Intraspecific Chloroplast Genome Genetic Polymorphism of Pinellia ternata (Xi Junecry) and Its Revelation of a Single Origin in Phylogeny
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Preservation of Materials
2.2. Chloroplast Genome Assembly and Comparative Analysis of Xi Junecry
2.2.1. DNA Extraction and Sequencing
2.2.2. Chloroplast Genome Assembly
2.2.3. Chloroplast Genome Annotation
2.2.4. Comparative Analysis of Chloroplast Genomes
2.2.5. Selective Pressure Analysis
2.2.6. Phylogenetic Tree Construction
3. Results
3.1. Basic Characteristics of Xi Junecry Chloroplast Genomes
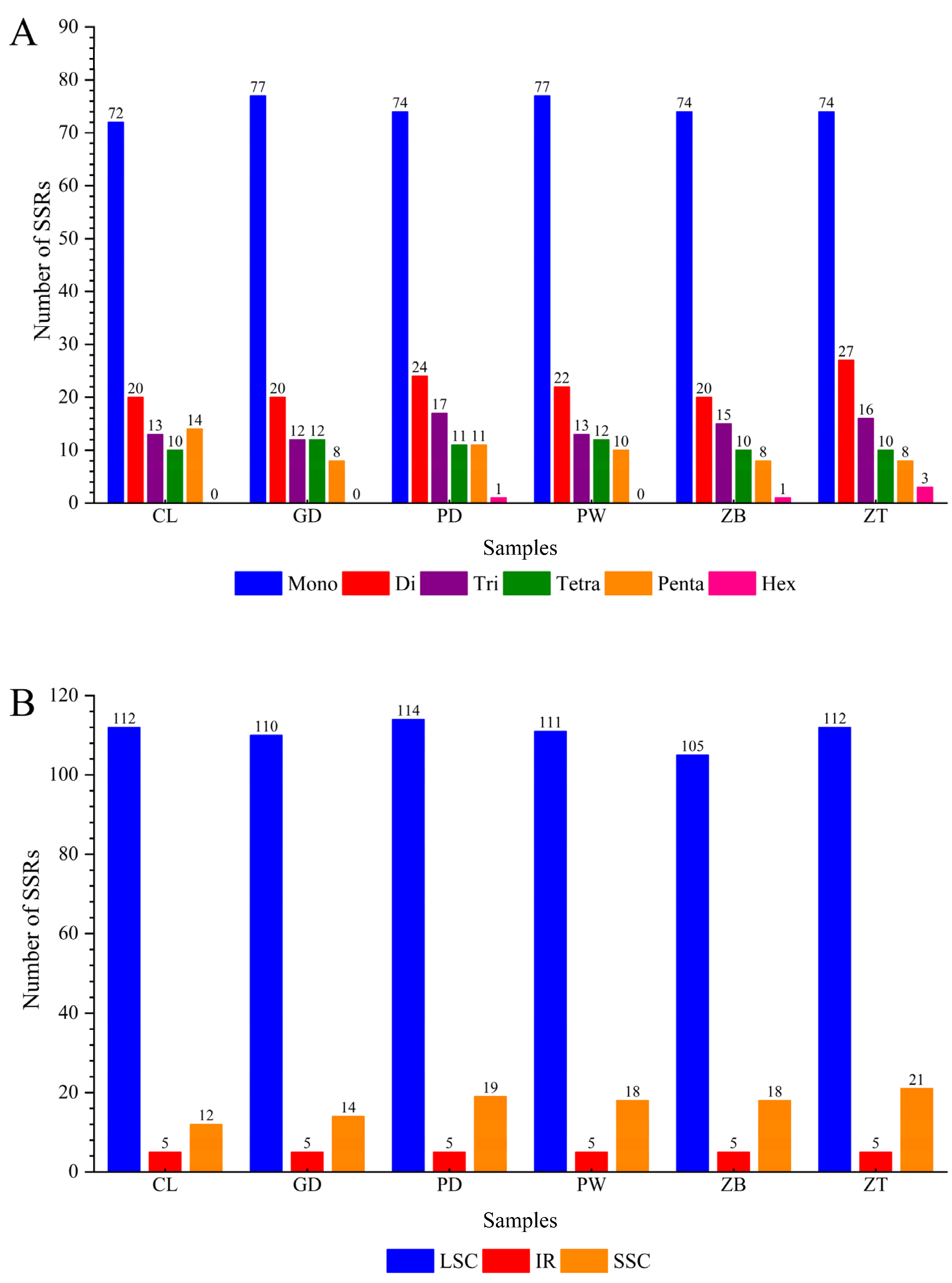
3.2. Simple Sequence Repeats (SSRs) Analysis
3.3. Genetic Polymorphism Analysis
3.4. Genome Variation Analysis
3.5. Adaptive Evolution Analysis
3.6. Phylogenetic Analysis
4. Discussion
4.1. The Intra-Specific Chloroplast Genome Nucleotide Mutation Patterns
4.2. Nucleotide Diversity of Chloroplast Genomes at the Species Level
4.3. Genetic Polymorphism Across Chloroplast Genome Regions
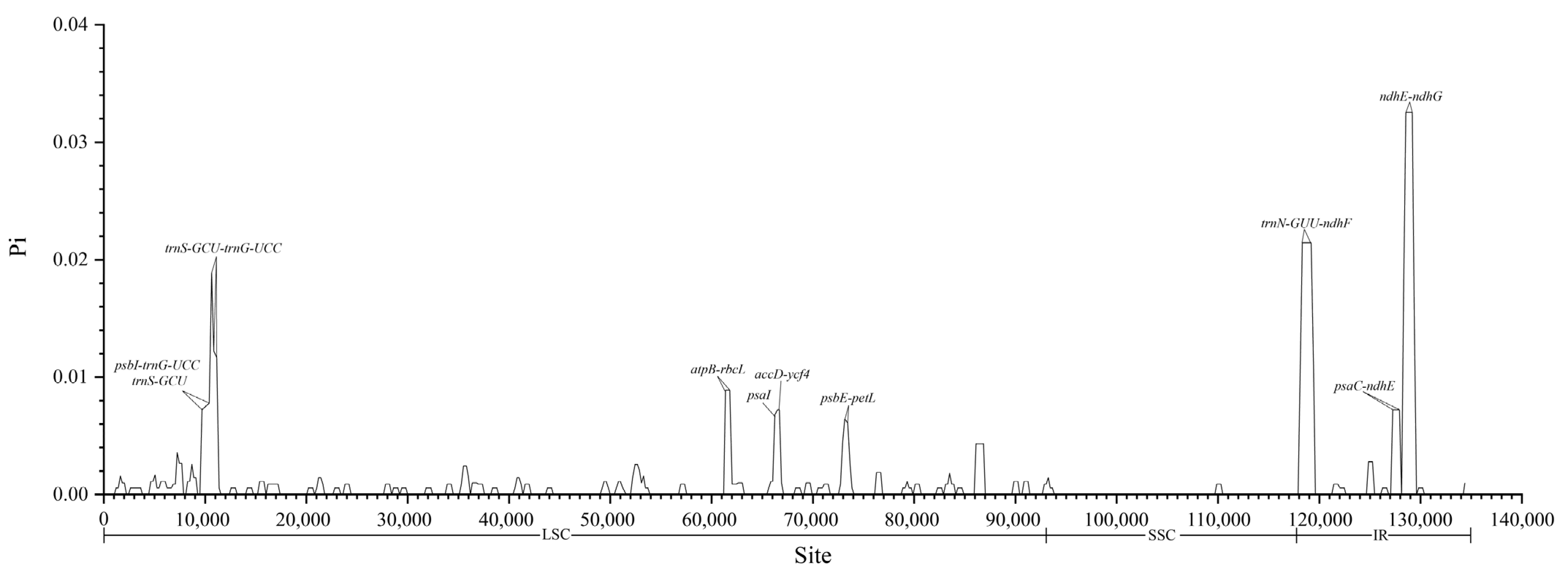
4.4. Highly Variable Regions of the Chloroplast Genome Within a Species
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, D.Y.; Wang, C.R.; Wang, L.; Ma, Z.Q. Observation on the chromosomes of Pinellia ternata of Zhaotong. J. Anhui Agric. Sci. 2006, 34, 2. [Google Scholar]
- Xue, T.; Jia, H.F.; Wang, M.; Zhang, Y.T.; Liu, X.; Chao, Q.J.; Zhao, F.L.; Meng, Z.; Xue, J.P.; Lin, J.S.; et al. A chromosome-level Pinellia ternata genome assembly provides insight into the evolutionary origin of ephedrine and acrid raphide formation. Med. Plant Biol. 2024, 3, e013. [Google Scholar] [CrossRef]
- Li, T.; Cheng, F.F.; Chen, J.H.; Ren, Z.L.; Chen, D.; Zheng, Y.X.; Wang, Q.G. Evolution of clinical dosage of Banxia (Pinelliae Rhizoma) through the ages and analysis of the causes. J. Tradit. Chin. Med. 2023, 25, 122–127. [Google Scholar]
- Zhang, M.F.; Shen, Y.Q. Research progress on pharmacologic actions of extract from Pinelliae rhizoma in respiratory and digestive systems. Anti-Infect. Pharm. 2017, 14, 1457–1462. [Google Scholar]
- Zuo, J.; Mou, J.G.; Hu, X.Y. Research progress in the chemical constituents and modern pharmacological effects of Pinellia ternata. J. Liaoning Univ. Tradit. Chin. Med. 2019, 21, 26–29. [Google Scholar]
- Xie, Q.L.; Geng, X.T.; Wang, G.; Zhou, B.; Yang, W.M.; Chen, Q. Quality analysis on wild and cultivated Xibanxia (Xi Pinellia ternata) in different regions. Acta Chin. Med. 2021, 36, 1296–1302. [Google Scholar]
- Ye, Z.W.; Ye, R.; He, D.X.; Ren, S.W.; Li, J.P.; Chen, Q. Optimization of polysaccharide extraction from Xi Pinellia ternate by response surface methodology and its antioxidant activity. China Food Addit. 2022, 33, 90–98. [Google Scholar]
- Tan, D.Q.; Zhang, B.T.; Li, X.H. A talk on Zhang Zhongjing’s magical use of Pinellia tuber. J. Tradit. Chin. Med. 2006, 24, 146–149. [Google Scholar]
- Gu, Q.Y.; Liu, L.D.; Wang, B.Y.; Haung, Y.Q.; Zhang, Y.Z.; Xiao, Y.; Chen, Q. Insecticidal activity of ethanol extract from peels of Pinellia ternate of Xi county against Plutella xylostella. Mod. Agric. Sci. Technol. 2022, 19, 108–111. [Google Scholar]
- Peng, C.Y.; Liang, J.S.; Pu, X.Y.; Yuan, G.; Wang, X.J. Genetic variation of medicinal active ingredient of Pinellia ternata in different provenances. Nat. Ecol. Evol. 2021, 36, 1296–1302. [Google Scholar]
- He, D.X.; Ye, Z.W.; Qiao, X.R.; Chen, Q. ISSR analysis of genetic diversity of Xi Pinellia ternata germplasm resources. J. Xinyang Norm. Univ. 2023, 36, 46–51. [Google Scholar]
- Wang, H.Y.; Li, J.P.; Zhou, B.; Dong, Z.Y.; Chen, Q. Study on quality of Pinellia ternate (Thunb.) Breit of Xixian county from different sampling points. Hubei Agric. Sci. 2020, 59, 126–129. [Google Scholar]
- Huotari, T.; Korpelainen, H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genome. Gene 2012, 508, 96–105. [Google Scholar] [CrossRef]
- Lilly, J.W.; Havey, M.J.; Jackson, S.A.; Jiang, J. Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell 2001, 13, 245–254. [Google Scholar] [CrossRef]
- Liu, C.; Zeng, X.F.; Yang, X.Y.; Zhao, Y.; Zhang, T.; Zhou, S.X. Comparative analysis and phylogeny of chloroplast genomes in Berchemia polyphylla. Chin. Tradit. Herb. Drugs 2023, 54, 2558–2565. [Google Scholar]
- Yang, S.H.; Wang, Y.L.; Zhang, Y.; Tian, Y.X.; Li, M.Y.; Chen, Q.; Zhang, F.; Sun, B. Analysis of the complete chloroplast genome of Basella alba. J. Sichuan Agric. Univ. 2024, 42, 529–539. [Google Scholar]
- Luo, Y.; Hu, B.X.; Zhang, H.; Shi, J.Z.; Ji, H.Y.; Jing, Y.Y.; Chen, X.Y.; Wang, B.Q.; Yan, Y.G.; Zhao, F.; et al. Chloroplast genome sequence characteristics and phylogenetic analysis of Polygala sibirica. Chin. Tradit. Herb. Drugs 2023, 54, 6065–6073. [Google Scholar]
- Fromm, H.; Galun, E.; Edelman, M. A novel site for streptomycin resistance in the “530 loop” of chloroplast 16s ribosomal RNA. Plant Mol. Biol. 1989, 12, 499–505. [Google Scholar] [CrossRef]
- Kang, B.C.; Bae, S.J.; Lee, S.; Lee, J.S.; Kim, A.; Lee, H.; Baek, G.; Seo, H.; Kim JKim, J.S. Chloroplast and mitochondrial DNA editing in plants. Nat. Plants 2021, 7, 899–905. [Google Scholar] [CrossRef]
- Nakazato, I.; Yamori, W.; Matsumura, H.; Qu, Y.C.; Okuno, M.; Tsutsumi, N.; Arimura, S.I. Resistance to the herbicide metribuzin conferred to Arabidopsis thaliana by targeted base editing of the chloroplast genome. Plant Biotechnol. J. 2024; early view. [Google Scholar]
- Fang, X.M.; Wang, H.W.; Cheng, Y.Q.; Ye, Y.Z.; Yang, C. Optimization of total DNA extraction and test of suitable molecular markers in Taihanggia rupestris. Chin. Agric. Sci. Bull. 2009, 25, 57–60. [Google Scholar]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.L.; Yan, N.; Song, Q.; Guo, J.Z. Complete chloroplast genome sequence and characteristics analysis of Morus multicaulis. Chin. Bull. Bot. 2018, 53, 94–103. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, E.D.; Durkin, M.; Wheat, L.J. Comparison of coccidioides antibody detection by the MVista enzyme immunoassay with immunodiffusion and complement fixation. Open Forum Infect. Dis. 2016, 3 (Suppl. S1), 1561. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Eduarda, D.A.B.D.S.; Dallynne, B.R.V.; Pierre, T.F.; Robson, D.S.R. Phylogenetic grouping and levels of genetic structuration in SARS-Cov-2, using FIGTREE v1.4.4 software. Biol. Syst. Open Access 2021, 10, 1–3. [Google Scholar]
- Jiang, D.; Zhao, Z.; Zhang, T.; Zhong, W.H.; Liu, C.S.; Yuan, Q.J.; Huang, L.Q. The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes 2017, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Ying, Z.; Yu, S.; Wang, Q.; Liao, G.; Ge, Y.; Cheng, R. Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: Insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships. BMC Genom. 2021, 22, 880–899. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, Y.; Garran, T.A.; Qiao, P.; Wang, M.; Yuan, Q.; Guo, L.; Huang, L. Heterogeneous genetic diversity estimation of a promising domestication medicinal motherwort Leonurus cardiaca based on chloroplast genome resources. Front. Genet. 2021, 12, 721022–721035. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Zhou, H.Y.; Liu, X.Q. Chloroplast genomic variation in Euonymus maackii Rupr. and its differentiation time in euonymus. Forests 2022, 13, 265. [Google Scholar] [CrossRef]
- Chen, S.F.; Ishizuka, W.; Hara, T.; Goto, S. Complete Chloroplast genome of Japanese Larch (Larix kaempferi): Insights into intraspecific variation with an isolated northern limit population. Forests 2020, 11, 884. [Google Scholar] [CrossRef]
- Tian, X.; Guo, J.; Song, Y.; Yu, Q.; Liu, C.; Fu, Z.X.; Shi, Y.H.; Shao, Y.Z.; Yuan, Z.L. Intraspecific differentiation of Lindera obtusiloba as revealed by comparative plastomic and evolutionary analyses. Ecol. Evol. 2024, 14, e11119–e11131. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Min, J.; Kim, Y.; Chung, Y. The comparative analyses of six complete chloroplast genomes of Morphologically diverse Chenopodium album L. (Amaranthaceae) collected in Korea. Int. J. Genom. 2021, 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.Q.; Zhang, Y.; Zhang, B.; Cheng, Y.Q.; Shang, F.D.; Wang, H.W. Intraspecific chloroplast genome variation and domestication origins of major cultivars of Styphnolobium japonicum. Genes 2023, 14, 1156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Q.; Wu, Y.X.; Wang, X.; Yang, A.S.; Zhang, X.Y.; Zhao, W.M.; Li, J.F.; Li, Z.H. Evolutionary history and population dynamics of a rare and endangered medicinal plant Bergenia scopulosa (Saxifragaceae): Evidences from chloroplast genomes and ecological niche analysis. Glob. Ecol. Conserv. 2024, 54, e03097–e03112. [Google Scholar] [CrossRef]
- Ferchichi, B.K.; Böhnert, T.; Ritter, B.; Harpke, D.; Stoll, A.; Morales, P.; Fiedler, S.; Mu, F.; Bechteler, J.; Münker, C.; et al. Genetic diversity of the Atacama Desert shrub Huidobria chilensis in the context of geography and climate. Glob. Planet. Change 2024, 234, 104385. [Google Scholar] [CrossRef]
- Pearman, P.B.; Zachos, F.E.; Paz-Vinas, I. European monitoring of genetic diversity must expand to detect impacts of climate change. Nat. Ecol. Evol. 2024, 8, 194–195. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Chauhan, B.S. Genetic diversity, conservation, and utilization of plant genetic resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.R.; Pinheiro, D.G.; Penha, H.A.; Płachno, B.J.; Michael, T.P.; Meer, E.J.; Miranda, V.F.; Varani, A.M. Intraspecific variation within the Utricularia amethystina species morphotypes based on chloroplast genomes. Int. J. Mol. Sci. 2019, 20, 6130. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.H.; Dai, W.; Xu, H.M.; Lin, Y.Y.; Zhu, X.L.; Long, H.; Cheng, L.L.; Xu, X.G. Intraspecific differentiation of Styrax japonicus (Styracaceae) as revealed by comparative chloroplast and evolutionary analyses. Genes 2024, 15, 940. [Google Scholar] [CrossRef]
- Ding, X.; Pan, H.; Shi, P.; Zhao, S.; Bao, S.; Zhong, S.; Dai, C.; Chen, J.; Gong, L.; Zhang, D.; et al. A comparative analysis of chloroplast genomes revealed the chloroplast heteroplasmy of Artemisia annua. Front. Pharmacol. 2024, 15, 1466578. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; He, N.; Peng, Y.; Fang, Y.M.; Zhang, X.W.; Zhang, F.L. Comparative chloroplast genome analysis of Chinese lacquer tree (Toxicodendron vernicifluum, Anacardiaceae): East-west divergence within its range in China. Forests 2023, 14, 818. [Google Scholar] [CrossRef]
- Zhang, R.S.; Yang, J.; Hu, H.L.; Xia, R.X.; Li, Y.P.; Su, J.F.; Li, Q.; Liu, Y.Q.; Qin, L. A high level of chloroplast genome sequence variability in the Sawtooth Oak Quercus acutissima. Int. J. Biol. Macromol. 2020, 152, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Hoppe, D.; Chitnis, V.P.; Odom, W.R.; Guikema, J.A.; Chitnis, P.R. Mutational analysis of photosystem I polypeptides in the cyanobacterium Synechocystis sp. PCC 6803. Targeted inactivation of psaI reveals the function of psaI in the structural organization of psaL. J. Biol. Chem. 1995, 270, 16243–16250. [Google Scholar] [CrossRef]
- Liu, M.M.; Liu, X.; You, Q.; Bo, C.; Zhu, Y.F.; Duan, Y.B.; Xue, J.P.; Wang, D.X.; Xue, T. Genome wide identification and expression analysis of the HD-Zip gene family in Pinellia ternate. J. Agric. Biotechnol. 2021, 12, 2540–2551. [Google Scholar]
- Shao, B.Y.; Wang, M.Z.; Chen, S.S.; Ya, J.D.; Jin, X.H. Habitat-related plastome evolution in the mycoheterotrophic Neottia listeroides complex (Orchidaceae, Neottieae). BMC Plant Biol. 2023, 23, 282–294. [Google Scholar] [CrossRef] [PubMed]


| Sample Name | Sample Location | |
|---|---|---|
| Changling (CL) | 115°6′9.9″ E | 32°22′57.2″ N |
| Guandian (GD) | 114°59′22.9″ E | 32°16′15.9″ N |
| Pengdian (PD) | 114°40′17.6″ E | 32°29′54.8″ N |
| Pangwan (PW) | 114°16′26.5″ E | 32°23′56.6″ N |
| Zhangbanqiao (ZB) | 114°53′39.6″ E | 32°31′20″ N |
| Zhangtao (ZT) | 114°52′49.7″ E | 32°28′27.1″ N |
| Sample ID | Genome Length/bp | LSC/bp | IR/bp | SSC/bp | Number of Gene | Number of CDS | Number of tRNA | Number of rRNA | GC % |
|---|---|---|---|---|---|---|---|---|---|
| CL | 158,318 | 92,345 | 25,174 | 15,625 | 129 | 85 | 36 | 8 | 36.0 |
| GD | 157,938 | 91,933 | 25,235 | 15,535 | 129 | 85 | 36 | 8 | 36.1 |
| PD | 157,673 | 91,657 | 25,235 | 15,546 | 129 | 85 | 36 | 8 | 36.1 |
| PW | 158,406 | 92,232 | 25,235 | 15,704 | 129 | 85 | 36 | 8 | 36.0 |
| ZB | 157,456 | 91,363 | 25,242 | 15,609 | 129 | 85 | 36 | 8 | 36.2 |
| ZT | 157,969 | 92,254 | 25,250 | 15,215 | 130 | 85 | 37 | 8 | 36.0 |
| Category of Genes | Group of Genes | Names of Genes |
|---|---|---|
| Self-replication | Transfer RNA genes | trnA-UGC *, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG **, trnI-CAU *, trnI-GAU *, trnK-UUU, trnL-UAA, trnL-CAA *, trnL-UAG, trnM-CAU, trnN-GUU *, trnP-GGG, trnQ-UUG, trnR-UCU, trnR-ACG *, trnS-GGA, trnS-UGA, trnS-GCU, trnT-GGU, trnT-UGU, trnV-UAC, trnV-GAC *, trnW-CCA, trnY-GUA |
| Ribosomal RNA genes | rRNA4.5S *, rRNA5S *, rRNA16S *, rRNA23S * | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7 *, rps8, rps11, rps12 *, rps14, rps15, rps16, rps18, rps19 | |
| Large subunit of ribosome | rpl2 *, rpl14, rpl16, rpl20, rpl22, rpl23 *, rpl32, rpl33, rpl36 | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis | Subunits of NADH-dehydrogenase | ndhA, ndhB *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Rubisco | rbcL | |
| cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Other genes | Envelop membrane protein | cemA |
| Acetyl-CoA-carboxylase c-type cytochrom synthesis gene | ccsA | |
| Subunit of Acetyl-CoA-carboxylase | accD | |
| Maturase | matK | |
| Protease | clpP | |
| Genes of unknown function | Conserved reading frames | ycf2 *, ycf3, ycf4, ycf68 * |
| Transition (S) | Transversion (V) | Other Type | Total | S/V | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A-G | T-C | Total | A-T | A-C | G-C | G-T | Total | ||||
| Whole genome | 28 | 33 | 61 | 135 | 32 | 4 | 27 | 198 | 6 | 265 | 0.31 |
| Coding region | 6 | 10 | 16 | 3 | 4 | 1 | 4 | 12 | 0 | 28 | 1.33 |
| Non-coding region | 22 | 23 | 45 | 132 | 28 | 3 | 23 | 186 | 6 | 237 | 0.24 |
| Structural Region | Coding Region | Non-Coding Region | ||||
|---|---|---|---|---|---|---|
| LSC | IR | SSC | Total | |||
| Total number of sites | 90,344 | 25,174 | 14,827 | 130,345 | 62,852 | 67,359 |
| Number of polymorphic sites | 160 | 3 | 102 | 265 | 28 | 237 |
| Pi values | 0.00075 | 0.00006 | 0.00273 | 0.00084 | 0.00018 | 0.00146 |
| Theta-W | 0.00078 | 0.00005 | 0.00301 | 0.00089 | 0.0002 | 0.00154 |
| Indels | 228 | 4 | 74 | 306 | 15 | 291 |
| Genes | lnL | df | p-Value | Positively Selected Sites | |||
|---|---|---|---|---|---|---|---|
| Null Model | Alternative Model | ||||||
| accD | −2070.689459 | −2053.870675 | 2 | 0.00000005 | 42 | R | 0.975 |
| 73 | F | 0.918 | |||||
| 75 | V | 0.959 | |||||
| 76 | S | 0.986 | |||||
| 77 | S | 0.975 | |||||
| 79 | M | 0.975 | |||||
| rbcL | −2008.916522 | −2002.544689 | 2 | 0.001709024 | 233 | F | 0.966 |
| 258 | M | 0.967 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, W.; Yu, W.; Kong, Y.; Ren, X.; Zhu, L.; Li, Q.; Yang, Y.; Cheng, Y.; Wang, H. Intraspecific Chloroplast Genome Genetic Polymorphism of Pinellia ternata (Xi Junecry) and Its Revelation of a Single Origin in Phylogeny. Genes 2024, 15, 1638. https://doi.org/10.3390/genes15121638
Xing W, Yu W, Kong Y, Ren X, Zhu L, Li Q, Yang Y, Cheng Y, Wang H. Intraspecific Chloroplast Genome Genetic Polymorphism of Pinellia ternata (Xi Junecry) and Its Revelation of a Single Origin in Phylogeny. Genes. 2024; 15(12):1638. https://doi.org/10.3390/genes15121638
Chicago/Turabian StyleXing, Wenlong, Weihan Yu, Yuanyuan Kong, Xian Ren, Liuying Zhu, Qingyang Li, Yujie Yang, Yueqin Cheng, and Hongwei Wang. 2024. "Intraspecific Chloroplast Genome Genetic Polymorphism of Pinellia ternata (Xi Junecry) and Its Revelation of a Single Origin in Phylogeny" Genes 15, no. 12: 1638. https://doi.org/10.3390/genes15121638
APA StyleXing, W., Yu, W., Kong, Y., Ren, X., Zhu, L., Li, Q., Yang, Y., Cheng, Y., & Wang, H. (2024). Intraspecific Chloroplast Genome Genetic Polymorphism of Pinellia ternata (Xi Junecry) and Its Revelation of a Single Origin in Phylogeny. Genes, 15(12), 1638. https://doi.org/10.3390/genes15121638






