12 DEGs between the SQUAMOUS and ADENO cervix cancer classes: boxplots showing their expression in the SQUAMOUS, ADENO, and ADENO_SQUAMOUS classes. TCGA-CESC vs. CGCI-HTMCP-CC samples after joint normalization of both datasets.
Figure 11.
Top 12 DEGs between the SQUAMOUS and ADENO cervix cancer classes: boxplots showing their expression in the SQUAMOUS, ADENO, and ADENO_SQUAMOUS classes. TCGA-CESC vs. CGCI-HTMCP-CC samples after joint normalization of both datasets.
Table 1.
Downloaded, randomly selected (through undersampling), and filtered samples of each class.
Table 1.
Downloaded, randomly selected (through undersampling), and filtered samples of each class.
| Class | Description | Project | Downloaded | Rand. Selected | Quality Samples |
|---|
| CERVIX_TUMOR | Cervix cancer | TCGA-CESC | 304 | 300 | 295 |
| CORPUS_TUMOR | Uterine corpus cancer | TCGA-UCEC/SARC | 552 | 300 | 284 |
| HEALTHY | Non-cancerous cervix or uterine corpus | TCGA-CESC/UCEC/SARC | 25 | 25 | 21 |
Table 2.
Number of samples of each class in training and test sets.
Table 2.
Number of samples of each class in training and test sets.
| Class | Training Samples | Test Samples |
|---|
| CERVIX_TUMOR | 233 | 62 |
| CORPUS_TUMOR | 230 | 54 |
| HEALTHY | 17 | 4 |
Table 3.
Number of downloaded cervical cancer samples that share the same primary diagnosis. All samples belonging to the first four subtypes in the table were labeled as ADENO; those belonging to the next 5 types as SQUAMOUS and those belonging to the last type were eliminated.
Table 3.
Number of downloaded cervical cancer samples that share the same primary diagnosis. All samples belonging to the first four subtypes in the table were labeled as ADENO; those belonging to the next 5 types as SQUAMOUS and those belonging to the last type were eliminated.
| Histological Cervical Cancer Subtype | Downloaded Samples |
|---|
| Adenocarcinoma, endocervical type | 21 |
| Adenocarcinoma, NOS | 7 |
| Endometrioid adenocarcinoma, NOS | 3 |
| Mucinous adenocarcinoma, endocervical type | 17 |
| Squamous cell carcinoma, NOS | 169 |
| Papillary squamous cell carcinoma | 1 |
| Squamous cell carcinoma, keratinizing, NOS | 30 |
| Basaloid squamous cell carcinoma | 1 |
| Squamous cell carcinoma, large cell, nonkeratinizing | 50 |
| Adenosquamous carcinoma | 5 |
Table 4.
Cancer cervix samples from TCGA-CESC (original dataset) and CGCI-HTMCP-CC (external dataset) projects. Downloaded, randomly selected (through undersampling), and filtered samples of each class.
Table 4.
Cancer cervix samples from TCGA-CESC (original dataset) and CGCI-HTMCP-CC (external dataset) projects. Downloaded, randomly selected (through undersampling), and filtered samples of each class.
| - | Class | Downloaded | Unders. | Quality Samples |
|---|
| CESC | ADENO | 48 | 48 | 47 |
| SQUAMOUS | 251 | 150 | 149 |
| CGCI | ADENO | 16 | 16 | 16 |
| SQUAMOUS | 174 | 174 | 170 |
Table 5.
Adjusted p-values (calculated by the t-statistic test moderated by an empirical Bayes method and adjusted using the BH method) for the differential expression of the first 12 extracted DEGs across each pair of uterine sample classes, using the training set.
Table 5.
Adjusted p-values (calculated by the t-statistic test moderated by an empirical Bayes method and adjusted using the BH method) for the differential expression of the first 12 extracted DEGs across each pair of uterine sample classes, using the training set.
| Gene | CERVIX_TUMOR-CORPUS_TUMOR | CERVIX_TUMOR-HEALTHY | CORPUS_TUMOR-HEALTHY |
|---|
| ADCYAP1R1 | | | |
| C20orf85 | | | |
| AC104596.1 | | | |
| S100A9 | | | |
| VWCE | | | |
| MTND2P26 | | | |
| AP000769.1 | | | |
| KPRP | | | |
| CIAO2B | | | |
| SHQ1 | | | |
| CCNK | | | |
| SERTM1 | | | |
Table 6.
Classification of CORPUS_TUMOR, CERVIX_TUMOR and HEALTHY uterine samples: top 10 MRMR-selected genes for each fold (train set) of the 5-fold cross-validation.
Table 6.
Classification of CORPUS_TUMOR, CERVIX_TUMOR and HEALTHY uterine samples: top 10 MRMR-selected genes for each fold (train set) of the 5-fold cross-validation.
| - | Gene 1 | Gene 2 | Gene 3 | Gene 4 | Gene 5 | Gene 6 | Gene 7 | Gene 8 | Gene 9 | Gene 10 |
|---|
| Fold 1 | VWCE | CLDN15 | ADCYAP1R1 | CCNK | KCNK10 | SHQ1 | SERTM1 | CCDC153 | CD34 | GDF11 |
| Fold 2 | CCNK | VWCE | SHQ1 | GDF11 | SERTM1 | CLDN15 | ADCYAP1R1 | CCDC153 | CD34 | CPOX |
| Fold 3 | S100A7 | CD34 | ADCYAP1R1 | CCNK | VWCE | SHQ1 | SERTM1 | KCNK10 | CLDN15 | CPOX |
| Fold 4 | VWCE | CLDN15 | SERTM1 | SHQ1 | CCNK | ADCYAP1R1 | KCNK10 | CD34 | CPOX | GDF11 |
| Fold 5 | VWCE | GDF11 | CCNK | ADCYAP1R1 | SHQ1 | CLDN15 | SERTM1 | CPOX | CD34 | SERPINB5 |
Table 7.
Adjusted p-values (calculated by the t-statistic test moderated by an empirical Bayes method and adjusted using the BH method) for the differential expression of VWCE, CLDN15, and ADCYAP1R1 genes across each pair of uterine sample classes, using all quality samples.
Table 7.
Adjusted p-values (calculated by the t-statistic test moderated by an empirical Bayes method and adjusted using the BH method) for the differential expression of VWCE, CLDN15, and ADCYAP1R1 genes across each pair of uterine sample classes, using all quality samples.
| Gene | CERVIX_TUMOR-CORPUS_TUMOR | CERVIX_TUMOR-HEALTHY | CORPUS_TUMOR-HEALTHY |
|---|
| VWCE | | | |
| CLDN15 | | | |
| ADCYAP1R1 | | | |
Table 8.
Classification of SQUAMOUS and ADENO cervix cancer samples: Top 10 MRMR-selected genes for each fold (training set) of the 5-fold cross-validation.
Table 8.
Classification of SQUAMOUS and ADENO cervix cancer samples: Top 10 MRMR-selected genes for each fold (training set) of the 5-fold cross-validation.
| - | Gene 1 | Gene 2 | Gene 3 | Gene 4 | Gene 5 | Gene 6 | Gene 7 | Gene 8 | Gene 9 | Gene 10 |
|---|
| Fold 1 | DYRK3 | ICA1L | GPR171 | THPO | AC091563.1 | RHOV | ZNF175 | TINCR | ANXA8L1 | POPDC3 |
| Fold 2 | ZNF812P | ICA1L | ZNHIT1 | GPR171 | TINCR | THPO | AC091563.1 | RHOV | EPCAM | ZNF175 |
| Fold 3 | GABRQ | ICA1L | GPR171 | THPO | AC091563.1 | RHOV | ZNF175 | SIPA1L3 | CARD16 | AC112907.2 |
| Fold 4 | SERPINB13 | ICA1L | GPR171 | THPO | AC091563.1 | AC012123.1 | RHOV | ZNF175 | SIPA1L3 | GPR89B |
| Fold 5 | LINC01679 | ICA1L | GPR171 | THPO | AC091563.1 | AC012123.1 | RHOV | ZNF175 | TINCR | ANKS4B |
Table 9.
LogFC and adjusted p-values (calculated by the t-statistic test moderated by an empirical Bayes method and adjusted using the BH method) for the differential expression of the top 12 DEGs, identified with the lowest logFC values, in the two major histological subtypes of cervical cancer using the combined dataset encompassing TCGA-CESC and CGCI-HTMCP-CC samples.
Table 9.
LogFC and adjusted p-values (calculated by the t-statistic test moderated by an empirical Bayes method and adjusted using the BH method) for the differential expression of the top 12 DEGs, identified with the lowest logFC values, in the two major histological subtypes of cervical cancer using the combined dataset encompassing TCGA-CESC and CGCI-HTMCP-CC samples.
| Gene | LogFC | adj.P.Val |
|---|
| DSG3 | 5.6369 | |
| MUC5B | | |
| MSMO1 | 5.6171 | |
| AC026725.1 | 5.4261 | |
| ICA1L | 5.3658 | |
| DSC3 | 5.1875 | |
| AP1G1 | 5.0555 | |
| CCDC89 | 4.7556 | |
| SNED1-AS1 | 4.6948 | |
| AC244034.3 | | |
| AL360182.1 | 4.6675 | |
| FBXL13 | | |