Involvement of Alfin-Like Transcription Factors in Plant Development and Stress Response
Abstract
1. Introduction
2. Survey Methodology
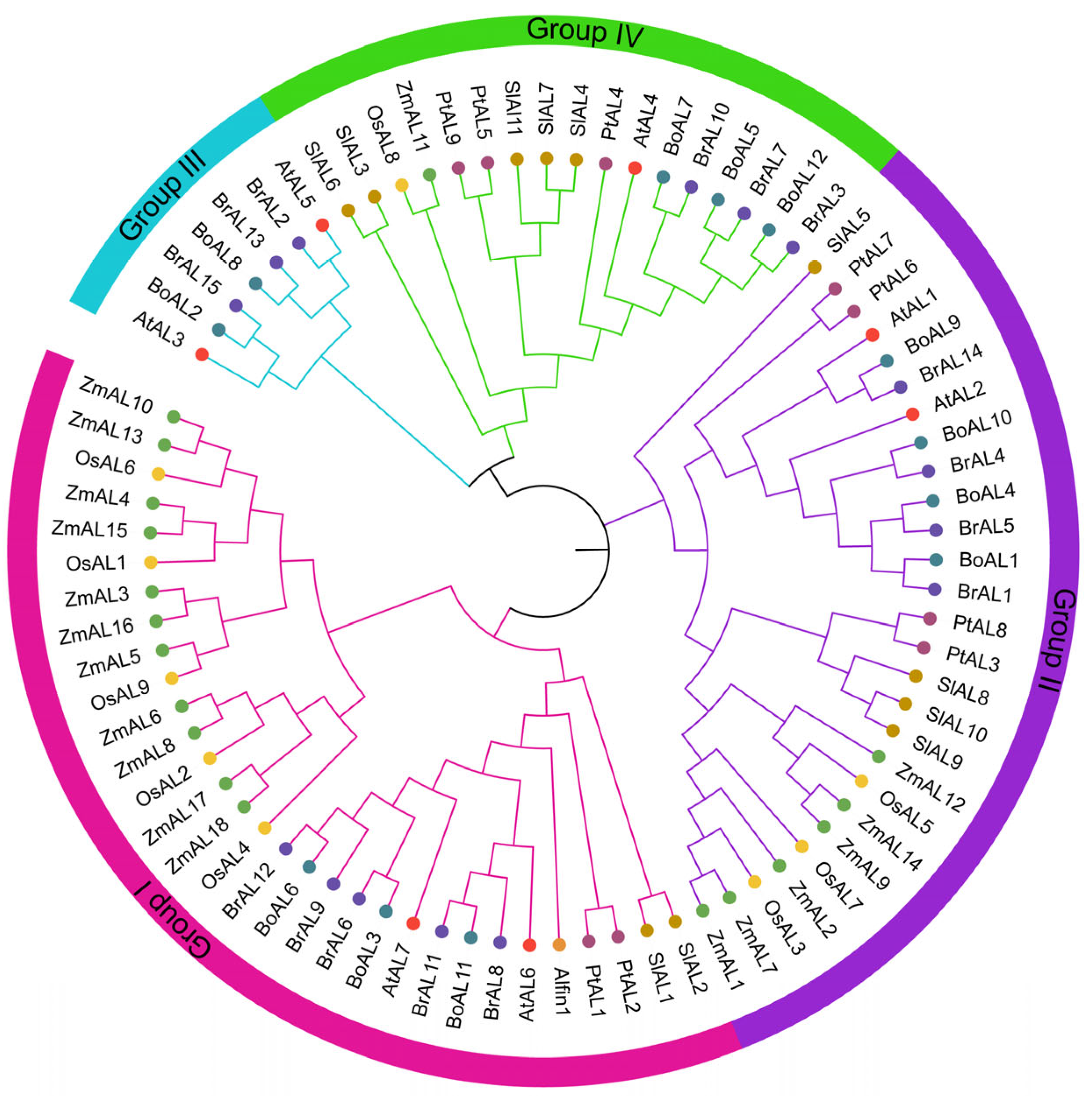
3. Structural Characteristics and Classification of the Alfin-Like Transcription Factors
4. Role of Alfin-Like Transcription Factors in Plant Growth and Development
4.1. Alfin-Like Transcription Factors Regulate Seed Shape and Seed Germination
4.2. Alfin-Like Transcription Factors Regulate Plant Root Development
5. Role of Alfin-Like Transcription Factors in Plant Biotic and Abiotic Stress
5.1. Role of Alfin-Like Transcription Factors in Biotic Stress
5.2. Role of Alfin-Like Transcription Factors in Abiotic Stress
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ashraf, M.Y. Heavy Metals-Environmental Pollutants or Potential Stress Ameliorants in Plants. View project Development of Nutrient Enrich Manure Phosphate (NEM-Phos) as efficient fertilizer for crop production on calcareous soils View project. Pak. J. Bot. 2009, 41, 647–654. [Google Scholar]
- Xu, Y.; Huang, B. Exogenous Ascorbic Acid Mediated Abiotic Stress Tolerance in Plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 233–253. ISBN 9783319740577. [Google Scholar]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Jaiswal, A.; Taj, G.; Jaiswal, J.P.; Qureshi, M.I.; Singh, N.K. DREB1/CBF Transcription Factors: Their Structure, Function and Role in Abiotic Stress Tolerance in Plants. J. Genet. 2012, 91, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Song, S.; Wang, Y.; Zeng, Y.; Dong, T.; Ge, X.; Duan, H. Genome-Wide Identification and Expression Analysis of DREB Family Genes in Cotton. BMC Plant Biol. 2023, 23, 169. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, M.; Daszkowska-Golec, A. Molecular Mechanisms of SNAC1 (Stress-Responsive NAC1) in Conferring the Abiotic Stress Tolerance. Plant Sci. 2023, 337, 111894. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Tang, N.; Zou, J.; Ran, J.; Chen, X. Rice MYB Transcription Factor OsMYB1R1 Negatively Regulates Drought Resistance. Plant Growth Regul. 2023, 99, 515–525. [Google Scholar] [CrossRef]
- Mahiwal, S.; Pahuja, S.; Pandey, G.K. Review: Structural-Functional Relationship of WRKY Transcription Factors: Unfolding the Role of WRKY in Plants. Int. J. Biol. Macromol. 2024, 257, 128769. [Google Scholar] [CrossRef]
- Zhang, B.; Feng, C.; Chen, L.; Li, B.; Zhang, X.; Yang, X. Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress. Int. J. Mol. Sci. 2022, 23, 14894. [Google Scholar] [CrossRef]
- Jin, R.; Wang, J.; Guo, B.; Yang, T.; Hu, J.; Wang, B.; Yu, Q. Identification and Expression Analysis of the Alfin-like Gene Family in Tomato and the Role of SlAL3 in Salt and Drought Stresses. Plants 2023, 12, 2829. [Google Scholar] [CrossRef]
- Zaynab, M.; Kanwal, S.; Furqan, M.; Islam, W.; Noman, A.; Ali, G.M.; Rehman, N.; Zafar, S.; Sughra, K.; Jahanzab, M. Proteomic Approach to Address Low Seed Germination in Cyclobalnopsis Gilva. Biotechnol. Lett. 2017, 39, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Zaynab, M.; Sharif, Y.; Fatima, M.; Afzal, M.Z.; Aslam, M.M.; Raza, M.F.; Anwar, M.; Raza, M.A.; Sajjad, N.; Yang, X.; et al. CRISPR/Cas9 to Generate Plant Immunity against Pathogen. Microb. Pathog. 2020, 141, 103996. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wu, J.; Zheng, Q.; Jiang, Y.; Zhang, M.; Zhu, S. Genome-Wide Identification and Comparative Analysis of Alfin-like Transcription Factors in Maize. Genes Genom. 2017, 39, 261–275. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to Drought and Salt Stress in Plants: Unraveling the Signaling Networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Carroll, S.B. Evolution at Two Levels: On Genes and Form. PLoS Biol. 2005, 3, e245. [Google Scholar] [CrossRef] [PubMed]
- Chandrika, N.; Sundaravelpandian, K.; Yu, S.M.; Schmidt, W. ALFIN-LIKE 6 Is Involved in Root Hair Elongation during Phosphate Deficiency in Arabidopsis. New Phytol. 2013, 198, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Winicov, I. Alfin1 Transcription Factor Overexpression Enhances Plant Root Growth under Normal and Saline Conditions and Improves Salt Tolerance in Alfalfa. Planta 2000, 210, 416–422. [Google Scholar] [CrossRef]
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W.-H. Arabidopsis AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes. PLoS Genet. 2014, 10, e1004091. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, Y.; Tao, J.; Chen, H.; Li, Q.; Zhang, W.; Ma, B.; Lin, Q.; Zhang, J.; Chen, S. The ALfin-like Homeodomain Finger Protein AL5 Suppresses Multiple Negative Factors to Confer Abiotic Stress Tolerance in Arabidopsis. Plant J. 2015, 81, 871–883. [Google Scholar] [CrossRef]
- Krochko, J.E.; Pramanik, S.K.; Bewley, J.D. Contrasting Storage Protein Synthesis and Messenger RNA Accumulation during Development of Zygotic and Somatic Embryos of Alfalfa (Medicago sativa L.). Plant Physiol. 1992, 99, 46–53. [Google Scholar] [CrossRef]
- Tao, J.J.; Wei, W.; Pan, W.J.; Lu, L.; Li, Q.T.; Ma, J.B.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. An Alfin-like Gene from Atriplex Hortensis Enhances Salt and Drought Tolerance and Abscisic Acid Response in Transgenic Arabidopsis. Sci. Rep. 2018, 8, 2707. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.Y.; Lee, D.; Chung, W.; Kwon, C.S. Arabidopsis ING and Alfin1-like Protein Families Localize to the Nucleus and Bind to H3K4me3/2 via Plant Homeodomain Fingers. Plant J. 2009, 58, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Winicov, I.; Valliyodan, B.; Xue, L.; Hoober, J.K. The MsPRP2 Promoter Enables Strong Heterologous Gene Expression in a Root-Specific Manner and Is Enhanced by Overexpression of Alfin1. Planta 2004, 219, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Guo, D.; Yan, J.; Zhang, H.; He, Z.; Wang, C.; Tang, W.; Zhou, Y.; Chen, J.; Xu, Z.; et al. The Alfin-like Transcription Factor GmAlfin09 Regulates 2 Endoplasmic Reticulum Stress in Soybean via a Peroxidase. bioRxiv 2023. [Google Scholar] [CrossRef]
- Bastola, D.R.; Pethe, V.V.; Winicov, I. Alfin1, a Novel Zinc-Finger Protein in Alfalfa Roots That Binds to Promoter Elements in the Salt-Inducible MsPRP2 Gene. Plant Mol. Biol. 1998, 38, 1123–1135. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Mylne, J.S.; Crevillen, P.; Geraldo, N.; An, H.; Gendall, A.R.; Dean, C. The PHD Finger Protein VRN5 Functions in the Epigenetic Silencing of Arabidopsis FLC. Curr. Biol. 2007, 17, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Schmitz, R.J.; Amasino, R.M. A PHD Finger Protein Involved in Both the Vernalization and Photoperiod Pathways in Arabidopsis. Genes Dev. 2006, 20, 3244–3248. [Google Scholar] [CrossRef]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis Thaliana Is Mediated by the PHD Finger Protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef]
- Saiga, S.; Furumizu, C.; Yokoyama, R.; Kurata, T.; Sato, S.; Kato, T.; Tabata, S.; Suzuki, M.; Komeda, Y. The Arabidopsis OBERON1 and OBERON2 Genes Encode Plant Homeodomain Finger Proteins and Required for Apical Meristem Maintenance. Development 2008, 135, 1751–1759. [Google Scholar] [CrossRef]
- Wu, T.; Pi, E.-X.; Tsai, S.-N.; Lam, H.-M.; Sun, S.-M.; Kwan, Y.W.; Ngai, S.-M. GmPHD5 Acts as an Important Regulator for Crosstalk between Histone H3K4 Di-Methylation and H3K14 Acetylation in Response to Salinity Stress in Soybean. BMC Plant Biol. 2011, 11, 178. [Google Scholar] [CrossRef]
- Ling, P.; Wang, L.; Zhang, Y.; Dong, A.; Shen, W.H.; Ying, H. Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight about Active-to-Repressive Chromatin State Switch. J. Mol. Biol. 2018, 430, 4245–4259. [Google Scholar]
- Winicov, I.; Bastola, D.R. Transgenic Overexpression of the Transcription Factor Alfin1 Enhances Expression of the Endogenous MsPRP2 Gene in Alfalfa and Improves Salinity Tolerance of the Plants. Plant Physiol. 1999, 120, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Schindler, U.; Beckmann, H.; Cashmore, A.R. HAT3. 1, a Novel Arabidopsis Homeodomain Protein Containing a Conserved Cysteine-rich Region. Plant J. 1993, 4, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Bienz, M. The PHD Finger, a Nuclear Protein-Interaction Domain. Trends Biochem. Sci. 2006, 31, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Gao, J.; Yang, F.; Kua, C.-S.; Liu, J.; Cannon, C.H. Molecular Evolutionary Analysis of the Alfin-like Protein Family in Arabidopsis Lyrata, Arabidopsis Thaliana, and Thellungiella Halophila. PLoS ONE 2013, 8, e66838. [Google Scholar] [CrossRef] [PubMed]
- Kayum, M.A.; Park, J.I.; Ahmed, N.U.; Jung, H.J.; Saha, G.; Kang, J.G.; Nou, I.S. Characterization and Stress-Induced Expression Analysis of Alfin-like Transcription Factors in Brassica Rapa. Mol. Genet. Genom. 2015, 290, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Kayum, M.A.; Park, J.-I.; Ahmed, N.U.; Saha, G.; Chung, M.-Y.; Kang, J.-G.; Nou, I.-S. Alfin-like Transcription Factor Family: Characterization and Expression Profiling against Stresses in Brassica Oleracea. Acta Physiol. Plant. 2016, 38, 127. [Google Scholar] [CrossRef]
- Rehman, S.; Sabir, I.A.; Wang, P.; Li, H.; Ahmad, Z.; Manzoor, M.A.; Zhuge, Q. Genome-Wide Identification of Alfin Like (AL) Transcription Factors and Their Regulatory Role in Abiotic Stress Responses in Poplar (Populus trichocarpa). Plant Stress 2023, 8, 100168. [Google Scholar] [CrossRef]
- Manzoor, M.A.; Li, G.; Xinya, W.; Wang, M.; Zhao, Y.; Sabir, I.A.; Shah, I.H.; Wang, H.; Abdullah, M.; Kim, G.T.; et al. The Alfin-like Transcription Factors: Identification, Characterization, and Expression Analysis in Pyrus Bretschenedri Provide Insight into Its Divergent Functions on Abiotic Response. Sci. Hortic. 2023, 321, 112320. [Google Scholar] [CrossRef]
- Wang, P.; Lu, S.; Li, W.; Ma, Z.; Mao, J.; Chen, B. Genome-Wide Characterization of Alfin-like (AL) Genes in Apple and Functional Identification of MdAL4 in Response to Drought Stress. Plant Cell Rep. 2023, 42, 395–408. [Google Scholar] [CrossRef]
- Sureshkumar, S.; Bandaranayake, C.; Lu, J.; Dent, C.I.; Atri, C.; York, H.M.; Tamizhselvan, P.; Shamaya, N.; Folini, G.; Bhagat, P.K.; et al. SUMO Protease FUG1, Histone Reader AL3 and the PRC1 Complex Are Integral to Repeat-Expansion Induced Epigenetic Silencing in Arabidopsis Thaliana. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, X.; Xia, H.; Wang, L.; Chen, S.; Xu, K.; Yang, F.; Zou, Y.; Wang, Y.; Zhu, J. Natural Variation of Alfin-like Family Affects Seed Size and Drought Tolerance in Rice. Plant J. 2022, 112, 1176–1193. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.-J.; Lin, S.-I. Signaling Network in Sensing Phosphate Availability in Plants. Annu Rev Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- López-Bucio, J.; Hernández-Abreu, E.; Sánchez-Calderón, L.; Nieto-Jacobo, M.F.; Simpson, J.; Herrera-Estrella, L. Phosphate Availability Alters Architecture and Causes Changes in Hormone Sensitivity in the Arabidopsis Root System. Plant Physiol. 2002, 129, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Brown, K.M. Topsoil Foraging—An Architectural Adaptation of Plants to Low Phosphorus Availability. Plant Soil 2001, 237, 225–237. [Google Scholar] [CrossRef]
- Williamson, L.C.; Ribrioux, S.P.C.P.; Fitter, A.H.; Leyser, H.M.O. Phosphate Availability Regulates Root System Architecture in Arabidopsis. Plant Physiol. 2001, 126, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Bates, T.R.; Lyncu, J.P. Stimulation of Root Hair Elongation in Arabidopsis Thaliana by Low Phosphorus Availability. Plant Cell Environ. 1996, 19, 529–538. [Google Scholar] [CrossRef]
- Sun, L.; Xing, S.; Zhang, J.; Yang, J.; Wang, X.; Dong, Y. Function of the Transcription Factors in Plant Domestication and Stress Resistance. Genom. Appl. Biol. 2009, 28, 569–577. [Google Scholar]
- Qu, L.-J.; Zhu, Y.-X. Transcription Factor Families in Arabidopsis: Major Progress and Outstanding Issues for Future Research. Curr. Opin. Plant Biol. 2006, 9, 544–549. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Wang, J.J.; Sun, N.; Liu, Z. Bioinformatics and Expression Analysis of Maize Alfin-like Transcription Factor ZmAL5a. Mol. Plant Breed. 2019, 17, 4859–4864. [Google Scholar]
- Yan, C.; Yang, N.; Li, R.; Wang, X.; Xu, Y.; Zhang, C.; Wang, X.; Wang, Y. Alfin-like Transcription Factor VqAL4 Regulates a Stilbene Synthase to Enhance Powdery Mildew Resistance in Grapevine. Mol. Plant Pathol. 2023, 24, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, Z.; Zhang, H.; Yang, Y.; Yang, X.; Zhao, X.; Guo, H.; Nagalakshmi, U.; Li, D.; Dinesh-Kumar, S.P.; et al. The MAPK-Alfin-like 7 Module Negatively Regulates ROS Scavenging Genes to Promote NLR-Mediated Immunity. Proc. Natl. Acad. Sci. USA 2023, 120, e2214750120. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Lei, M.; Li, F.; Yang, X.; Zhou, M.; Li, B.; Cao, Y.; Gong, S.; Liu, K.; Liu, J.; et al. Family-Wide Characterization of Histone Binding Abilities of PHD Domains of AL Proteins in Arabidopsis Thaliana. Protein J. 2018, 37, 531–538. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, L.; Liu, B.-Y.; Tan, C.-F.; Chen, D.-H.; Shen, W.-H.; Ruan, Y. Evolution and Conservation of Polycomb Repressive Complex 1 Core Components and Putative Associated Factors in the Green Lineage. BMC Genom. 2019, 20, 533. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Wang, P.; He, H.; Cao, X.; Mao, J.; Chen, B. Identification and Expression Analysis of Alfin-like Transcription Factor Family in Vitis vinifera. Acta Bot. Boreali-Occident. Sin. 2020, 40, 1467–1474. [Google Scholar]
- Quiroz-Iturra, L.F.; Simpson, K.; Arias, D.; Silva, C.; González-Calquin, C.; Amaza, L.; Handford, M.; Stange, C. Carrot DcALFIN4 and DcALFIN7 Transcription Factors Boost Carotenoid Levels and Participate Differentially in Salt Stress Tolerance When Expressed in Arabidopsis Thaliana and Actinidia Deliciosa. Int. J. Mol. Sci. 2022, 23, 12157. [Google Scholar] [CrossRef]



| Species | Gene | Function | References |
|---|---|---|---|
| Alfalfa | Alfin1 | salt stress | [18,26,33] |
| A.thaliana | AL5 | salt, drought, and cold stress | [20] |
| AL7 | salt stress | [36] | |
| AL6 | control root hair elongation under phosphate deficient conditions | [17] | |
| A. hortensis | AL1 | drought stress, mediated ABA pathway, consistent seed germination, reduced initial rooting | [22] |
| O.sativa | AL7.1, AL11 | Regulation of seed shape, responds to ABA and mannite | [43] |
| Z.mays | AL5a | salt, drought stress, and fungal pathogens | [52] |
| AL1-18 | salt, drought, and cold stress | [14] | |
| B.oleracea | AL8, AL12 | salt, drought, cold stress, and fungal stress | [38] |
| B.rapa | AL1-15 | salt, drought, cold stress, and fungal stress | [37] |
| M.domestica | AL4 | drought stress | [41] |
| S.lycopersicum | AL3 | salt, drought stress | [11] |
| D.carota | AL4, AL7 | salt stress | [58] |
| V.quinquangularis | AL4 | Promotion of stilbene accumulation, activation of SA signaling, and enhancement of resistance to powdery mildew | [53] |
| V.vinifera | AL1-6 | salt stress, phytohormones | [57] |
| P. bretschenedri | AL1-15 | response to indole acetic acid, gibberellin, melatonin, and abscisic acid | [40] |
| P. trichocarpa | AL1-9 | salt, drought, cold, and high temperature stress | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, R.; Yang, H.; Muhammad, T.; Li, X.; Tuerdiyusufu, D.; Wang, B.; Wang, J. Involvement of Alfin-Like Transcription Factors in Plant Development and Stress Response. Genes 2024, 15, 184. https://doi.org/10.3390/genes15020184
Jin R, Yang H, Muhammad T, Li X, Tuerdiyusufu D, Wang B, Wang J. Involvement of Alfin-Like Transcription Factors in Plant Development and Stress Response. Genes. 2024; 15(2):184. https://doi.org/10.3390/genes15020184
Chicago/Turabian StyleJin, Ruixin, Haitao Yang, Tayeb Muhammad, Xin Li, Diliaremu Tuerdiyusufu, Baike Wang, and Juan Wang. 2024. "Involvement of Alfin-Like Transcription Factors in Plant Development and Stress Response" Genes 15, no. 2: 184. https://doi.org/10.3390/genes15020184
APA StyleJin, R., Yang, H., Muhammad, T., Li, X., Tuerdiyusufu, D., Wang, B., & Wang, J. (2024). Involvement of Alfin-Like Transcription Factors in Plant Development and Stress Response. Genes, 15(2), 184. https://doi.org/10.3390/genes15020184






