Abstract
Torreya grandis, an economically significant evergreen tree species exclusive to subtropical China, is highly valued for its seeds. However, the seed development process of T. grandis remains relatively unexplored. Given the pivotal role WRKY transcription factors (TFs) play in coordinating diverse cellular and biological activities, as well as crucial signaling pathways essential for plant growth and development, and the lack of comprehensive investigation into their specific functions in T. grandis, our study investigated its genome and successfully isolated 78 WRKY genes and categorized them into three distinct clades. A conserved motif analysis unveiled the presence of the characteristic WRKY domain in each identified TgWRKY protein. The examination of gene structures revealed variable numbers of introns (ranging from zero to eight) and exons (ranging from one to nine) among TgWRKY genes. A chromosomal distribution analysis demonstrated the presence of TgWRKY across eight chromosomes in T. grandis. Tissue-specific expression profiling unveiled distinctive patterns of these 78 TgWRKY genes across various tissues. Remarkably, a co-expression analysis integrating RNA-seq data and morphological assessments pinpointed the pronounced expression of TgWRKY25 during the developmental stages of T. grandis seeds. Moreover, a KEGG enrichment analysis, focusing on genes correlated with TgWRKY25 expression, suggested its potential involvement in processes such as protein processing in the endoplasmic reticulum, starch, and sucrose metabolism, thereby modulating seed development in T. grandis. These findings not only underscore the pivotal role of WRKY genes in T. grandis seed development but also pave the way for innovative breeding strategies.
1. Introduction
Transcription factors (TFs), renowned for their ability to selectively bind to specific sequences in the upstream promoter regions of genes, play a crucial role in regulating gene expression involved in numerous cellular processes. This regulatory function is significant not only in plants but also across all living organisms [1,2]. Within this context, the WRKY family of transcription factors is particularly noteworthy for its critical role in the complex network of plant development under diverse environmental conditions.
Initially discovered in Ipomoea batatas [3], WRKY proteins are now recognized as crucial regulators of plant growth, development, and response to environmental stresses. These TFs are characterized by a distinctive WRKYGQK sequence at their N-terminus, the hallmark of the WRKY domain. This domain typically comprises around 60 amino acids, featuring a four-stranded β-sheet and a zinc-finger motif, which enables specific interactions with DNA sequences known as W-box elements (5’-[T]TGAC[C/T]-3’) [4]. Although the structural elements of WRKY TFs are highly conserved, the family exhibits considerable diversity in the plant kingdom. Categorized into three primary groups—Group I, Group II, and Group III—differentiated by the number of WRKY domains and the zinc-finger motif structures [5], each group and subgroup of WRKY TFs may have unique functions and roles in plant biology, playing significant parts in responses to biotic and abiotic stresses and developmental processes. Group I proteins contain two WRKY domains, while those in Groups II and III have one. The distinguishing factor between Group II and III proteins is their zinc-finger motifs; Group II resembles Group I with the motif C-X4-5-C-X22-23-H-X1-H, whereas Group III has a C-X7-C-X23-HX1-C motif [6]. Unique to plants, WRKY genes have been identified in various species, including Arabidopsis thaliana with 74 WRKY proteins, Glycine max, and Oryza sativa, which has 109 WRKY proteins [7,8,9].
Recent advances in molecular biology and genomics have led to the identification of an increasing number of WRKY genes, uncovering their varied functional roles. These genes are expressed in a range of plant tissues, including roots, stems, leaves, flowers, and fruits, and are integral in regulating diverse growth and development stages. Notably, specific WRKY genes like WRKY13, WRKY33, and WRKY71 are essential in processes such as root development, stem node differentiation, leaf expansion, and fruit maturation, forming a critical molecular regulatory base for plant growth [10,11,12,13,14,15]. Additionally, WRKY genes are central to plant responses to a variety of stresses [16]. Studies have shown their inducible response to both abiotic stresses (such as drought, high temperature, and salinity) and biotic stresses (including pathogen infections and insect infestations). By modulating stress-responsive gene cascades, WRKY genes enhance plant resilience and adaptability [17]. For instance, in wheat (Triticum aestivum), 47 WRKY genes are upregulated under salt stress, and in rice (O. sativa) [18,19], 17 WRKY genes respond to drought conditions [20]. WRKY genes play a crucial role in the complex interplay of plant growth, development, and stress responses by regulating downstream target genes through DNA binding [21]. They also interact with other transcription factors and hormone signaling pathways, creating intricate regulatory networks [22]. A deeper understanding of WRKY genes’ regulatory mechanisms and associated signaling pathways promises to offer significant theoretical and practical contributions to plant biology.
T. grandis, a notable tree species indigenous to the subtropical mountains of China and belonging to the Torreya genus of the Taxaceae family, is distinguished by its seeds’ high fatty acid content, including linoleic acid, oleic acid, and other unsaturated fatty acids. These bioactive compounds are known for their cholesterol-lowering effects in human blood, offering potential benefits in preventing or mitigating cardiovascular and cerebrovascular diseases such as atherosclerosis and hypertension [23]. This highlights the species’ significant economic and health-related value. However, previous research on T. grandis seeds has largely concentrated on their composition and health benefits [24,25,26], while the process of seed development and the underlying regulatory mechanisms, crucial for seed yield, have not yet been fully understood.
In this study, we aimed to elucidate the regulatory role of the WRKY gene family in T. grandis seed development by undertaking a comprehensive identification and analysis of WRKY genes in the T. grandis genome. Our analysis included examining their chromosomal distribution, phylogenetic relationships, and conserved domains. Furthermore, a transcriptomic analysis provided insights into the expression patterns of the WRKY gene family across different seed development stages. This research advances our understanding of the WRKY gene family’s function in T. grandis seed development and contributes to the broader knowledge of WRKY functions.
2. Material and Methods
2.1. Plant Materials
The study utilized seeds of T. grandis obtained from the Botanical Garden of Zhejiang Agriculture and Forestry University in Linan City, Zhejiang Province, China. Collection of the seeds occurred monthly from May to September. Subsequently, the seeds were carefully de-scaled using a manual peeling method based on established protocols for T. grandis seed kernels [26,27,28], ensuring clean and intact seeds, then rapidly frozen in liquid nitrogen and preserved at −80 °C for subsequent analyses.
2.2. Identification of WRKY Genes in T. grandis
Genomic sequences of T. grandis, encompassing both nucleotide and amino acid data, were retrieved from the Figshare database [26]. An HMM search, utilizing Pfam ID PF03106, was conducted to identify the WRKY-DNA-binding domain, employing an E-value threshold of ≤e−10. This process initially identified 90 putative WRKY sequences in T. grandis. A comparative analysis was subsequently performed by juxtaposing these sequences against 72 WRKY gene sequences from Arabidopsis sourced from Phytozome V13 [23], using the blastp tool. We applied a strict e-value cutoff of ≤e−5 to pinpoint potential WRKY genes. Finally, utilizing NCBI-CDD results, we definitively identified 78 WRKY genes in T. grandis, confirming the presence of the WRKY domain in each.
2.3. Phylogenetic Analysis of TgWRKY and AtWRKY Gene Families
The full-length WRKY amino acid sequences from both T. grandis and A. thaliana were aligned using MAFFT version 7.520 with the default settings [29]. Maximum-likelihood phylogenetic trees were constructed with IQ-TREE v2.2.0 [30], employing the model automatically selected by the software (IQ-TREE v2.2.0, ‘Auto’ option) and 5000 ultrafast bootstraps for robustness [31]. The resulting evolutionary trees were modified and visualized using Evolview v2 [32].
2.4. Gene Structure Analysis of TgWRKY Gene Family
For the WRKY gene family in T. grandis, MEME Suite version 5.5.4 was employed to analyze the conserved motif structures and assess the functional significance. Functional annotations of these motifs were verified against the NCBI-CDD database [33,34]. The gene feature format file (gff3) from the T. grandis genome facilitated the verification of the chromosomal positions of the TgWRKY genes. The visualization of the findings was achieved using TBtools-Ⅱ version 2.016 [35].
2.5. RNA Extraction and Transcriptome Analysis
Total RNA extraction from T. grandis seeds was performed using the RNAprep Pure Plant Kit (DP441, Tiangen, Beijing, China). Approximately 100 mg of each sample type was immediately frozen in liquid nitrogen and subsequently pulverized into a fine dust using a mortar and pestle. Subsequently, 500 μL of pre-β-ME-added Buffer SL was introduced to the powder and vigorously vortexed. After centrifugation, the supernatant was moved to a fresh tube, followed by the addition of ethanol and another centrifugation step to remove the supernatant. The spin column was then treated with a Buffer RW1 and DNase I solution and incubated at an ambient temperature for 15 min. Following several centrifugation steps and buffer exchanges, the membrane of the spin column was dried, and the RNA was finally eluted using RNase-free water, then preserved at −70 °C for future analysis. The purity, concentration, and integrity of the extracted RNA were assessed using a NanoDrop 2000 spectrophotometer.
mRNA libraries were subsequently prepared for each sample and sequenced on an Illumina Novaseq platform. The quality assessment of the raw data was performed by discarding reads with adapter sequences, poly-N segments, and those of inferior quality. Subsequently, FastQC was employed to determine the Q20 and Q30 scores, and the high-quality reads were aligned to the T. grandis reference genome using HISAT2 version 2.2.1 [26,36]. The mapped reads for each gene were quantified using feature-counts software (version 1.5.0), and gene expression levels were quantified using fragments per kilobase million (FPKM) as the unit.
2.6. Weighted Gene Coexpression Network Analysis (WGCNA)
The R-package WGCNA was utilized to identify gene co-expression modules among the 24,548 genes obtained after FPKM normalization and filtering [37]. A soft-thresholding power (β = 10) was applied to the gene expression matrix to facilitate this analysis. The resulting modules consisted of clusters of highly interconnected genes, characterized by substantial correlation coefficients among them. To ensure the robustness of our findings, the minimum module size was set to 30, and modules with eigengenes exhibiting high correlations (a threshold of 0.25) were merged.
3. Results
3.1. Identification and Phylogenetic Analysis of WRKY Proteins in T. grandis
To elucidate the WRKY gene family in T. grandis, a genome-wide search was conducted using HMMER, employing A. thaliana WRKY sequences as reference queries. A conserved domain analysis substantiated that all identified WRKY genes in T. grandis exhibited either single or double WRKY domains at their N-termini, a defining characteristic of the WRKY gene family. This search resulted in the identification of 78 potential WRKY genes within the T. grandis genome (Table S1).
To analyze the evolutionary relationships of TgWRKY and AtWRKY, a phylogenetic analysis was conducted using IQ-TREE. The amino acid sequences of TgWRKY from T. grandis were aligned with those of AtWRKY from A. thaliana. The analysis revealed that the 78 predicted TgWRKY proteins could be classified into three primary groups: WRKY-I, WRKY-II, and WRKY-III. Specifically, Group I contained 6, Group II comprised 70, and Group III encompassed 2 TgWRKY proteins (Figure 1).
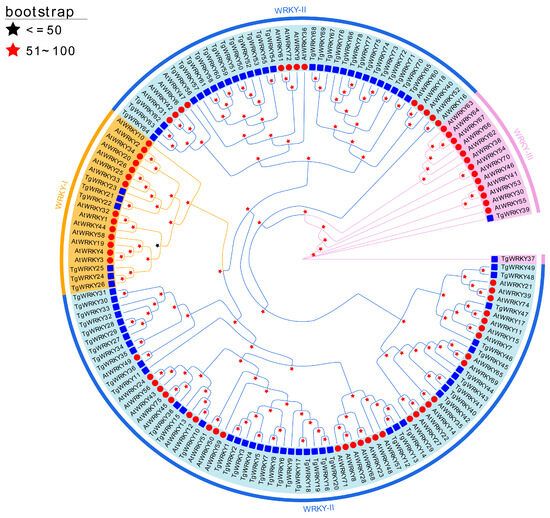
Figure 1.
Phylogenetic relationships among WRKY proteins in T. grandis and A. thaliana. Phylogenies were deduced using a maximum-likelihood inference via IQ-TREE v2.2.0, applying the model automatically selected by IQ-TREE (‘Auto’ option) and 5000 ultrafast bootstraps.
3.2. Gene Structure and Synteny Analysis of TgWRKY Gene Family
The gene structure diversity of the TgWRKY gene family was investigated using annotation files from the T. grandis reference genome, with visualizations generated via TBtools-Ⅱ. A MEME (version 5.5.4) analysis of the conserved motifs within TgWRKY proteins identified seven distinct motifs across the 78 TgWRKY proteins. These motifs varied in number within individual proteins, ranging from one in TgWRKY38 to eight in proteins such as TgWRKY53 and TgWRKY55 (Figure 2A). It was observed that all TgWRKY proteins possessed the WRKY domain (Figure 2B).
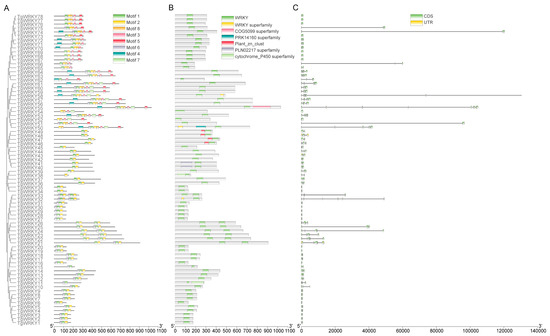
Figure 2.
Phylogenetic relationship and gene structure of TgWRKY. (A) Phylogenetic relationship and motifs of TgWRKY. (B) The conserved structural domains of TgWRKY. (C) The distribution of coding sequences (CDS) in TgWRKY, with green boxes indicating CDS and yellow boxes denoting untranslated regions (UTRs).
The analysis of exon–intron structural diversity, a crucial aspect of gene family evolution, also supported phylogenetic classifications. This study’s intron and exon structure analysis aimed to enhance understanding of the phylogenetic relationships and classifications within the WRKY family. The findings indicated a variation in the number of introns (ranging from zero to eight) and exons (ranging from one to nine) across the gene family (Figure 2C).
As depicted in Figure 3, the 78 identified TgWRKY genes were distributed across eight chromosomes of T. grandis. Notably, chromosomes 1, 2, and 7 lacked TgWRKY genes. Chromosomes 10 and 4 had the highest numbers of TgWRKY genes, with 28 and 22 genes, respectively. In contrast, chromosomes 11 and 3 had only one or two TgWRKY genes. The distribution of the remaining TgWRKY genes spanned the other four chromosomes, with chromosomes 6, 9, 5, and 8 hosting 10, 6, 5, and 4 TgWRKY genes, respectively.
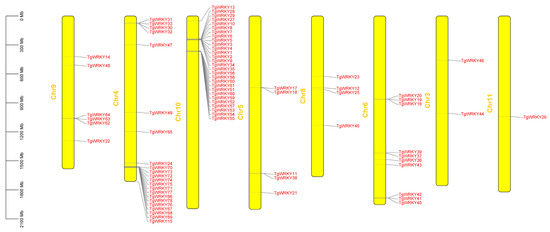
Figure 3.
Chromosomal distribution of TgWRKY in T. grandis.
3.3. Expression Pattern Analysis of TgWRKY in Various Tissues of T. grandis
To elucidate the functions of TgWRKY genes in the seed development of T. grandis, we analyzed the expression profiles of these genes across different organs, including stems, leaves, roots, and seeds, using transcriptomic data (Table S2). Our findings revealed that 51.3% (40 out of 78), 50.0% (39 out of 78), 51.3% (40 out of 78), and 33.3% (26 out of 78) of the TgWRKY genes were expressed in roots, stems, leaves, and seeds, respectively (Figure 4B). Notably, 15 TgWRKY genes exhibited expression in all these tissues, while the expression patterns of the other TgWRKY genes significantly varied across stem, leaf, root, and seed tissues. In stems, TgWRKY67 showed the highest expression levels, while TgWRKY63 was characterized by the lowest expression (Figure 4A). In leaves, TgWRKY66 was highly expressed, as opposed to TgWRKY48, which showed low expression. TgWRKY68 displayed high expression in roots, while TgWRKY19 had a lower expression level than the average. Additionally, in seed tissue, TgWRKY18 exhibited the highest expression, contrasting with TgWRKY47, which showed the lowest expression.
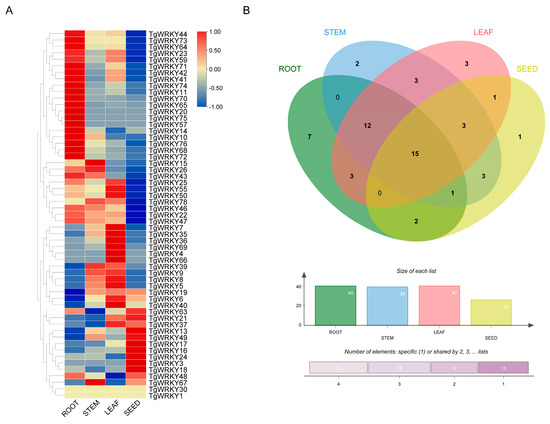
Figure 4.
Expression patterns of TgWRKY in various tissues of T. grandis. (A) Heatmap displaying expression levels of TgWRKY in different tissues. (B) Venn diagram showing the number of TgWRKY expressed in various tissues.
3.4. Expression Pattern Analysis of TgWRKY during the Seed Development of T. grandis
To elucidate the role of TgWRKY genes in the seed development of T. grandis, seeds were collected at five developmental stages, designated S1–S5, from May to September. A morphological analysis identified distinct patterns of development: a phase of rapid expansion from S1 to S2, followed by a relatively stable period from S3 to S5 (Figure 5A). Supporting these morphological findings, statistical analyses of seed length and width further illustrated the developmental progression of T. grandis seeds (Figure 5B,C). These stages formed the basis for the subsequent RNA-seq analysis.
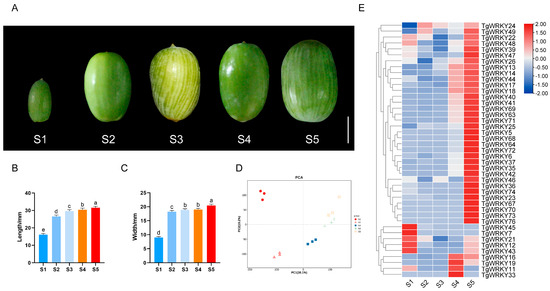
Figure 5.
Transcriptomic and morphological characteristics of T. grandis seeds at different developmental stages. (A) Seed photographs. The scale bar is 1 cm. Seed length (B) and width (C) measurements, with error bars representing the standard deviation (SD) for 15 samples from three trees. Different letters denote significant differences (p < 0.05) as determined by a one-way ANOVA with Tukey’s post hoc test. (D) A principal component analysis (PCA) of the gene expression dataset in ovulate strobilus and (E) a heatmap of TgWRKY expression during seed development.
A principal component analysis (PCA) confirmed a strong correlation among the three replicates for each sample, affirming data reliability (Figure 5D). Notably, PCA segregated the samples into three distinct clusters: S1, S2, and S3–S5, aligning with the developmental stages. As illustrated in Figure 5E and Table S3, 55.1% (43 out of 78) of TgWRKY genes were expressed across the various developmental stages. Among these, 20 TgWRKY genes, such as TgWRKY5, TgWRKY13, TgWRKY14, TgWRKY18, and TgWRKY25, exhibited a marked increase in expression. Conversely, only two TgWRKY genes, TgWRKY45 and TgWRKY7, showed limited expression during these stages, suggesting their potential roles in seed development in T. grandis.
3.5. Construction of the Regulatory Network Associated with Seed Development in T. grandis
A weighted gene co-expression network analysis (WGCNA) is a systems-biology approach that focuses on networks of strongly associated genes rather than individual genes. This method has been effectively employed in various genomic studies. After filtering the data, a total of 24,548 genes with FPKM values were subjected to analysis using the R-package WGCNA. This analysis revealed fifteen distinct modules (Figure 6A), visualized as tree branches in a hierarchical clustering dendrogram, with each leaf representing a gene (Figure 6B). The modules were displayed in various colors (Figure 6B). A correlation analysis, illustrated in Figure 6C, was conducted between seed length, width, and the fifteen modules. The chart’s color shading (red for positive and blue for negative correlations) indicated significant correlations between these modules and the seed dimensions. A module-trait association analysis identified the ‘red’ module, comprising 1203 genes, as potentially involved in seed size increases due to its strong correlation with length and width. Notably, TgWRKY25 was part of the ‘red’ module, suggesting its potential role in T. grandis seed development.
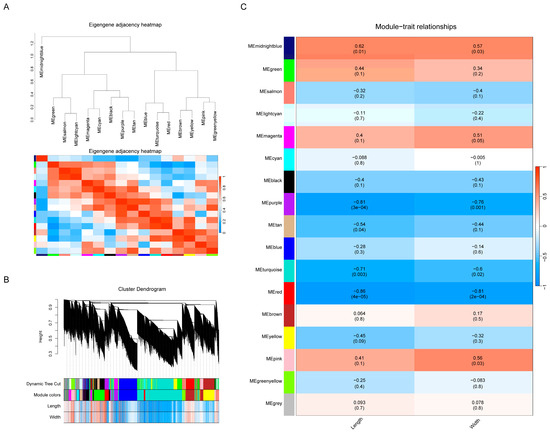
Figure 6.
Construction and analysis of a weighted gene co-expression network. (A) Eigengene adjacency heatmap. (B) Cluster dendrogram. (C) Heatmap showing module-trait correlations.
Furthermore, to explore the potential regulatory network of TgWRKY25 in seed development, we conducted a KEGG enrichment analysis on the genes within the ‘red’ module where TgWRKY25 is located. The KEGG enrichment analysis of these genes, as depicted in Figure 7A, highlighted enriched pathways including those for protein processing in the endoplasmic reticulum, starch and sucrose metabolism, pyrimidine metabolism, among others. Among these pathways, 21 of 1203 genes involved in starch and sucrose metabolism, such as TgPYG (evm.model.PTG005492L.44) and Tgbata-glucosidase (evm.model.PTG0066099L.20), exhibited a significant increase during seed development (Figure 7B and Table S4). This suggested that these genes might play essential roles in the seed development of T. grandis.
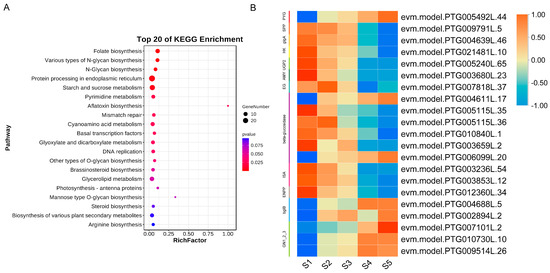
Figure 7.
DEGs related to TgWRKY25 during the seed development. (A) KEGG analysis of DEGs related to TgWRKY25 expression. (B) Heatmap of DEGs related to starch and sucrose metabolism.
4. Discussion
T. grandis seeds offer significant dietary benefits, with their yield and nutritional value varying depending on their developmental stage [38]. The molecular regulation of seed development involves a complex interplay of various factors and signaling pathways, encompassing plant hormone regulation (such as gibberellins, auxins, abscisic acid, ethylene, etc.), transcription factor control (such as LEC1, LEC2, FUSCA3, etc.), nutrient transport and storage, cell division and expansion, and signaling transduction pathways [39,40,41,42,43]. These regulatory factors and pathways interact with each other to collectively regulate the process of seed development, ensuring the normal development of seeds in morphology, structure, and function. There exists a notable correlation between these molecular regulatory pathways and the WRKY gene family. Previous research has highlighted the involvement of WRKY transcription factors in crucial processes of seed development, such as seed morphogenesis, endosperm development, and the differentiation of endosperm and seed coat tissues [44]. WRKY TFs achieve this by modulating plant hormone synthesis, signal transduction pathways, and responsive gene expression. For instance, WRKY TFs interact with various plant hormone signaling pathways, including gibberellins, auxins, abscisic acid, and ethylene, to regulate downstream gene expression, thereby influencing processes such as nutrient accumulation in seeds, endosperm development, and seed coat rupture [45]. Moreover, WRKY TFs regulate genes associated with cell cycle regulation, impacting the rate, direction, and pattern of cell division and expansion, ultimately affecting seed size, quality, and morphology [46]. Therefore, the WRKY family plays a pivotal role in the molecular regulatory pathways of seed development.
The number of identified WRKY genes varies among different plant species, such as A. thaliana, O. sativa, Arachis hypogaea, Cucumis sativus, Liriodendron chinense, Eucommia ulmoides, and others [11,12,16,47,48,49,50,51]. Despite their ubiquitous presence across different plants, the quantity and classification of WRKY families vary among species, reflecting the diversity of plant genomes. For example, A. thaliana has approximately 74 WRKY members, instrumental in key plant processes like growth, development, and environmental stress responses [52], whereas rice (O. sativa) has over 100 known WRKY members [53]. These variations may result from environmental pressures and evolutionary forces, leading to distinct evolutionary trajectories within the gene family [54]. These differences suggest gene amplifications or reductions in specific lineages [5]. In our study, 78 TgWRKY family members were identified in T. grandis, categorized into three groups, WRKY-I, WRKY-II, and WRKY-III (Figure 1), and distributed across eight chromosomes (Figure 3). Although similar to that in Arabidopsis, the number of WRKY genes in T. grandis is fewer than in rice, suggesting potential gene amplifications or reductions in specific lineages. Furthermore, our findings revealed closely related orthologous WRKY genes between T. grandis and A. thaliana (e.g., TgWRKY10 and AtWRKY112; TgWRKY36 and AtWRKY49; TgWRKY38 and AtWRKY45/AtWRKY75; TgWRKY45 and AtWRKY65/AtWRKY69; TgWRKY47 and AtWRKY11/AtWRKY17), indicating a common set of ancestral WRKY genes before their divergence, highlighting a close relationship between T. grandis WRKY proteins and those of Arabidopsis. Despite this, the variations in WRKY gene quantity and classification among different species emphasize the diversity and adaptability of plant genomes.
WRKY proteins, prevalent in plants, play pivotal roles in vital biological processes, including growth, development, and stress responses [52,55]. In Taxus, another genus within the Taxaceae family, 61 WRKY transcripts were identified from Taxus chinensis transcriptome datasets, with certain WRKY genes significantly enhancing the expression levels of taxol-biosynthesis-related genes [56]. In our study, 26 WRKY members were detected in T. grandis seeds, with the expression levels of 20 TgWRKY genes (including TgWRKY5, TgWRKY13, TgWRKY14, TgWRKY18, TgWRKY25, etc.) varying during seed development (Figure 5). It is notable that the number of WRKY family members is relatively small in the T. grandis seed. This may be attributed to the fact that different plant tissues may possess distinct biological functions and metabolic demands, thus resulting in varying requirements for WRKY family members [57]. Certain tissues may necessitate a greater number of WRKY genes to regulate specific biological processes, while others may require fewer. Additionally, gene expression levels are regulated by genetic and environmental factors, which may lead to differential expression levels of WRKY genes across different tissues [58]. Nevertheless, a KEGG enrichment analysis indicated the WRKY family’s significant role in regulating key biological processes, underscoring the important impact of WRKY genes on seed growth and development.
Additionally, through a co-expression analysis and morphological assessments, it was found that TgWRKY25 was closely related to the seed development of T. grandis (Figure 6). These results strongly suggested the crucial role of WRKY genes in regulating seed development in T. grandis. WRKY transcription factors (TFs) regulate gene expression by binding to the DNA of target genes via the WRKY domain [59]. For example, in Arabidopsis, AtWRKY12 and AtWRKY13 regulate the expression of the downstream gene AtFUL, thus influencing flowering [60]. Here, our study found that 21 genes belonging to the ‘red’ module, such as TgPYG (evm.model.PTG005492L.44), TgEG (evm.model.PTG007818L.37), TgGN1_2_3 (evm.model.PTG007101L.2), etc., which were involved in starch and sucrose metabolism (two crucial carbon sources playing pivotal roles in seed maturation), correlated with TgWRKY25 expression. These suggested the potential role of TgWRKY25 in regulating genes related to starch and sucrose biosynthesis/metabolism, thereby affecting seed development in T. grandis. Future research should elucidate the function and regulatory mechanism of WRKY in the seed development of T. grandis.
5. Conclusions
In this study, we conducted a comprehensive analysis of the WRKY gene family in the T. grandis genome, resulting in the identification of 78 TgWRKY genes distributed across eight chromosomes and classified into three distinct clades. Through a conserved motif analysis, we confirmed the presence of the characteristic WRKY domain in each TgWRKY gene. Additionally, a gene structure analysis revealed variability in the number of introns and exons among TgWRKY genes, which ranged from zero to eight and one to nine, respectively, highlighting structural diversity within the gene family. A chromosomal distribution analysis indicated the dispersed localization of TgWRKYs across T. grandis chromosomes. Moreover, our investigation into tissue-specific expression patterns unveiled differential expressions of these 78 TgWRKYs across various tissues, providing insights into their potential roles in different physiological processes. Furthermore, a co-expression analysis, integrating RNA-seq data with morphological assessments, identified TgWRKY25 as closely associated with seed development in T. grandis. These findings underscored the crucial role of WRKY genes in the seed development of T. grandis, shedding light on their regulatory mechanisms and functional significance. Furthermore, our study paves the way for future research avenues, offering new prospects for the breeding and genetic improvement of T. grandis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15030267/s1, Table S1. The ID numbers and names of WRKY genes in T. grandis and A. thaliania. Table S2. The FPKM values of TgWRKY genes in various tissues of T. grandis. Table S3. The FPKM values of TgWRKY genes in various seed development stages in T. grandis. Table S4. DEGs related to sucrose and starch metabolism.
Author Contributions
Conceptualization, R.Z., N.G., and W.S.; methodology, R.Z. and W.S.; software, R.Z. and N.G.; validation, R.Z., N.G. and W.S.; formal analysis, R.Z. and N.G.; investigation, R.Z., N.G. and J.L.; resources, R.Z. and N.G.; data curation, R.Z. and N.G.; writing—original draft preparation, R.Z. and N.G.; writing—review and editing, R.Z., N.G., J.L. and W.S.; visualization, R.Z.; supervision, W.S.; project administration, W.S.; funding acquisition, W.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (32271971, 31901369).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to thank Biotree Technology (Shanghai, China) for their technical assistance with DNA sequencing. Our special thanks go to the executive editor and anonymous reviewers for their insightful comments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jeon, B.W.; Kim, M.J.; Pandey, S.K.; Oh, E.; Seo, P.J.; Kim, J. Recent Advances in Peptide Signaling during Arabidopsis Root Development. J. Exp. Bot. 2021, 72, 2889–2902. [Google Scholar] [CrossRef] [PubMed]
- El Yahyaoui, F.; Küster, H.; Ben Amor, B.; Hohnjec, N.; Pühler, A.; Becker, A.; Gouzy, J.; Vernié, T.; Gough, C.; Niebel, A.; et al. Expression Profiling in Medicago Truncatula Identifies More than 750 Genes Differentially Expressed during Nodulation, Including Many Potential Regulators of the Symbiotic Program. Plant Physiol. 2004, 136, 3159–3176. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Nakamura, K. Characterization of a CDNA Encoding a Novel DNA-Binding Protein, SPF1, That Recognizes SP8 Sequences in the 5′ Upstream Regions of Genes Coding for Sporamin and β-Amylase from Sweet Potato. MGG Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its Structure, Evolutionary Relationship, DNA-Binding Selectivity, Role in Stress Tolerance and Development of Plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY Superfamily of Plant Transcription Factors. Trends Plant Sci. 2000, 5, 1360–1385. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmüller, R. WRKY Transcription Factors: Jack of Many Trades in Plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and Functional Analyses of the Rice WRKY Gene Superfamily Reveal Positive and Negative Regulators of Abscisic Acid Signaling in Aleurone Cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY Transcription Factors: From DNA Binding towards Biological Function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Bencke-Malato, M.; Cabreira, C.; Wiebke-Strohm, B.; Bücker-Neto, L.; Mancini, E.; Osorio, M.B.; Homrich, M.S.; Turchetto-Zolet, A.C.; De Carvalho, M.C.C.G.; Stolf, R.; et al. Genome-Wide Annotation of the Soybean WRKY Family and Functional Characterization of Genes Involved in Response to Phakopsora pachyrhizi Infection. BMC Plant Biol. 2014, 14, 236. [Google Scholar] [CrossRef]
- Yue, M.; Jiang, L.; Zhang, N.; Zhang, L.; Liu, Y.; Wang, Y.; Li, M.; Lin, Y.; Zhang, Y.; Zhang, Y.; et al. Importance of FaWRKY71 in Strawberry (Fragaria × Ananassa) Fruit Ripening. Int. J. Mol. Sci. 2022, 23, 12483. [Google Scholar] [CrossRef]
- Bi, C.; Xu, Y.; Ye, Q.; Yin, T.; Ye, N. Genome-Wide Identification and Characterization of WRKY Gene Family in Salix suchowensis. PeerJ 2016, 4, e2437. [Google Scholar] [CrossRef]
- Chen, D.; Lu, X.; Wu, X.; Ying, X.; Long, W.; Su, H.; Liu, H.; Lin, X.; Xu, C.; Cai, Q. Transcriptome Analysis of Axillary Bud Differentiation in a New Dual-Axillary Bud Genotype of Sugarcane. Genet. Resour. Crop Evol. 2020, 67, 685–701. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-Wide Analysis of the WRKY Gene Family in the Cucumber Genome and Transcriptome-Wide Identification of WRKY Transcription Factors That Respond to Biotic and Abiotic Stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Rosado, D.; Ackermann, A.; Spassibojko, O.; Rossi, M.; Pedmale, U.V. WRKY Transcription Factors and Ethylene Signaling Modify Root Growth during the Shade-Avoidance Response. Plant Physiol. 2022, 188, 1294–1311. [Google Scholar] [CrossRef]
- Li, W.; Tian, Z.; Yu, D. WRKY13 Acts in Stem Development in Arabidopsis thaliana. Plant Sci. 2015, 236, 205–213. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY Transcription Factors and Plant Defense Responses: Latest Discoveries and Future Prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Javed, T.; Gao, S.J. WRKY Transcription Factors in Plant Defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef]
- Zhou, S.; Zheng, W.J.; Liu, B.H.; Zheng, J.C.; Dong, F.S.; Liu, Z.F.; Wen, Z.Y.; Yang, F.; Wang, H.B.; Xu, Z.S.; et al. Characterizing the Role of TaWRKY13 in Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 5712. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Lethin, J.; Blomberg, R.; Mousavi, H.; Aronsson, H. In Silico Based Screening of WRKY Genes for Identifying Functional Genes Regulated by WRKY under Salt Stress. Comput. Biol. Chem. 2019, 83, 107131. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating Transcription Factor OsWRKY5 Enhances Drought Tolerance through Abscisic Acid Signaling Pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef]
- Wu, H.; Ni, Z.; Yao, Y.; Guo, G.; Sun, Q. Cloning and Expression Profiles of 15 Genes Encoding WRKY Transcription Factor in Wheat (Triticum aestivem L.). Prog. Nat. Sci. 2008, 18, 697–705. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY Transcription Factors in Plant Responses to Stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Yan, J.; Zeng, H.; Chen, W.; Zheng, S.; Luo, J.; Jiang, H.; Yang, B.; Farag, M.A.; Lou, H.; Song, L.; et al. Effects of Tree Age on Flavonoids and Antioxidant Activity in Torreya grandis Nuts via Integrated Metabolome and Transcriptome Analyses. Food Front. 2023, 4, 358–367. [Google Scholar] [CrossRef]
- Lou, H.; Yang, Y.; Zheng, S.; Ma, Z.; Chen, W.; Yu, C.; Song, L.; Wu, J. Identification of Key Genes Contributing to Amino Acid Biosynthesis in Torreya grandis Using Transcriptome and Metabolome Analysis. Food Chem. 2022, 379, 132078. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Song, L.; Li, X.; Zi, H.; Chen, W.; Gao, Y.; Zheng, S.; Fei, Z.; Sun, X.; Wu, J. The Torreya grandis Genome Illuminates the Origin and Evolution of Gymnosperm-Specific Sciadonic Acid Biosynthesis. Nat. Commun. 2023, 14, 1315. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, L.; Gao, L.; Gao, Y.; Suo, J.; Yu, W.; Hu, Y.; Wei, C.; Farag, M.A.; Wu, J.; et al. Transcription Factors TgbHLH95 and TgbZIP44 Cotarget Terpene Biosynthesis Gene TgGPPS in Torreya Grandis Nuts. Plant Physiol. 2023, 193, 1161–1176. [Google Scholar] [CrossRef]
- Suo, J.; Ma, Z.; Zhao, B.; Ma, S.; Zhang, Z.; Hu, Y.; Yang, B.; Yu, W.; Wu, J.; Song, L. Metabolomics Reveal Changes in Flavor Quality and Bioactive Components in Post-Ripening Torreya grandis Nuts and the Underlying Mechanism. Food Chem. 2023, 406, 134987. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An Online Visualization and Management Tool for Customized and Annotated Phylogenetic Trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Shi, H.; Wang, H.; Wang, M.; Li, X. Antioxidant Activity and Chemical Composition of Torreya grandis Cv. Merrillii Seed. Nat. Prod. Commun. 2009, 4, 1565–1570. [Google Scholar] [PubMed]
- Kong, L.; Guo, H.; Sun, M. Signal Transduction during Wheat Grain Development. Planta 2015, 241, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Dante, R.A.; Larkins, B.A.; Sabelli, P.A. Cell Cycle Control and Seed Development. Front. Plant Sci. 2014, 5, 93. [Google Scholar] [CrossRef]
- Kozaki, A.; Aoyanagi, T. Molecular Aspects of Seed Development Controlled by Gibberellins and Abscisic Acids. Int. J. Mol. Sci. 2022, 23, 1876. [Google Scholar] [CrossRef] [PubMed]
- Coello, P.; Martínez-Barajas, E. Changes in Nutrient Distribution Are Part of the Mechanism That Promotes Seed Development under Severe Nutrient Restriction. Plant Physiol. Biochem. 2016, 99, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Attuluri, V.P.S.; Robert, H.S. Transcriptional Control of Arabidopsis Seed Development. Planta 2022, 255, 90. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, N.; Yoshioka, H. Post-Translational Regulation of WRKY Transcription Factors in Plant Immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [PubMed]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY Family Gene, and HAIKU2 (IKU2), a Leucine-Rich Repeat (LRR) KINASE Gene, Are Regulators of Seed Size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nam, K.H. Regulation of Brassinosteroid Signaling by a GSK3/SHAGGY-like Kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.; Chen, Y.; Liu, Y.; Wu, Y.; Ren, S.; Li, L. Identification, Evolution and Expression Analysis of WRKY Gene Family in Eucommia ulmoides. Genomics 2021, 113, 3294–3309. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Xu, L.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Hao, Z.; Lu, Y.; Yang, L.; et al. Genome-Wide Identification of the Liriodendron Chinense WRKY Gene Family and Its Diverse Roles in Response to Multiple Abiotic Stress. BMC Plant Biol. 2022, 22, 25. [Google Scholar] [CrossRef]
- Song, H.; Guo, Z.; Duan, Z.; Li, M.; Zhang, J. WRKY Transcription Factors in Arachis hypogaea and Its Donors: From Identification to Function Prediction. Plant Physiol. Biochem. 2023, 204, 108131. [Google Scholar] [CrossRef]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome- Wide Identification and Expression Analysis of the WRKY Gene Family in Cassava. Front. Plant Sci. 2016, 7, 25. [Google Scholar] [CrossRef]
- Song, X.; Hou, X.; Zeng, Y.; Jia, D.; Li, Q.; Gu, Y.; Miao, H. Genome-Wide Identification and Comprehensive Analysis of WRKY Transcription Factor Family in Safflower during Drought Stress. Sci. Rep. 2023, 13, 16955. [Google Scholar]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY Transcription Factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A Comprehensive Transcriptional Profiling of the WRKY Gene Family in Rice under Various Abiotic and Phytohormone Treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef]
- Innan, H.; Kim, Y. Pattern of Polymorphism after Strong Artificial Selection in a Domestication Event. Proc. Natl. Acad. Sci. USA 2004, 101, 10667–10672. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Y.; Nie, L.; Jin, X.; Liao, W.; Zhao, S.; Fu, C.; Yu, L. Transcriptome-Wide Identification and Screening of WRKY Factors Involved in the Regulation of Taxol Biosynthesis in Taxus chinensis. Sci. Rep. 2018, 8, 55197. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Guo, D.; Yang, Z.P.; Tang, X.; Peng, S.Q. Genome-Wide Identification and Characterization of WRKY Gene Family in Hevea brasiliensis. Genomics 2014, 104, 14–23. [Google Scholar] [CrossRef]
- Shu, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-Wide Analysis of the AP2/ERF Superfamily Genes and Their Responses to Abiotic Stress in Medicago truncatula. Front. Plant Sci. 2016, 6, 1247. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Kracher, B.; Somssich, I.E. Induced Genome-Wide Binding of Three Arabidopsis WRKY Transcription Factors during Early MAMP-Triggered Immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, H.; Yu, D. Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short-Day Conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).