Genetic Mapping by 55K Single-Nucleotide Polymorphism Array Reveals Candidate Genes for Tillering Trait in Wheat Mutant dmc
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotypic Investigations and Genetic Analysis
2.3. Cytological Karyotype Analysis
2.4. Wheat 55K SNP Array Analysis
2.5. Expression Analysis
3. Results
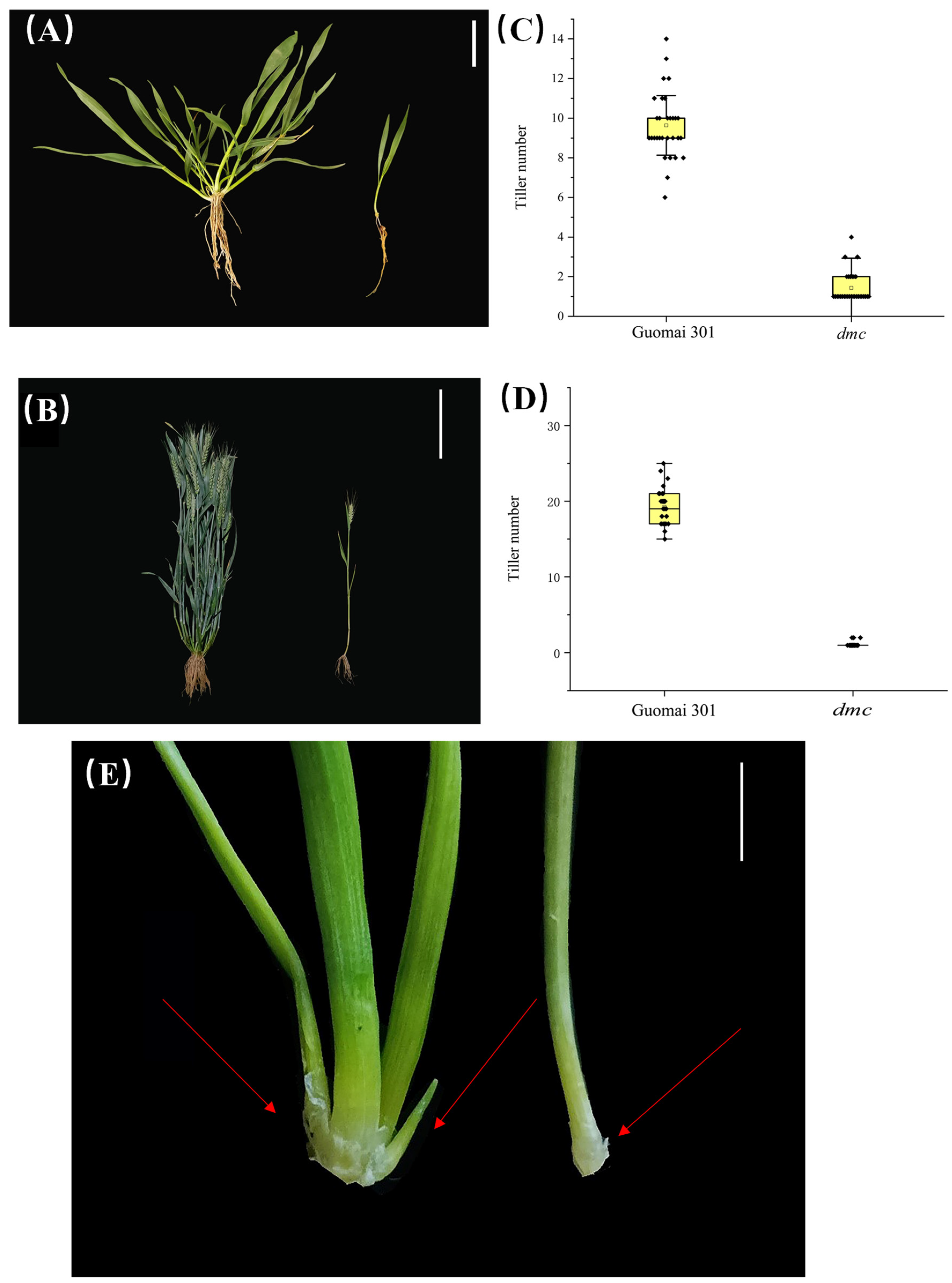
3.1. Identification of dmc Mutants
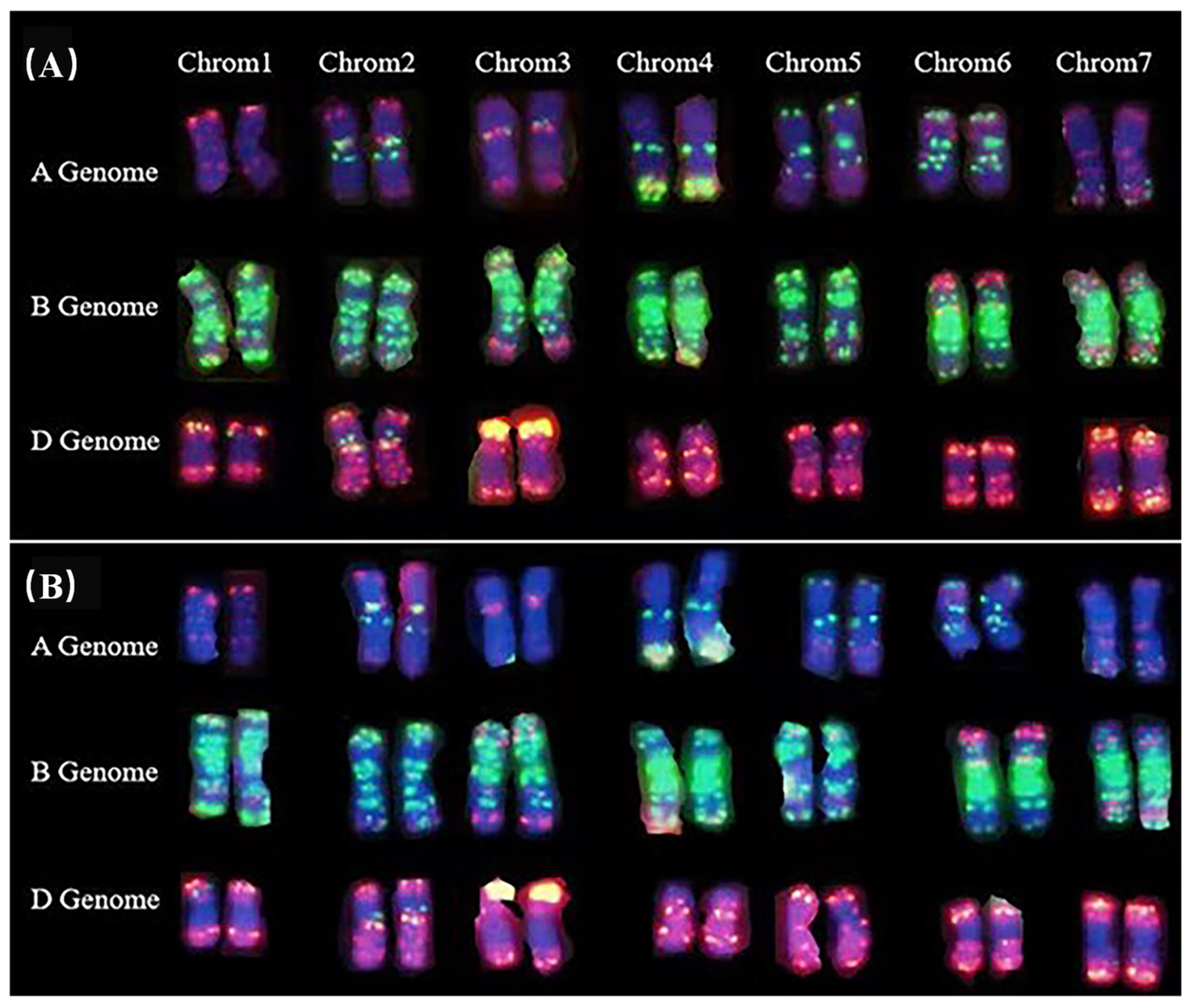
3.2. Fluorescence In Situ Hybridization Analysis of dmc Mutant Chromosomes
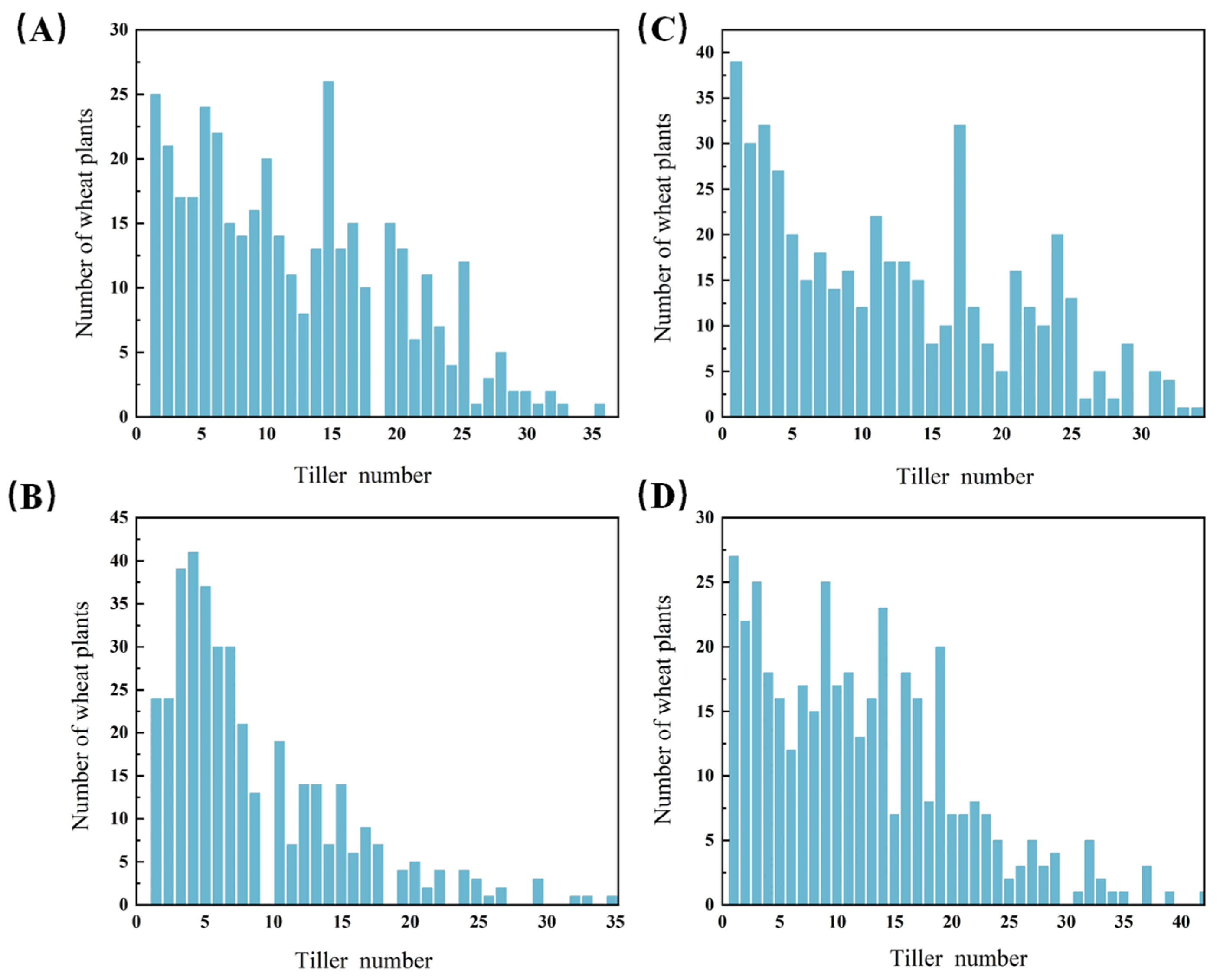
3.3. Genetic Analysis of Tiller Number of dmc Mutant
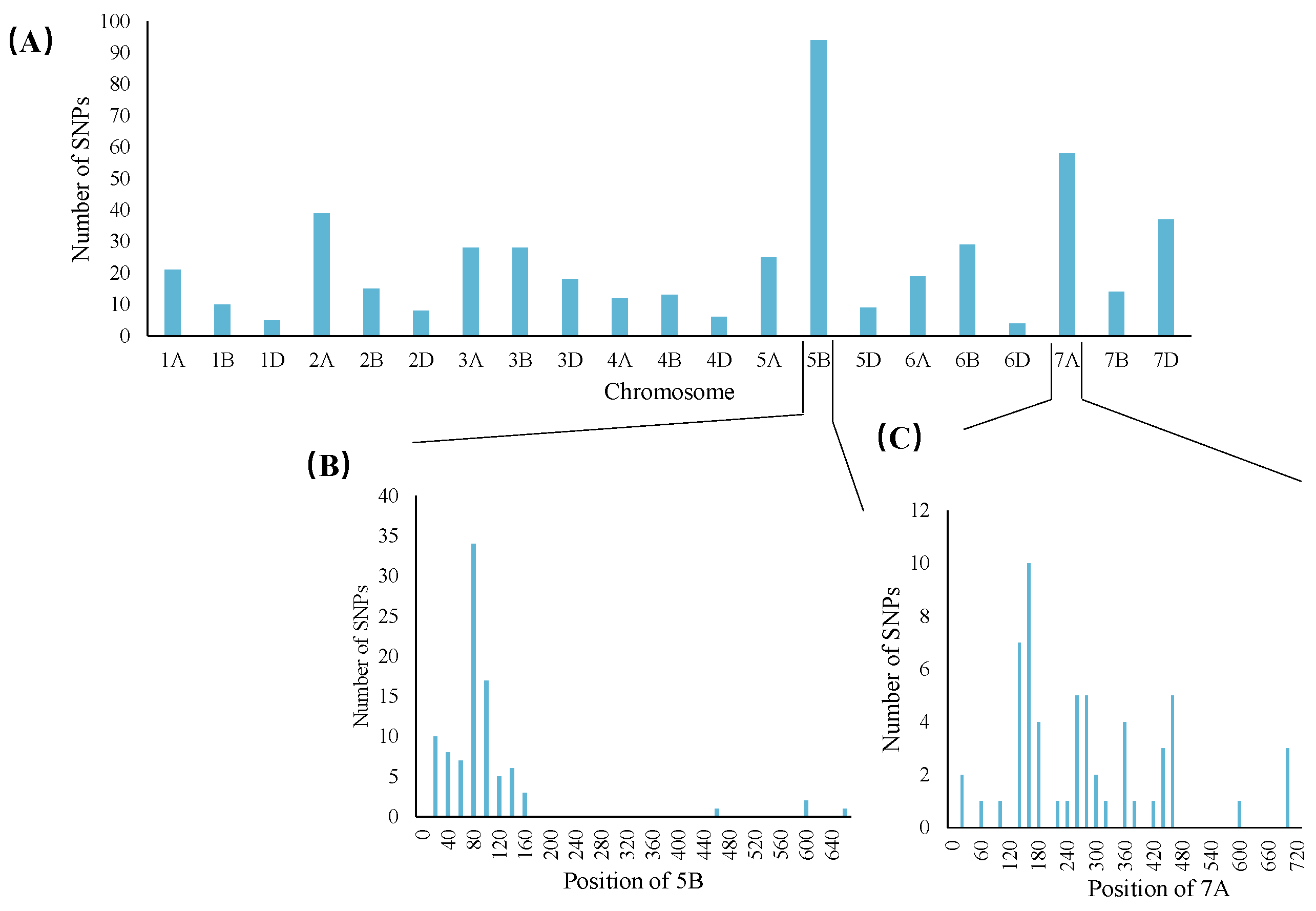
3.4. Analysis of Gene Microarrays
3.5. Candidate Gene Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, X.M.; Yang, Y.M.; Lin, X.L.; Xiao, J. Deciphering spike architecture formation towards yield improvement in wheat. J. Genet. Genom. 2023, 50, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Sareen, S.; Dadrasi, A.; Chaichi, M.; Nehbandani, A.; Sheikhi, A.; Salmani, F.; Nemati, A. Addressing food insecurity: An exploration of wheat production expansion. PLoS ONE 2023, 18, e0290684. [Google Scholar]
- Luo, L. Is strigolactone signaling a key player in regulating tiller formation in response to nitrogen? Front. Plant Sci. 2022, 13, 1081740. [Google Scholar] [CrossRef] [PubMed]
- Almeshal, A.M.; Almazrouee, A.I.; Alenizi, M.R.; Alhajeri, S.N. Forecasting the spread of COVID-19 in Kuwait using compartmental and logistic regression model. Appl. Sci. 2020, 10, 3402. [Google Scholar] [CrossRef]
- Sun, H.W.; Ma, J.H.; Wang, L. Changes in per capita wheat production in China in the context of climate change and population growth. Food Secur. 2023, 15, 597–612. [Google Scholar] [CrossRef]
- Dadrasi, A.; Chaichi, M.; Nehbandani, A.; Soltani, E.; Nemati, A.; Salmani, F.; Heydari, M.; Yousefi, A.R. Global insight into understanding wheat yield and production through Agro-Ecological Zoning. Sci. Rep. 2023, 13, 15898. [Google Scholar] [CrossRef]
- Yang, B.; Qiao, L.; Zheng, X.; Zheng, J.; Wu, B.; Li, X.; Zhao, J. Quantitative Trait Loci Mapping of Heading Date in Wheat under Phosphorus Stress Conditions. Genes 2024, 15, 1150. [Google Scholar] [CrossRef]
- Shang, Q.S.; Wang, Y.P.; Tang, H.; Sui, N.; Zhang, X.S.; Wang, F. Genetic, hormonal, and environmental control of tillering in wheat. Crop J. 2021, 9, 986–991. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Rameau, C.; Wijerathna-Yapa, A.; Lunn, J. Lessons from a century of apical dominance research. J. Exp. Bot. 2023, 74, 3903–3922. [Google Scholar] [CrossRef]
- Schneider, A.; Godin, C.; Boudon, F.; Demotes-Mainard, S.; Sakr, S.; Bertheloot, J. Light Regulation of Axillary Bud Outgrowth Along Plant Axes: An Overview of the Roles of Sugars and Hormones. Front. Plant Sci. 2019, 10, 1296. [Google Scholar] [CrossRef]
- Zhu, M.Q.; Jiang, S.; Huang, J.Q.; Li, Z.H.; Xu, S.; Liu, S.J.; He, Y.G.; Zhang, Z.H. Biochemical and Transcriptome Analyses Reveal a Stronger Capacity for Photosynthate Accumulation in Low-Tillering Rice Varieties. Int. J. Mol. Sci. 2024, 25, 1648. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Huang, L.Y.; Zhang, S.L.; Zhang, Y.S.; He, W.M.; Zeng, P.; Zeng, Y.; Huang, G.F.; Zhang, J.; Ning, M.; et al. Neo-functionalization of a Teosinte branched 1 homologue mediates adaptations of upland rice. Nat. Commun. 2020, 11, 725. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.Y.; Luo, Q.; Shen, Y.H.; Wei, L.; Song, X.; Liao, H.Q.; Ni, L.; Shen, T.; Du, X.L.; Han, J.Y.; et al. Coordinated regulation of vegetative phase change by brassinosteroids and the age pathway in Arabidopsis. Nat. Commun. 2023, 14, 2608. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Richards, R.A. Physiological perspectives of reduced tillering and stunting in the tiller inhibition (tin) mutant of wheat. Funct. Plant Biol. 2013, 40, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Bauer, B.; von Wirén, N. Modulating tiller formation in cereal crops by the signalling function of fertilizer nitrogen forms. Sci. Rep. 2020, 10, 20504. [Google Scholar] [CrossRef]
- Chen, Y.P.; Dan, Z.W.; Li, S.Q. Rice GROWTH-REGULATING FACTOR 7 controls tiller number by regulating strigolactone synthesis. Plant Signal Behav. 2020, 15, 1804685. [Google Scholar] [CrossRef]
- Dong, C.H.; Zhang, L.C.; Zhang, Q.; Yang, Y.; Li, D.; Xie, Z.; Cui, G.; Chen, Y.; Wu, L.; Li, Z.; et al. Tiller Number1 encodes an ankyrin repeat protein that controls tillering in bread wheat. Nat. Commun. 2023, 14, 836. [Google Scholar] [CrossRef]
- Li, L.; Xie, C.M.; Zong, J.Q.; Guo, H.L.; Li, D.D.; Liu, J.X. Physiological and Comparative Transcriptome Analyses of the High-Tillering Mutant mtn1 Reveal Regulatory Mechanisms in the Tillering of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.). Int. J. Mol. Sci. 2022, 23, 11580. [Google Scholar] [CrossRef]
- Feng, G.Y.; Xu, X.H.; Liu, W.W.; Hao, F.X.; Yang, Z.F.; Nie, G.; Huang, L.K.; Peng, Y.; Bushman, S.; He, W.; et al. Transcriptome Profiling Provides Insights into the Early Development of Tiller Buds in High- and Low-Tillering Orchardgrass Genotypes. Int. J. Mol. Sci. 2023, 24, 16370. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Ni, Y.; Jiang, Y.; Jiao, Z.; Li, H.; Wang, T.; Zhang, P.; Han, M.; Li, L.; et al. Key wheat GRF genes constraining wheat tillering of mutant dmc. PeerJ 2021, 9, e11235. [Google Scholar] [CrossRef]
- Luo, Z.W.; Janssen, B.J.; Snowden, K.C. The molecular and genetic regulation of shoot branching. Plant Physiol. 2021, 187, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Q.; Yuan, C.Q.; Cong, T.C.; Zhang, Q.X. The Secrets of Meristems Initiation: Axillary Meristem Initiation and Floral Meristem Initiation. Plants 2023, 12, 1879. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, W.; Tsuda, K.; Hirano, H. Class I KNOX Gene OSH1 is Indispensable for Axillary Meristem Development in Rice. Cytologia 2019, 84, 343–346. [Google Scholar] [CrossRef]
- Mach, J. Rice Axillary Meristem Formation Requires Directional Movement of LAX PANICLE1 Protein. Plant Cell 2009, 21, 1027. [Google Scholar] [CrossRef][Green Version]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of Rice Encodes a Novel Nuclear Protein and Regulates the Formation of Axillary Meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef]
- Li, X.Y.; Qian, Q.; Fu, Z.M.; Wang, Y.H.; Xiong, G.S.; Zeng, D.L.; Wang, X.Q.; Liu, X.F.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, X.; Xu, W.N.; Chang, J.Z.; Li, A.; Mao, X.G.; Zhang, X.Y.; Jing, R.L. Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Sci. Rep.-UK 2015, 5, 12211. [Google Scholar] [CrossRef]
- Liao, Z.G.; Yu, H.; Duan, J.B.; Yuan, K.; Yu, C.J.; Meng, X.B.; Kou, L.Q.; Chen, M.J.; Jing, Y.H.; Liu, G.F.; et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 2019, 10, 2738. [Google Scholar] [CrossRef]
- Sun, F.L.; Zhang, W.P.; Xiong, G.S.; Yan, M.X.; Qian, Q.; Li, J.Y.; Wang, Y.H. Identification and functional analysis of the MOC1 interacting protein 1. J. Gene Genom. 2010, 37, 69–77. [Google Scholar] [CrossRef]
- Xu, C.; Wang, Y.G.; Yu, Y.C.; Duan, J.B.; Liao, Z.G.; Xiong, G.S.; Meng, X.B.; Liu, G.F.; Qian, Q.; Li, J.Y. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat. Commun. 2012, 3, 750. [Google Scholar] [CrossRef]
- Shao, G.N.; Lu, Z.F.; Xiong, J.S.; Wang, B.; Jing, Y.H.; Meng, X.B.; Liu, G.F.; Ma, H.Y.; Liang, Y.; Chen, F.; et al. Tiller Bud Formation Regulators MOC1 and MOC3 Cooperatively Promote Tiller Bud Outgrowth by Activating FON1 Expression in Rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, G.M.; Ramakrishna, K.; Chongloi, G.L.; Vijayraghavan, U. Functions for rice RFL in vegetative axillary meristem specification and outgrowth. J. Exp. Bot. 2015, 66, 2773–2784. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bao, J.L.; Zhou, B.B.; Li, M.; Li, X.Z.; Jin, J. The osa-miR164 target OsCUC1 functions redundantly with OsCUC3 in controlling rice meristem/organ boundary specification. New Phytol. 2020, 229, 1566–1581. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.F.; Huang, Y.; Hu, Y.; Liu, H.Y.; Zhang, B.; Smaczniak, C.; Hu, G.; Han, Z.M.; Xing, Y.Z. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 2017, 3, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Varshney, K.; Gutjahr, C. KAI2 Can Do: Karrikin Receptor Function in Plant Development and Response to Abiotic and Biotic Factors. Plant Cell Physiol. 2023, 64, 984–995. [Google Scholar] [CrossRef]
- Wang, Y.X.; Shang, L.G.; Yu, H.; Zeng, L.J.; Hu, J.; Ni, S.; Rao, Y.C.; Li, S.F.; Chu, J.F.; Meng, X.B.; et al. A Strigolactone Biosynthesis Gene Contributed to the Green Revolution in Rice. Mol. Plant 2020, 13, 923–932. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.B.; Zhu, L.H.; Ren, Y.L.; Zhou, K.N.; Shabek, N.; Wu, F.Q.; Mao, H.B.; Dong, W.; Gan, L.; et al. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Lin, H.; Wang, R.X.; Qian, Q.; Yan, M.X.; Meng, X.B.; Fu, Z.M.; Yan, C.Y.; Jiang, B.; Su, Z.; Li, J.Y.; et al. DWARF27, an Iron-Containing Protein Required for the Biosynthesis of Strigolactones, Regulates Rice Tiller Bud Outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef]
- Zhao, B.; Wu, T.T.; Ma, S.S.; Jiang, D.J.; Bie, X.M.; Sui, N.; Zhang, X.S.; Wang, F. TaD27-B gene controls the tiller number in hexaploid wheat. Plant Biotechnol. J. 2020, 18, 513–525. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.L.; Lu, Z.F.; Meng, X.B.; Wang, Y.H.; Smith, S.M.; Li, J.Y. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142. [Google Scholar] [CrossRef]
- Chen, L.P.; Zhao, Y.; Xu, S.J.; Zhang, Z.Y.; Xu, Y.Y.; Zhang, J.Y.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Duan, E.; Wang, Y.H.; Li, X.H.; Lin, Q.B.; Zhang, T.; Wang, Y.P.; Zhou, C.L.; Zhang, H.; Jiang, L.; Wang, J.L.; et al. OsSHI1 Regulates Plant Architecture Through Modulating the Transcriptional Activity of IPA1 in Rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Han, T.W.; Song, Q.X.; Ye, W.X.; Song, X.G.; Chu, J.F.; Li, J.Y.; Chen, Z.J. The Rice Circadian Clock Regulates Tiller Growth and Panicle Development Through Strigolactone Signaling and Sugar Sensing. Plant Cell 2020, 32, 3124–3138. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Tachibana, C.; Tamaki, S.; Taoka, K.; Kyozuka, J.; Shimamoto, K. Hd3a promotes lateral branching in rice. Plant J. 2015, 82, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Moritoh, S.; Eun, C.; Ono, A.; Asao, H.; Okano, Y.; Yamaguchi, K.; Shimatani, Z.; Koizumi, A.; Terada, R. Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 2012, 71, 85–98. [Google Scholar] [CrossRef]
- Zhang, Q.; Xie, J.Y.; Zhu, X.Y.; Ma, X.Q.; Yang, T.; Khan, N.U.; Zhang, S.Y.; Liu, M.S.; Li, L.; Liang, Y.T.; et al. Natural variation in Tiller Number 1 affects its interaction with TIF1 to regulate tillering in rice. Plant Biotechnol. J. 2023, 21, 1044–1057. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Wu, F.K.; Chen, X.D.; Zhou, W.L.; Shi, H.R.; Lin, Y.; Hou, S.; Yu, S.F.; Zhou, H.; Li, C.X.; et al. Fine mapping of the tiller inhibition gene TIN4 contributing to ideal plant architecture in common wheat. Theor. Appl. Genet. 2022, 135, 527–535. [Google Scholar] [CrossRef]
- Si, Y.; Lu, Q.; Tian, S.; Niu, J.; Cui, M.; Liu, X.; Gao, Q.; Shi, X.; Ling, H.; Zheng, S. Fine mapping of the tiller inhibition gene TIN5 in Triticum urartu. Theor. Appl. Genet. 2022, 135, 2665–2673. [Google Scholar] [CrossRef]
- Zhang, J.P.; Wu, J.; Liu, W.H.; Lu, X.; Yang, X.M.; Gao, A.N.; Li, X.Q.; Lu, Y.Q.; Li, L.H. Genetic mapping of a fertile tiller inhibition gene, ftin, in wheat. Mol. Breed. 2012, 31, 441–449. [Google Scholar] [CrossRef]
- Schoen, A.; Yadav, I.; Wu, S.Y.; Poland, J.; Rawat, N.; Tiwari, V. Identification and high-resolution mapping of a novel tiller number gene (tin6) by combining forward genetics screen and MutMap approach in bread wheat. Funct. Integr. Genom. 2023, 23, 157. [Google Scholar] [CrossRef]
- Kuraparthy, V.; Sood, S.; Dhaliwal, H.S.; Chhuneja, P.; Gill, B.S. Identification and mapping of a tiller inhibition gene (tin3) in wheat. Theor. Appl. Genet. 2007, 114, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.X.; Guo, H.J.; Xiong, H.C.; Xie, Y.D.; Gu, J.Y.; Zhao, L.S.; Zhao, S.R.; Ding, Y.P.; Liu, L.X. Strigolactone and abscisic acid synthesis and signaling pathways are enhanced in the wheat oligo-tillering mutant ot1. Mol. Breed. 2024, 44, 12. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.A. A tiller inhibitor gene in wheat and its effect on plant growth. Crop Pasture Sci. 1988, 39, 749–757. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Chandler, P.M.; Swain, S.M.; King, R.W.; Richards, R.A.; Spielmeyer, W. Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 2012, 160, 308–318. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Richards, R.A. Comparative mapping of wheat chromosome 1AS which contains the tiller inhibition gene (tin) with rice chromosome 5S. Theor. Appl. Genet. 2004, 109, 1303–1310. [Google Scholar] [CrossRef]
- He, R.S.; Ni, Y.J.; Li, J.C.; Jiao, Z.X.; Zhu, X.X.; Jiang, Y.M.; Li, Q.Y.; Niu, J.S. Quantitative Changes in the Transcription of Phytohormone-Related Genes: Some Transcription Factors Are Major Causes of the Wheat Mutant dmc Not Tillering. Int. J. Mol. Sci. 2018, 19, 1324. [Google Scholar] [CrossRef]
- An, J.H.; Niu, H.; Ni, Y.J.; Jiang, Y.M.; Zheng, Y.X.; He, R.S.; Li, J.C.; Jiao, Z.X.; Zhang, J.; Li, H.J.; et al. The miRNA-mRNA Networks Involving Abnormal Energy and Hormone Metabolisms Restrict Tillering in a Wheat Mutant dmc. Int. J. Mol. Sci. 2019, 20, 4586. [Google Scholar] [CrossRef]
- Li, Q.Y.; Qin, Z.; Jiang, Y.M.; Shen, C.C.; Duan, Z.B.; Niu, J.S. Screening wheat genotypes for resistance to black point and the effects of diseased kernels on seed germination. J. Plant Dis. Prot. 2014, 121, 79–88. [Google Scholar] [CrossRef]
- Du, P.; Zhuang, L.F.; Wang, Y.Z.; Yuan, L.; Wang, Q.; Wang, D.R.; Dawadondup; Tan, L.J.; Shen, J.; Xu, H.B.; et al. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 2017, 60, 93–103. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, J.C.; Jiao, Z.X.; He, R.S.; Sun, Y.L.; Xu, Q.Q.; Zhang, J.; Jiang, Y.M.; Li, Q.Y.; Niu, J.S. Gene Expression Profiles and microRNA Regulation Networks in Tiller Primordia, Stem Tips, and Young Spikes of Wheat Guomai 301. Genes 2019, 10, 686. [Google Scholar] [CrossRef] [PubMed]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson Scott, A.; Leal-Bertioli Soraya, C.M.; Tester, M.; Gao, C.X.; Godwin Ian, D.; Hayes Ben, J.; Wulff Brande, B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Hatta, M.A.M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef]
- Yu, H.; Li, J.Y. Breeding future crops to feed the world through de novo domestication. Nat. Commun. 2022, 13, 23–29. [Google Scholar] [CrossRef]
- Wolde, G.M.; Mascher, M.; Schnurbusch, T. Genetic modification of spikelet arrangement in wheat increases grain number without significantly affecting grain weight. Mol. Genet. Genom. 2018, 294, 457–468. [Google Scholar] [CrossRef]
- Liu, J.J.; Luo, W.; Qin, N.N.; Ding, P.Y.; Zhang, H.; Yang, C.C.; Mu, Y.; Tang, H.P.; Liu, Y.X.; Li, W.; et al. A 55 K SNP array-based genetic map and its utilization in QTL mapping for productive tiller number in common wheat. Theor. Appl. Genet. 2018, 131, 2439–2450. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Liu, Y.X.; Shi, H.R.; Mo, H.J.; Wu, F.K.; Lin, Y.; Gao, S.; Wang, J.R.; Wei, Y.M.; Liu, C.J.; et al. Identification and validation of novel low-tiller number QTL in common wheat. Theor. Appl. Genet. 2016, 129, 603–612. [Google Scholar] [CrossRef]
- Naruoka, Y.; Talbert, L.E.; Lanning, S.P.; Blake, N.K.; Martin, J.M.; Sherman, J.D. Identification of quantitative trait loci for productive tiller number and its relationship to agronomic traits in spring wheat. Theor. Appl. Genet. 2011, 123, 1043–1053. [Google Scholar] [CrossRef]
- Kato, K.; Miura, H.; Sawada, S. Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor. Appl. Genet. 2000, 101, 1114–1121. [Google Scholar] [CrossRef]
- Hu, Y.S.; Ren, T.H.; Li, Z.; Tang, Y.Z.; Ren, Z.L.; Yan, B.J. Molecular mapping and genetic analysis of a QTL controlling spike formation rate and tiller number in wheat. Gene 2017, 634, 15–21. [Google Scholar] [CrossRef]
- Liu, J.J.; Tang, H.P.; Qu, X.R.; Liu, H.; Li, C.; Tu, Y.; Li, S.Q.; Habib, A.; Mu, Y.; Dai, S.F.; et al. A novel, major, and validated QTL for the effective tiller number located on chromosome arm 1BL in bread wheat. Plant Mol. Biol. 2020, 104, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.H.; Hu, Y.S.; Tang, Y.Z.; Li, C.S.; Yan, B.J.; Ren, Z.L.; Tan, F.Q.; Tang, Z.X.; Fu, S.L.; Li, Z. Utilization of a Wheat55K SNP Array for Mapping of Major QTL for Temporal Expression of the Tiller Number. Front. Plant Sci. 2018, 9, 333. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Li, X.; Wang, H.; Song, Y.N.; Li, Z.Q.; Li, X.G.; Gao, X.Q.; Zhang, X.S.; Bie, X.M. Down-expression of TaPIN1s Increases the Tiller Number and Grain Yield in Wheat. Bmc Plant Biol. 2021, 21, 443. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Hao, X.; Zhang, J.; Tang, H.; Wang, F. Reducing expression of TaOTUB1s decreases tiller number in wheat. Plant Signal Behav. 2021, 16, 201–217. [Google Scholar] [CrossRef]
- Jian, C.; Pan, Y.; Liu, S.; Guo, M.; Huang, Y.; Cao, L.; Zhang, W.; Yan, L.; Zhang, X.; Hou, J.; et al. The TaGW2-TaSPL14 module regulates the trade-off between tiller number and grain weight in wheat. J. Integr. Plant Biol. 2024, 66, 1953–1965. [Google Scholar] [CrossRef]
- Li, J.C.; Jiang, Y.M.; Zhang, J.; Ni, Y.J.; Jiao, Z.X.; Li, H.J.; Wang, T.; Zhang, P.P.; Guo, W.L.; Li, L.; et al. Key auxin response factor (ARF) genes constraining wheat tillering of mutant dmc. PeerJ 2021, 9, e12221. [Google Scholar] [CrossRef]
- Zhao, L.; Tan, L.B.; Zhu, Z.F.; Xiao, L.T.; Xie, D.X.; Sun, C.Q. PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 2015, 83, 528–536. [Google Scholar] [CrossRef]
- Gallavotti, A. The role of auxin in shaping shoot architecture. J. Exp. Bot. 2013, 64, 2593–2608. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, X.L.; Liu, P.; Sun, J.Q. miR156-Targeted SBP-Box Transcription Factors Interact with DWARF53 to Regulate TEOSINTE BRANCHED1 and BARREN STALK1 Expression in Bread Wheat. Plant Physiol. 2017, 174, 1931–1948. [Google Scholar] [CrossRef]
- Zhang, L.C.; He, G.H.; Li, Y.P.; Yang, Z.Y.; Liu, T.Q.; Xie, X.Z.; Kong, X.Y.; Sun, J.Q. Transcription factors PILs directly interact with SPLs and repress tillering/branching in plants. New Phytol. 2021, 233, 1414–1425. [Google Scholar] [CrossRef]
- Liu, R.Z. Creation of Tillcring Germplasm Using TaPAY1 Gene in Wheat; Shandong Agriculture University: Tai’an, China, 2021. [Google Scholar]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; GerJürgens, G.; et al. AtPIN4 Mediates Sink-Driven Auxin Gradients and Root Patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Bilanovičová, V.; Rýdza, N.; Koczka, L.; Hess, M.; Feraru, E.; Friml, J.; Nodzyński, T. The Hydrophilic Loop of Arabidopsis PIN1 Auxin Efflux Carrier Harbors Hallmarks of an Intrinsically Disordered Protein. Int. J. Mol. Sci. 2022, 23, 6352. [Google Scholar] [CrossRef] [PubMed]
- Mikihisa, U.; Atsushi, H.; Satoko, Y.; Kohki, A.; Tomotsugu, A.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar]
- Sun, J.; Bie, X.M.; Chu, X.L.; Wang, N.; Zhang, X.S.; Gao, X.Q. Genome-edited TaTFL1-5 mutation decreases tiller and spikelet numbers in common wheat. Front. Plant Sci. 2023, 14, 1142779. [Google Scholar] [CrossRef]

| Hybrid Combination | Number of Individual F1 Plants | Number of Tillers of Multi-Tiller Parent | Number of Tillers in the F1 Generation 1 |
|---|---|---|---|
| dmc × Zhengmai 9405 | 4 | 13 ± 1.48 | 9.75 ± 5.58 |
| Zhengmai 9405 × dmc | 17 | 13 ± 1.48 | 4.64 ± 1.93 |
| dmc × Aikang 58 | 3 | 22 ± 2.23 | 5 ± 2 |
| Aikang 58 × dmc | 13 | 22 ± 2.24 | 5.53 ± 1.39 |
| dmc × Zhengmai 379 | 17 | 14 ± 1.14 | 8.9 ± 3.73 |
| Zhengmai 379 × dmc | 10 | 14 ± 1.14 | 5.23 ± 2.07 |
| dmc × Guomai 301 | 11 | 23 ± 2.22 | 11.75 ± 2.06 |
| Guomai 301 × dmc | 6 | 23 ± 2.22 | 6.18 ± 4.52 |
| Hybrid Combination | Year | Multiple Tillers | No Tiller | Expected Ratio | χ2 | p-Value 1 |
|---|---|---|---|---|---|---|
| χ2 (15:1) | P *0.05 | |||||
| dmc × Aikang 58 | 2023 | 424 | 36 | 15:1 | 1.95 | 3.84 |
| dmc × Chinese Spring | 2023 | 396 | 27 | 15:1 | 0.0013 | 3.84 |
| 2024 | 368 | 20 | 15:1 | 0.795 | 3.84 | |
| dmc × Guomai 301 | 2023 | 380 | 21 | 15:1 | 0.702 | 3.84 |
| 2024 | 585 | 50 | 15:1 | 2.858 | 3.84 |
| # | Gene | Annotation | Expression in dmc |
|---|---|---|---|
| 1 | TraesCS5B02G058800 | Auxin-responsive protein IAA31 | up |
| 2 | TraesCS5B02G059300 | CBL-interacting protein kinase 4 | up |
| 3 | TraesCS5B02G059800 | Tuliposide A-converting enzyme 2, chloroplastic | up |
| 4 | TraesCS5B02G060200 | Dolabradiene monooxygenase | up |
| 5 | TraesCS5B02G060300 | Dolabradiene monooxygenase | up |
| 6 | TraesCS5B02G061600 | 11-beta-hydroxysteroid dehydrogenase A | up |
| 7 | TraesCS5B02G062500 | Methylcrotonoyl-CoA carboxylase subunit alpha | up |
| 8 | TraesCS7A02G184200 | Protein containing a Bric-a-Brac/Tramtrack/Broad (BTB) complex and an NPH3 domain (BTBN), regulation of auxin transport | down |
| 9 | TraesCS7A02G184600 | Shikimate kinase domain-containing protein | up |
| 10 | TraesCS7A02G187800 | Aquaporin NIP III subfamily protein | down |
| 11 | TraesCS7A02G188100 | Amino acid transporter | down |
| 12 | TraesCS7A02G188400 | Plant regulator RWP-RK domain-containing protein | down |
| 13 | TraesCS7A02G191400 | Small GTP-binding protein OsRac3 | down |
| 14 | TraesCS7A02G191700 | Domain of unknown function DUF23 | down |
| 15 | TraesCS7A02G196100 | TGF-beta receptor, type I/II extracellular region family protein | down |
| 16 | TraesCS7A02G197900 | Similar to CEL5=CELLULASE 5 (fragment) | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, K.; Xia, G.; Zhou, Y.; Zhao, C.; Yan, H.; Qi, M.; Xie, P.; Ni, Y.; Zhao, J.; Niu, J.; et al. Genetic Mapping by 55K Single-Nucleotide Polymorphism Array Reveals Candidate Genes for Tillering Trait in Wheat Mutant dmc. Genes 2024, 15, 1652. https://doi.org/10.3390/genes15121652
Jiao K, Xia G, Zhou Y, Zhao C, Yan H, Qi M, Xie P, Ni Y, Zhao J, Niu J, et al. Genetic Mapping by 55K Single-Nucleotide Polymorphism Array Reveals Candidate Genes for Tillering Trait in Wheat Mutant dmc. Genes. 2024; 15(12):1652. https://doi.org/10.3390/genes15121652
Chicago/Turabian StyleJiao, Kemeng, Guojun Xia, Yuan Zhou, Chenyu Zhao, Huiyuan Yan, Menglei Qi, Pingfan Xie, Yongjing Ni, Jingxue Zhao, Jishan Niu, and et al. 2024. "Genetic Mapping by 55K Single-Nucleotide Polymorphism Array Reveals Candidate Genes for Tillering Trait in Wheat Mutant dmc" Genes 15, no. 12: 1652. https://doi.org/10.3390/genes15121652
APA StyleJiao, K., Xia, G., Zhou, Y., Zhao, C., Yan, H., Qi, M., Xie, P., Ni, Y., Zhao, J., Niu, J., Chao, Z., Ren, J., & Li, L. (2024). Genetic Mapping by 55K Single-Nucleotide Polymorphism Array Reveals Candidate Genes for Tillering Trait in Wheat Mutant dmc. Genes, 15(12), 1652. https://doi.org/10.3390/genes15121652






