Endurance Effort Affected Expression of Actinin 3 and Klotho Different Isoforms Basing on the Arabian Horses Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Gene Selection
2.2. RNA Isolation
2.3. Real-Time PCR Gene Expression Measurements
2.4. Data Analysis
3. Results
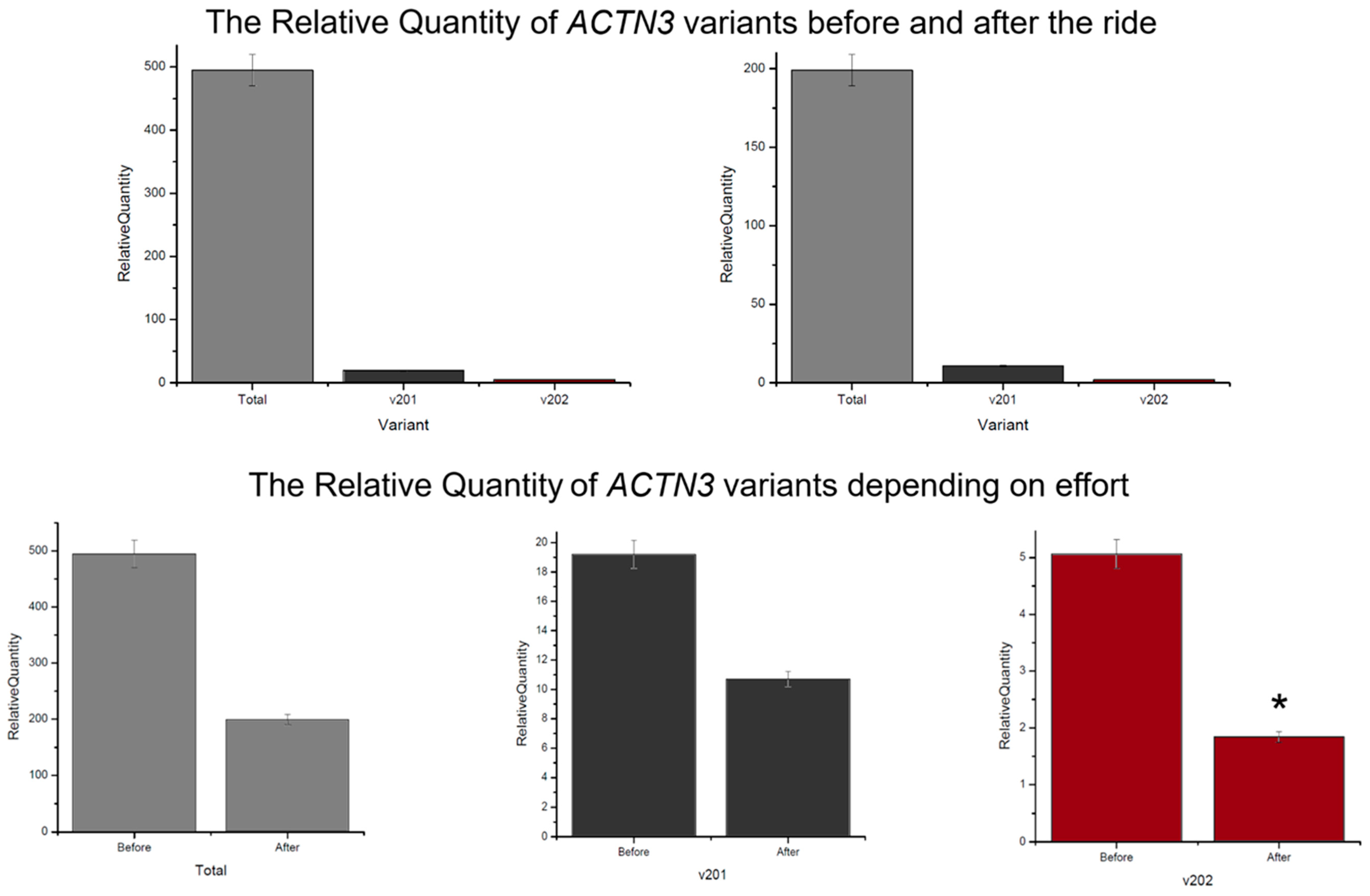
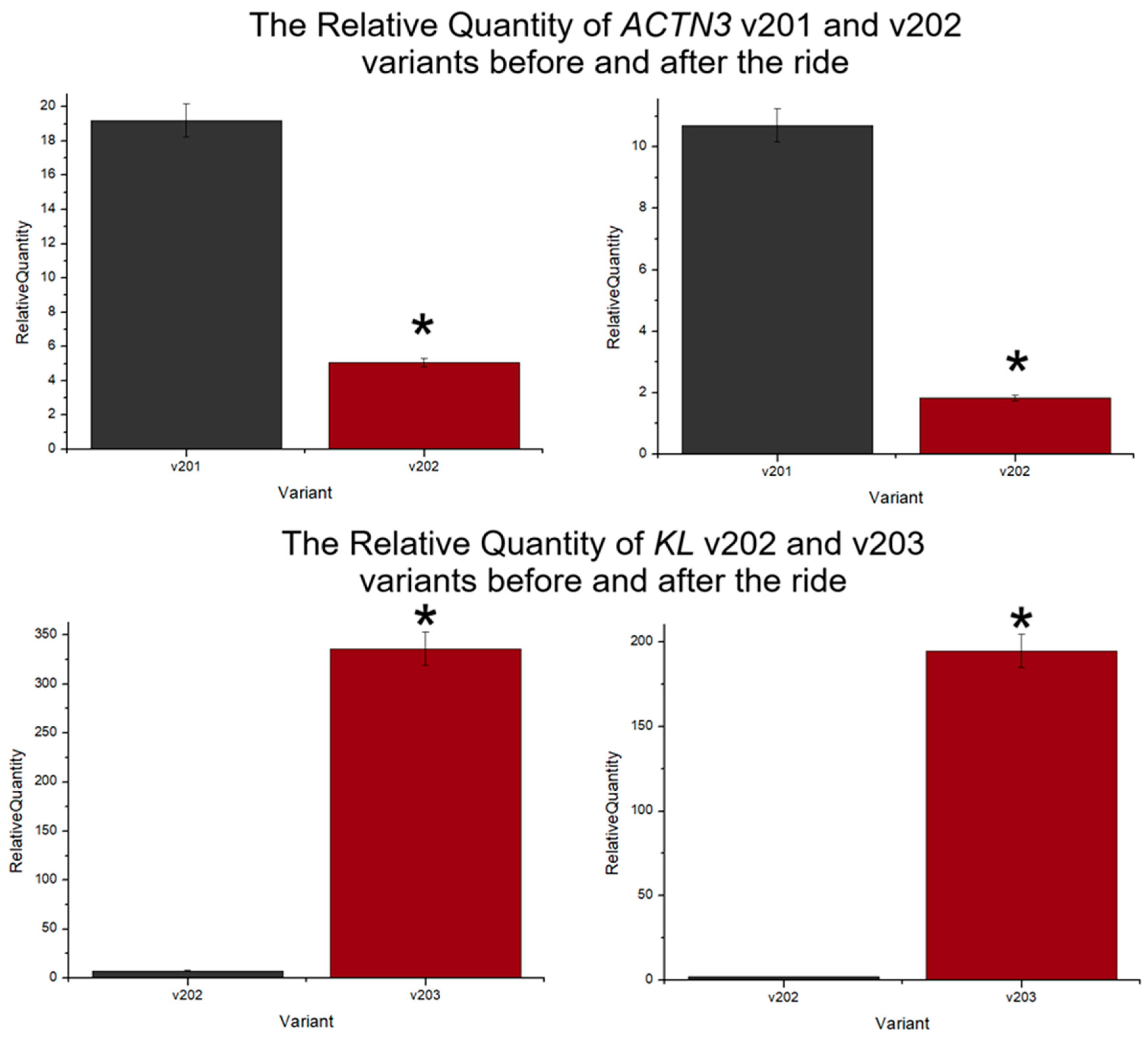
3.1. Differences in ACTN3 Transcript Expression Level in the Context of Endurance Ride
3.2. Differences in KL Transcript Expression Level in the Context of Endurance Ride
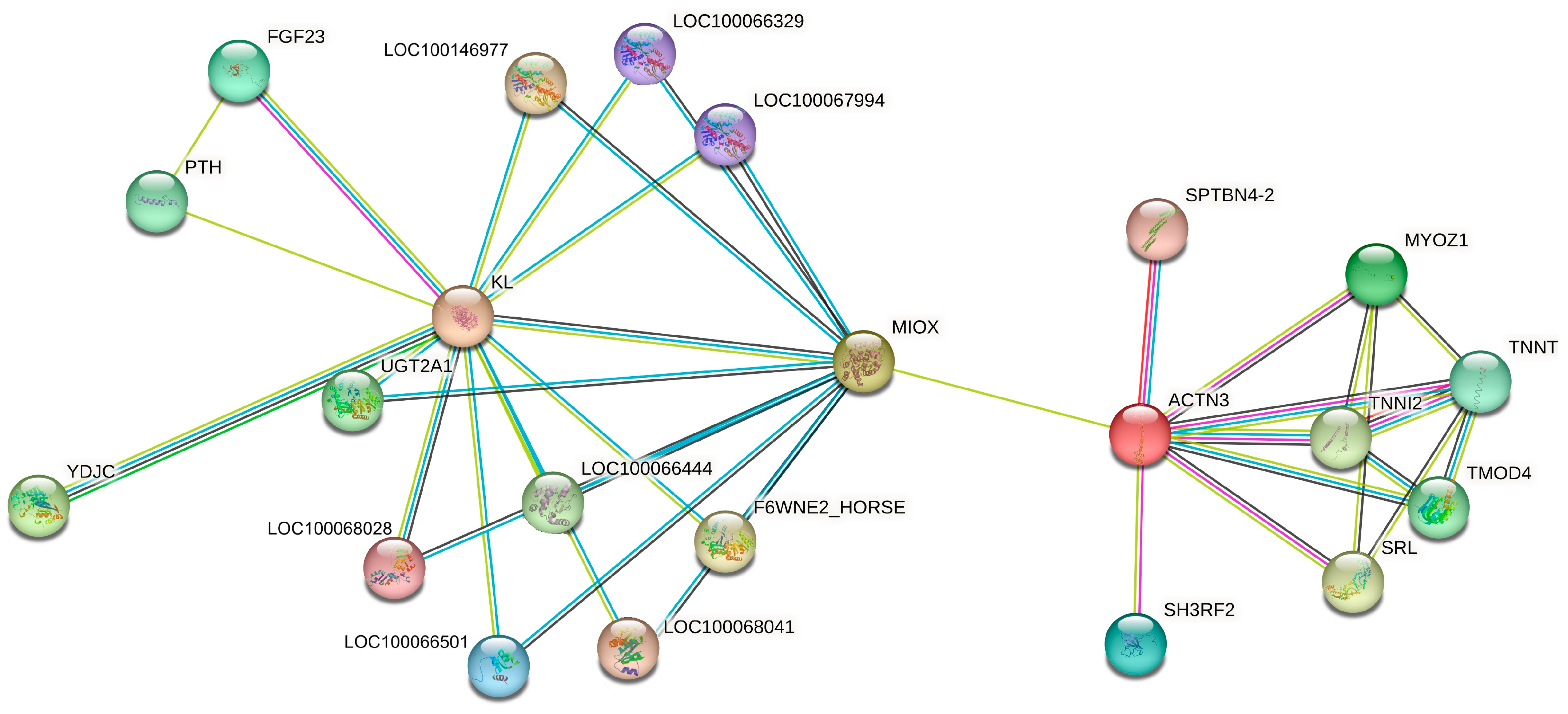
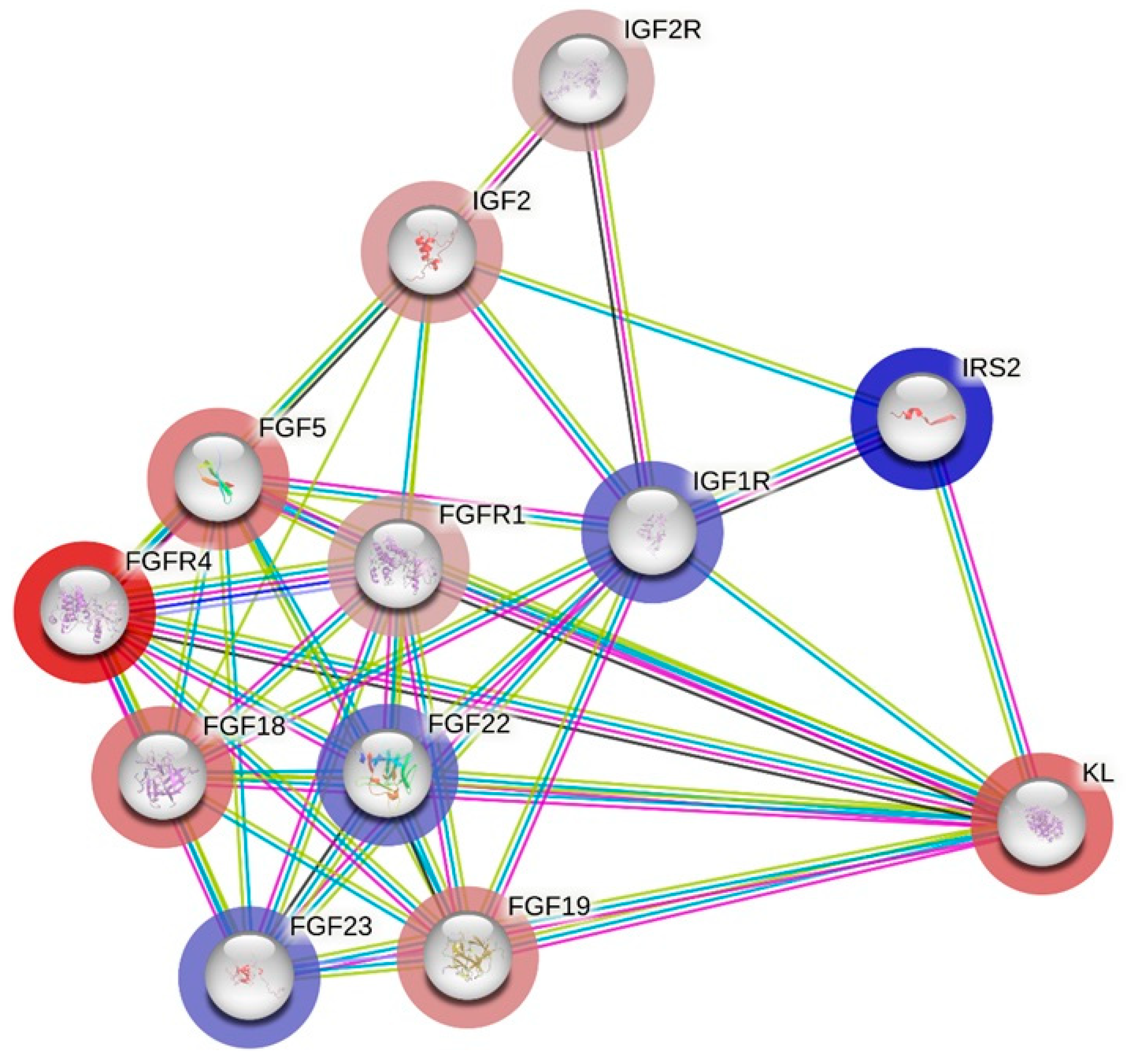
3.3. Molecular Interaction Network Between ACTN3, KL and Most Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, P.; Ahi, E.P. The importance of alternative splicing in adaptive evolution. Mol. Ecol. 2022, 31, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Innocenzi, E.; Cariati, I.; De Domenico, E.; Tiberi, E.; D’Arcangelo, G.; Verdile, V.; Paronetto, M.P.; Tancredi, V.; Barchi, M.; Rossi, P.; et al. Aerobic Exercise Induces Alternative Splicing of Neurexins in Frontal Cortex. J. Funct. Morphol. Kinesiol. 2021, 6, 48. [Google Scholar] [CrossRef]
- Nakka, K.; Ghigna, C.; Gabellini, D. Diversification of the muscle proteome through alternative splicing. Skelet. Muscle 2018, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Z. Molecular basis of Klotho: From gene to function in aging. Endocr. Rev. 2015, 36, 174–193. [Google Scholar] [CrossRef] [PubMed]
- Phelps, M.; Pettan-Brewer, C.; Ladiges, W.; Yablonka-Reuveni, Z. Decline in muscle strength and running endurance in Klotho deficient C57BL/6 mice. Biogerontology 2013, 14, 729–739. [Google Scholar] [CrossRef]
- Dittmer, K.E.; Heathcott, R.W.; Marshall, J.C.; Azarpeykan, S. Expression of Phosphatonin-Related Genes in Sheep, Dog and Horse Kidneys Using Quantitative Reverse Transcriptase PCR. Animals 2020, 10, 1806. [Google Scholar] [CrossRef]
- Myćka, G.; Ropka-Molik, K.; Cywińska, A.; Szmatoła, T.; Stefaniuk-Szmukier, M. Modifications to the longevity regulating pathway and related network resulting from endurance effort in Arabian horses. Ann. Anim. Sci. 2024, 24, 1161–1170. [Google Scholar] [CrossRef]
- Roig-Soriano, J.; Griñán-Ferré, C.; Espinosa-Parrilla, J.F.; Abraham, C.R.; Bosch, A.; Pallàs, M.; Chillón, M. AAV-mediated expression of secreted and transmembrane αKlotho isoforms rescues relevant aging hallmarks in senescent SAMP8 mice. Aging Cell 2022, 21, e13581. [Google Scholar] [CrossRef]
- Ma, F.; Yang, Y.; Li, X.; Zhou, F.; Gao, C.; Li, M. The Association of Sport Performance with ACE and ACTN3 Genetic Polymorphisms: A Systematic Review and Meta-Analysis. PLoS ONE 2013, 8, e54685. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.; Kiely, J. ACTN3: More than Just a Gene for Speed. Front. Physiol. 2017, 8, 1080. [Google Scholar] [CrossRef]
- Baltazar-Martins, G.; Gutiérrez-Hellín, J.; Aguilar-Navarro, M.; Ruiz-Moreno, C.; Moreno-Pérez, V.; López-Samanes, Á.; Domínguez, R.; Del Coso, J. Effect of ACTN3 Genotype on Sports Performance, Exercise-Induced Muscle Damage, and Injury Epidemiology. Sports 2020, 8, 99. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Zukowski, K.; Piórkowska, K.; Bugno-Poniewierska, M. Exercise-induced modification of the skeletal muscle transcriptome in Arabian horses. Physiol. Genom. 2017, 49, 318–326. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Musiał, A.D.; Piórkowska, K.; Szmatoła, T. Sequence analysis and expression profiling of the equine ACTN3 gene during exercise in Arabian horses. Gene 2019, 685, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Knych, H.K.; Harrison, L.M.; Steinmetz, S.J. Differential expression of skeletal muscle genes following administration of clenbuterol to exercised horses. BMC Genom. 2016, 17, 596. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Piórkowska, K. Molecular characterization of the apoptosis-related SH3RF1 and SH3RF2 genes and their association with exercise performance in Arabian horses. BMC Vet. Res. 2018, 14, 237. [Google Scholar] [CrossRef]
- Sheng, J.; Jin, J.P. TNNI1, TNNI2 and TNNI3: Evolution, regulation, and protein structure–function relationships. Gene 2016, 576 Pt 3, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Norman, B.; Esbjörnsson, M.; Rundqvist, H.; Österlund, T.; Glenmark, B.; Jansson, E. ACTN3 genotype and modulation of skeletal muscle response to exercise in human subjects. J. Appl. Physiol. 2014, 116, 1197–1203. [Google Scholar] [CrossRef]
- Almarzook, S.; Said Ahmed, A.; Reissmann, M.; Brockmann, G. The Genetic variation of ACTN3 and MSTN genes in a cohort of endurance Arabian horses. Glob. J. Anim. Sci. Res. 2019, 7, 10–12. [Google Scholar]
- Turki, W.Z.; Al-Anbari, N.N. Genetic Diversity in Original Arabian-WAHO, Local and Thoroughbred Horses in Iraq from ACTN3-EXON19 Gene Polymorphism. IOP Conf. Ser. Earth Environ. Sci. 2023, 1214, 012033. [Google Scholar] [CrossRef]
- Yang, B.; Hodgkinson, A.; Millward, B.A.; Demaine, A.G. Polymorphisms of myo-inositol oxygenase gene are associated with Type 1 diabetes mellitus. J. Diabetes Its Complicat. 2010, 24, 404–408. [Google Scholar] [CrossRef]
- Kuro-o, M.; Matsumura, Y.; Aizawa, H. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Z. Current understanding of klotho. Ageing Res. Rev. 2009, 8, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hwang, K.H.; Park, K.S.; Kong, I.D.; Cha, S.K. Biological Role of Anti-aging Protein Klotho. J. Lifestyle Med. 2015, 5, 1–6. [Google Scholar] [CrossRef]
- Ochi, E.; Barrington, A.; Wehling-Henricks, M.; Avila, M.; Kuro-o, M.; Tidball, J.G. Klotho regulates the myogenic response of muscle to mechanical loading and exercise. Exp. Physiol. 2023, 108, 1531–1547. [Google Scholar] [CrossRef]
- Soykan, M.N.; Gunes, S. Overexpression of Klotho gene using CRISPR/Cas9 induces apoptosis and inhibits cell motility in the human colorectal cancer cells. Biotechnol. J. 2024, 19, e2300496. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Gene Name | References (Ensembl Database) | Primers Sequence | Amplicon Length |
|---|---|---|---|---|
| ACTN3 | Actinin alpha 3, total variant | ENSECAG00000018961 | F: CAGGCCTTCATCGACTTCAT R: GGAGAAGGCCACATAATCCA | 243 |
| ACTN3-201 | Actinin alpha 3 v 201 | ENSECAT00000021149.1 | F: TTTCAGAGATCGTCGACGGG R: AGATGTCCTGGATGGCAAAG | 78 |
| ACTN3-202 | Actinin alpha 3 v 202 | ENSECAT00000021249.1 | F: TTGCTCCTGGAGGTCATTTC R: CCGATGGACACCAGCTTAAC | 135 |
| KL | Klotho total variant | ENSECAG00000008185 | F: ACCAAGAGAGATGACGCCAA R: GGCTCAGGAAGTCGACATAGA | 306 |
| KL-202 | Klotho v 202 | ENSECAT00000008327.3 | F: CTCACATGAAGTTCCGCCAA R: GGCTCAGGAAGTCGACATAGA | 184 |
| KL-203 | Klotho v 203 | ENSECAT00000108964.1 | F: CCAAGACAGGCTGAGAGTGT R: TCATGGAAGGTTTGGGCTCA | 179 |
| B2M | Beta-2-Microglobulin | ENSECAG00000000685 | F: TGTCTTTCAGCAAGGACTGG R: CAAGCCTTCATGATGCTGGT | 159 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Myćka, G.; Ropka-Molik, K.; Cywińska, A.; Stefaniuk-Szmukier, M. Endurance Effort Affected Expression of Actinin 3 and Klotho Different Isoforms Basing on the Arabian Horses Model. Genes 2024, 15, 1618. https://doi.org/10.3390/genes15121618
Myćka G, Ropka-Molik K, Cywińska A, Stefaniuk-Szmukier M. Endurance Effort Affected Expression of Actinin 3 and Klotho Different Isoforms Basing on the Arabian Horses Model. Genes. 2024; 15(12):1618. https://doi.org/10.3390/genes15121618
Chicago/Turabian StyleMyćka, Grzegorz, Katarzyna Ropka-Molik, Anna Cywińska, and Monika Stefaniuk-Szmukier. 2024. "Endurance Effort Affected Expression of Actinin 3 and Klotho Different Isoforms Basing on the Arabian Horses Model" Genes 15, no. 12: 1618. https://doi.org/10.3390/genes15121618
APA StyleMyćka, G., Ropka-Molik, K., Cywińska, A., & Stefaniuk-Szmukier, M. (2024). Endurance Effort Affected Expression of Actinin 3 and Klotho Different Isoforms Basing on the Arabian Horses Model. Genes, 15(12), 1618. https://doi.org/10.3390/genes15121618






