Genome-Wide Identification and Expression Analysis of PaNRT Gene Family Under Various Nitrogen Conditions in Avocado (Persea americana Mill.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Characterization of NRT Genes in Avocado
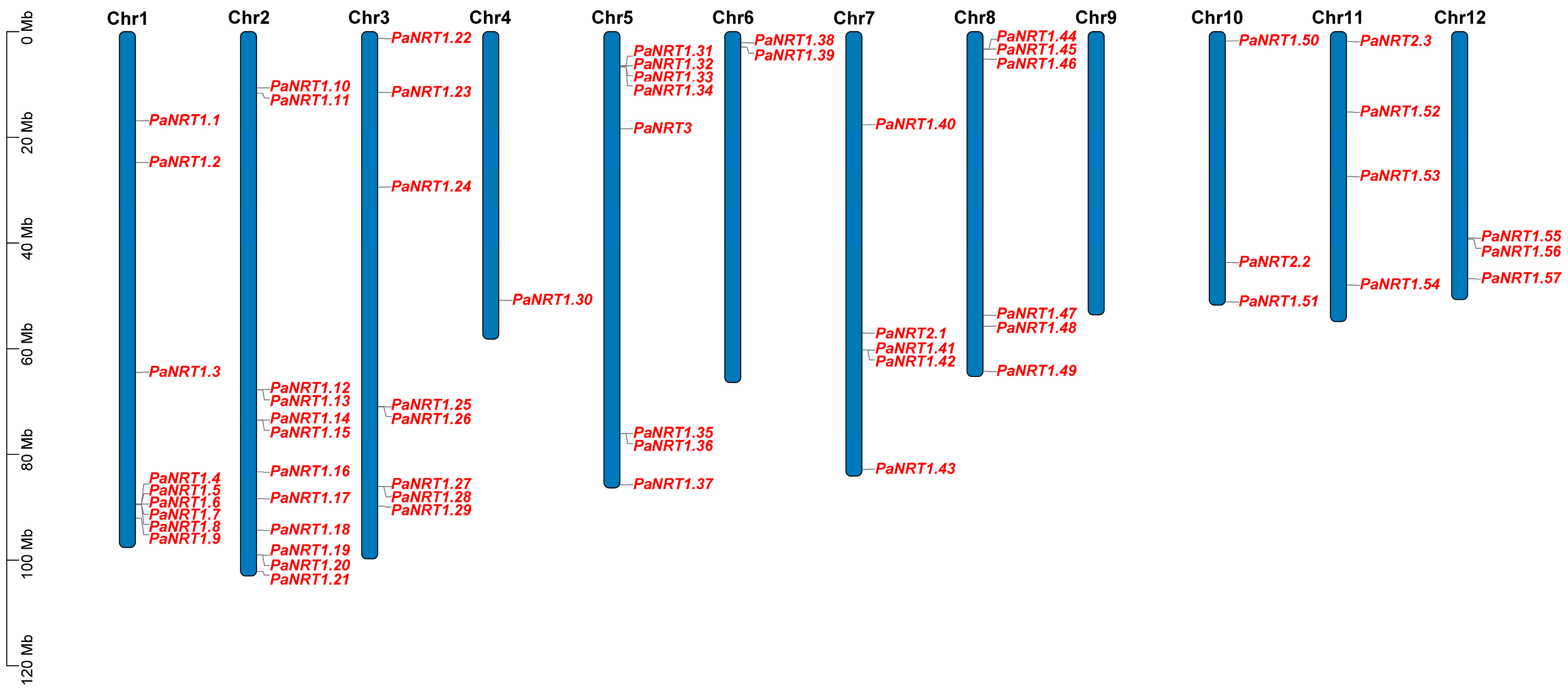
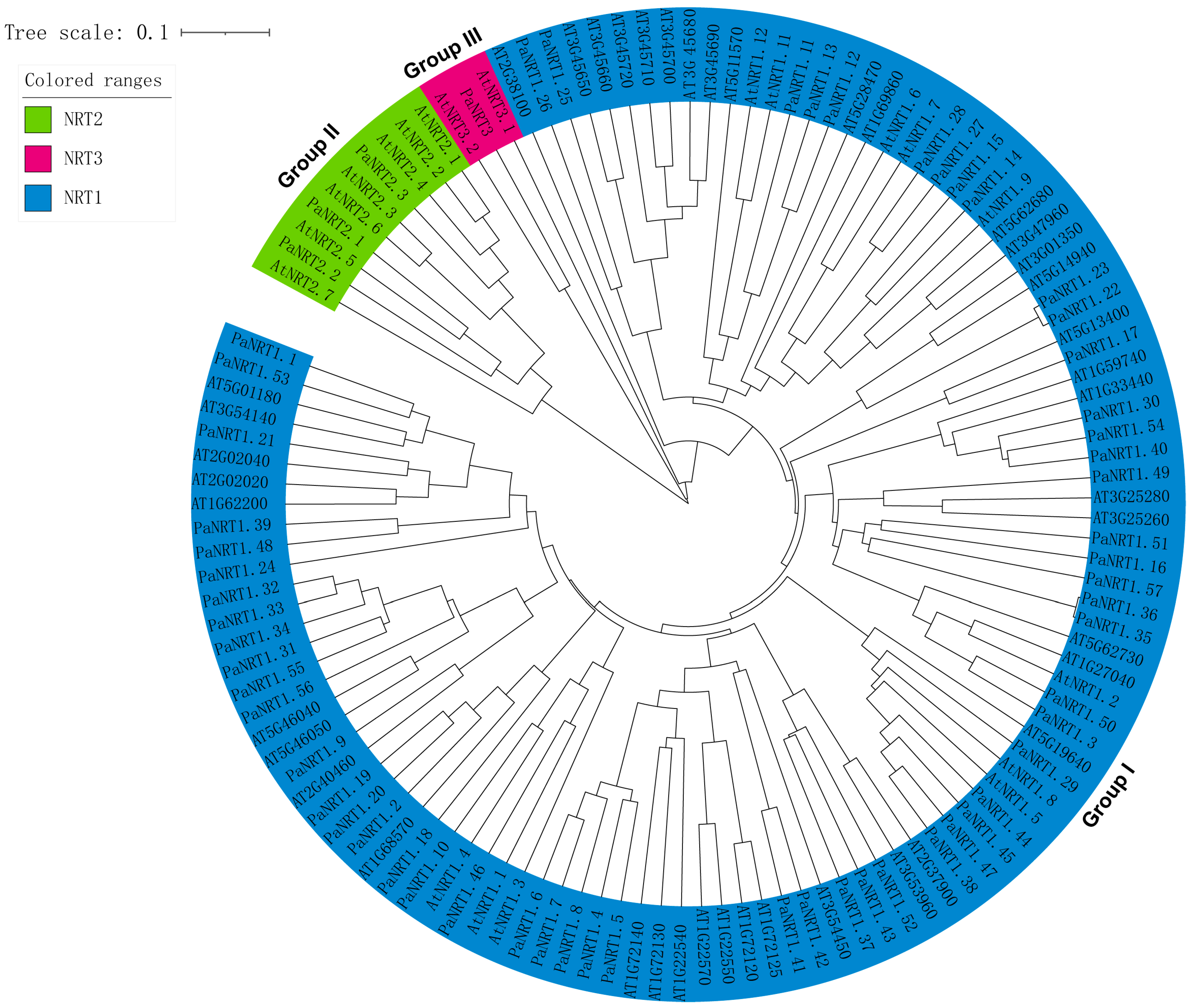
2.3. Chromosomal Localization and Phylogenetic Analysis of PaNRT Proteins
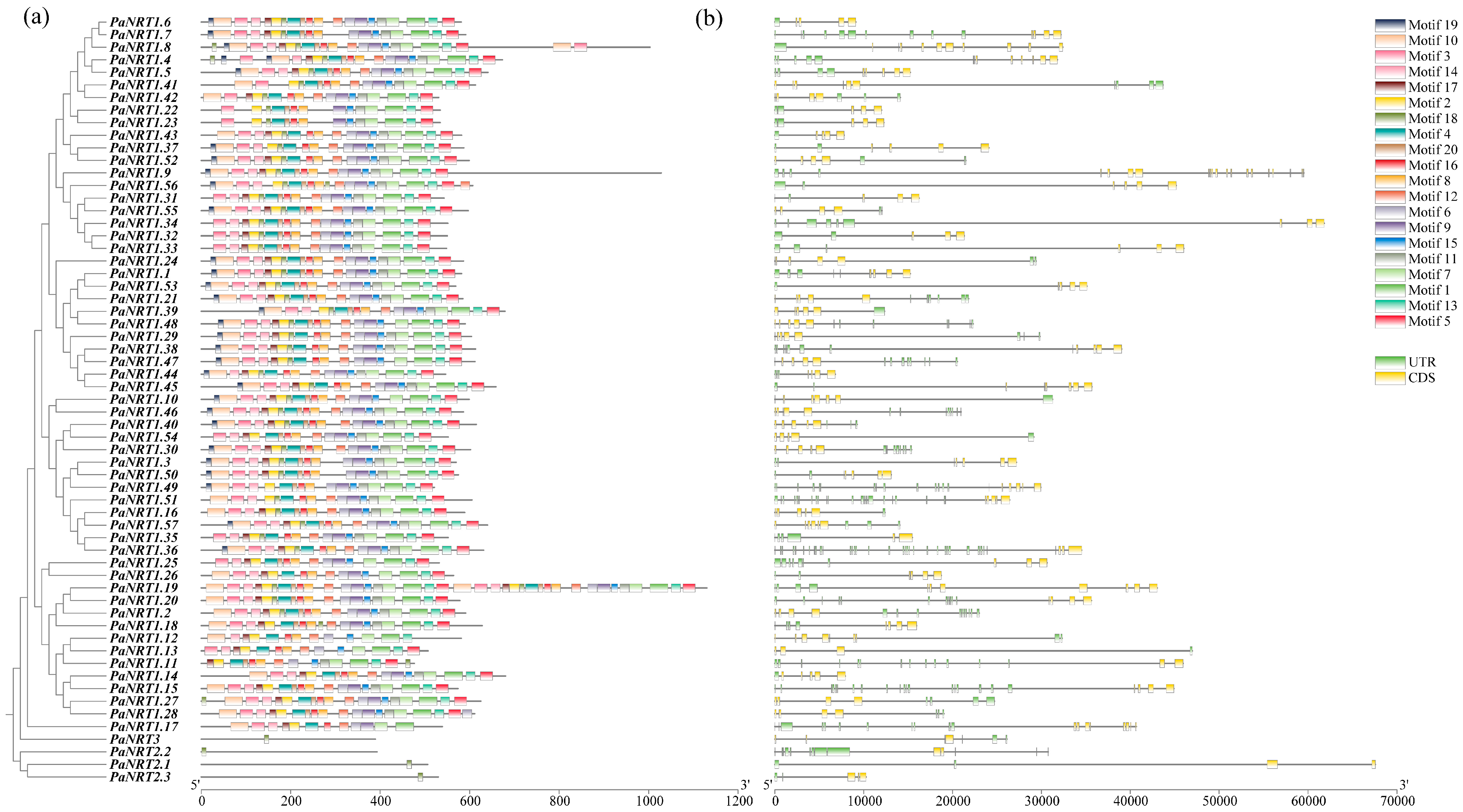
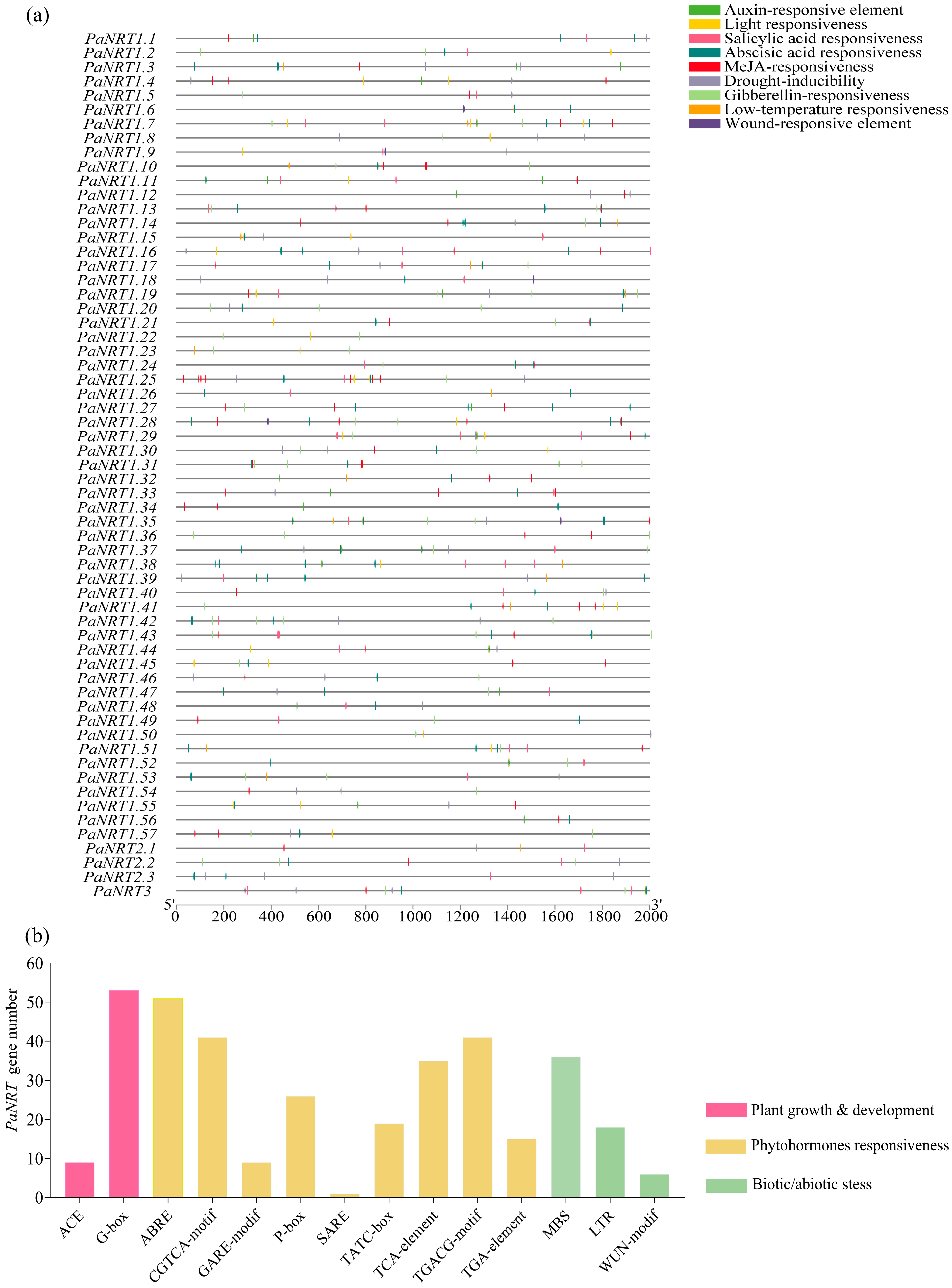
2.4. Gene Structure, Conserved Motif, and Cis-Regulatory Element (CRE) Analysis of PaNRT Proteins
2.5. Measurement of the Physiological Indexes
2.6. RNA Sequencing and Gene Expression Analysis
2.7. Statistical Analysis
3. Results
3.1. Characterization of the PaNRT Gene Family
3.2. Phylogenetic Relationship and Conserved Motifs of PaNRTs
3.3. Characterization of CREs in the Promoter Regions of PaNRTs
3.4. Physiological Indexes in Avocado Leaves Under Different N Supply
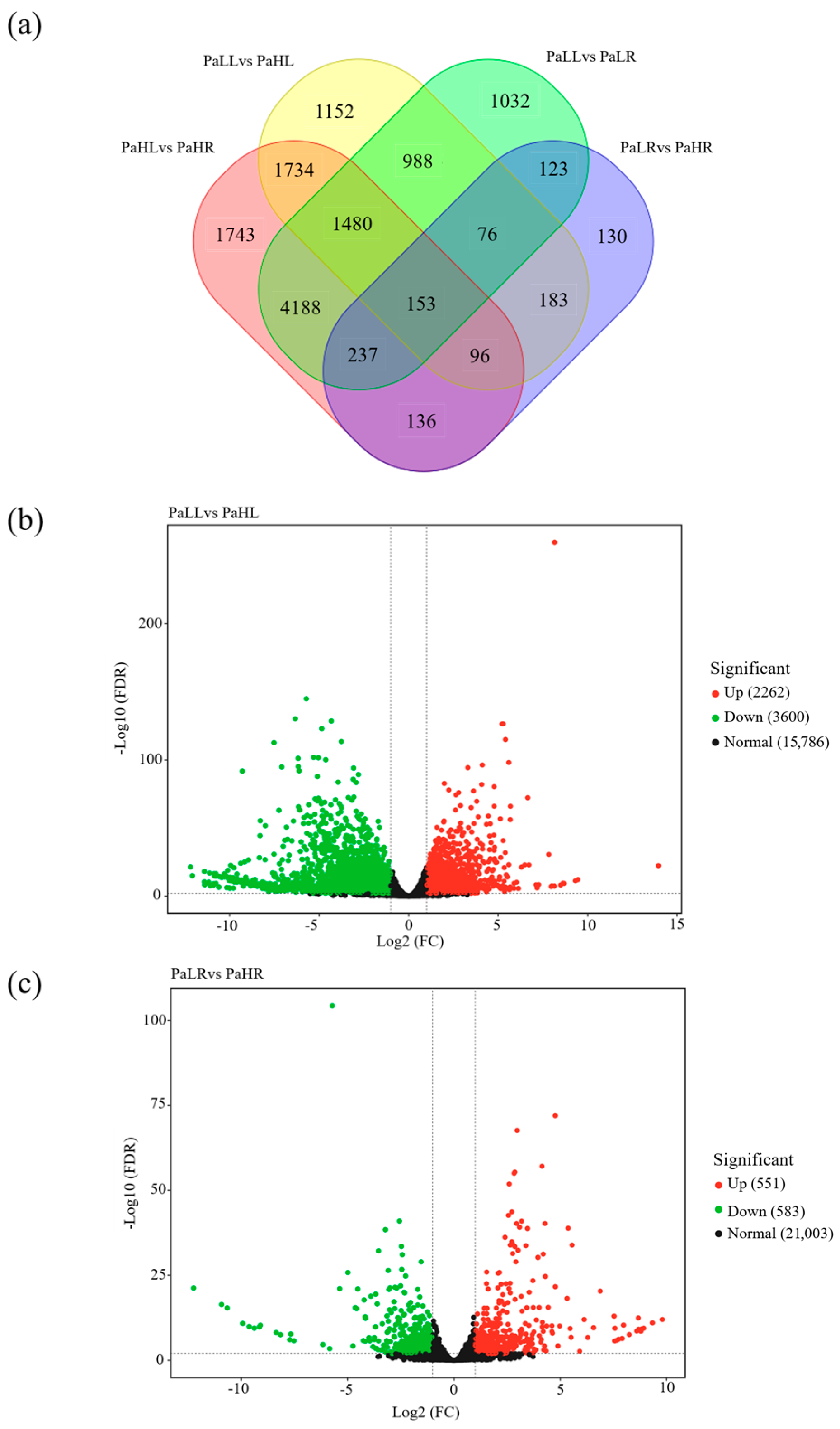
3.5. RNA-Seq Analysis in Avocado Under Different N Supply
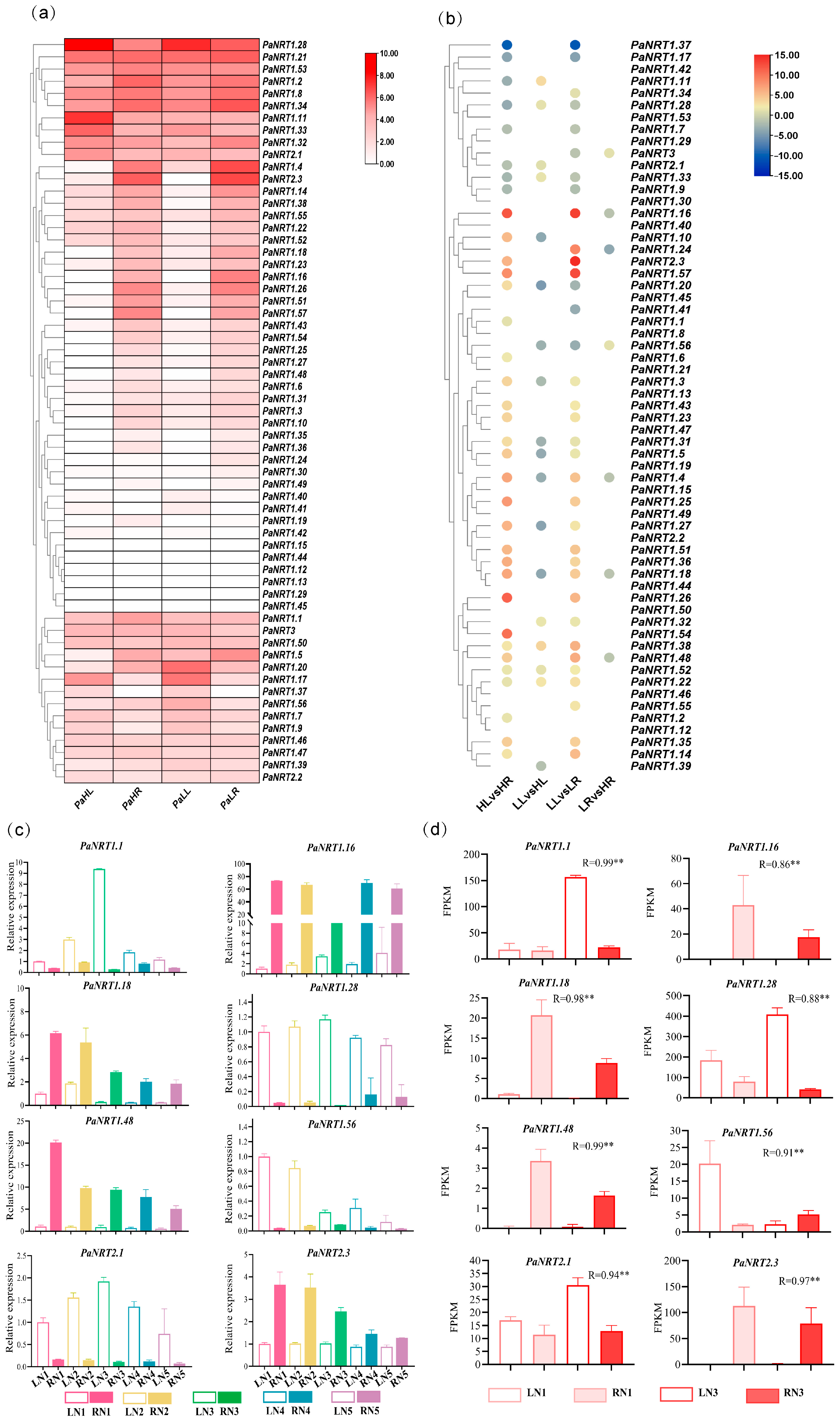
3.6. Expression Profiles of PaNRT Genes Under Different N Supply
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qin, J.; Yue, X.L.; Shang, X.L.; Fang, S.Z. Response of nitrogen use efficiency, N-assimilating enzymes and growth in Cyclocarya paliurus seedlings to different nitrogen nutrition. J. Plant Nutr. 2023, 46, 3547–3556. [Google Scholar] [CrossRef]
- Ding, M.C.; He, M.; Zhang, W.L.; Han, Y.; Zhang, X.Y.; Zhang, X.L.; Zhu, Y.L.; Wang, Y.; Liu, L.W.; Xu, L. Genome-wide identification and expression analysis of RsNRT gene family reveals their potential roles in response to low-nitrogen condition in radish (Raphanus sativus L.). Sci. Hortic. 2023, 321, 112273. [Google Scholar] [CrossRef]
- Swarbreck, S.M.; Wang, M.; Wang, Y.; Kindred, D.; Sylvester-Bradley, R.; Shi, W.M.; Varinderpal-Singh; Bentley, A.R.; Griffiths, H. A roadmap for lowering crop nitrogen requirement. Trends Plant Sci. 2019, 24, 892–904. [Google Scholar] [CrossRef]
- Stevens, C.J. Nitrogen in the environment. Science 2019, 363, 578–580. [Google Scholar] [CrossRef] [PubMed]
- Gaudinier, A.; Rodriguez-Medina, J.; Zhang, L.; Olson, A.; LiseronMonfils, C.; Bagman, A.M.; Foret, J.; Abbitt, S.; Tang, M.; Li, B.; et al. Transcriptional regulation of nitrogen associated metabolism and growth. Nature 2018, 563, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Gojon, A.; Krouk, G.; Perrine, W.F.; Laugier, E. Nitrate transceptor(s) in plants. J. Exp. Bot. 2011, 62, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.X.; Ling, F.L.; Jia, D.S.; Cui, J.J.; Zhang, Z.A.; Xu, C.; Yu, L.T.; Guan, C.L.; Wang, Y.; Zhang, M.R.; et al. Effects of low nitrogen on seedling growth, photosynthetic characteristics and antioxidant system of rice varieties with different nitrogen efficiencies. Sci. Rep. 2023, 13, 19780. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Yan, M.K.; Hu, B.Y.; Priyadarshani, S.V.G.N.; Hou, Z.M.; Ojolo, P.S.; Xiong, J.J.; Zhao, H.M.; Qin, Y. Characterization and the expression analysis of nitrate transporter (NRT) gene family in pineapple. Trop. Plant Biol. 2018, 11, 177–191. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, P.F.; Liu, P.; Song, Y.P.; Zhang, D.Q. Genetic effects and expression patterns of the nitrate transporter (NRT) gene family in Populus tomentosa. Front. Plant Sci. 2021, 12, 661635. [Google Scholar] [CrossRef] [PubMed]
- Bellegarde, F.; Gojon, A.; Martin, A. Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2553–2565. [Google Scholar] [PubMed]
- Wang, Y.; Cheng, Y.; Chen, K.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [PubMed]
- You, L.L.; Wang, Y.; Zhang, T.T.; Zhu, Y.F.; Ren, N.; Jiang, X.Y.; Zhou, Y. Genome-wide identification of nitrate transporter 2 (NRT2) gene family and functional analysis of MeNRT2.2 in cassava (Manihot esculenta Crantz). Gene 2022, 809, 146038. [Google Scholar] [CrossRef]
- Okamoto, M.; Kumar, A.; Li, W.; Wang, Y.; Siddiqi, M.Y.; Crawford, N.M.; Glass, A.D.M. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006, 140, 1036–1046. [Google Scholar] [PubMed]
- Kotur, Z.; Mackenzie, N.; Ramesh, S.; Tyerman, S.D.; Kaiser, S.N.; Glass, A.D. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012, 194, 724–731. [Google Scholar] [PubMed]
- Gallart, M.; Paungfoo-Lonhienne, C.; Trueman, S.J. Effects of a growth-promoting Paraburkholderia species on nitrogen acquisition by avocado seedlings. Sci. Hortic. 2022, 295, 110767. [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAOSTAT). Cultivos. 2024. Available online: http://www.fao.org/faostat/es/#data/QC (accessed on 5 June 2024).
- Selladurai, R.; Awachare, C.M. Nutrient management for avocado (Persea americana miller). J. Plant Nutr. 2020, 43, 138–147. [Google Scholar] [CrossRef]
- Lovatt, C.J. Properly timed soil applied nitrogen fertilizer increases yield and fruit size of ‘Hass’ avocado. J. Am. Soc. Hortic. Sci. 2001, 126, 555–559. [Google Scholar]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Masouleh, A.K.; Furtado, A.; Henry, R.J.; Mitter, N. A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Hortic. Res. 2022, 9, uhac157. [Google Scholar] [CrossRef]
- Li, G.Y.; Yang, D.M.; Hu, Y.; Xu, J.M.; Lu, Z.H. Genome-wide identification and expression analysis of nitrate transporter (NRT) gene family in Eucalyptus grandis. Genes 2024, 15, 930. [Google Scholar] [CrossRef] [PubMed]
- Landrein, B.; Formosa-Jordan, P.; Malivert, A.; Schuster, C.; Melnyk, W.C.; Yang, W.B.; Turnbull, C.; Meyerowitz, M.E.; Locke, C.W.J.; Jönsson, H. Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. USA 2018, 115, 201718670. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Chen, X.; Tang, X.H.; Shao, X.H.; Lai, D.; Xiao, W.Q.; Zhuang, Q.L.; Wang, W.L.; Dong, T. Near-freezing temperature suppresses avocado (Persea americana Mill.) fruit softening and chilling injury by maintaining cell wall and reactive oxygen species metabolism during storage. Plant Physiol. Biochem. 2024, 210, 108621. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinform. 2017, 60, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Rangel, A.; Li, R.; Mauk, P.; Santiago, L.; Lovatt, C.J. Effects of temperature, soil moisture and light intensity on the temporal pattern of floral gene expression and flowering of avocado buds (Persea americana cv. Hass). Sci. Hortic. 2021, 280, 109940. [Google Scholar] [CrossRef]
- Tahir, M.M.; Wang, H.; Ahmad, B.; Liu, Y.; Fan, S.; Li, K.; Lei, C.; Shah, K.; Li, S.H.; Zhang, D. Identification and characterization of NRT gene family reveals their critical response to nitrate regulation during adventitious root formation and development in apple rootstock. Sci. Hortic. 2021, 275, 109642. [Google Scholar] [CrossRef]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The nitrate transporter (NRT) gene family in poplar. PLoS ONE 2013, 8, e72126. [Google Scholar] [CrossRef]
- Li, G.Y.; Yang, D.M.; Hu, Y.; Xu, J.M.; Li, J.; Lu, Z.H. Genome-wide identification, expression analysis, and transcriptome analysis of the NPF gene family under various nitrogen conditions in Eucalyptus grandis. Forests 2024, 15, 1697. [Google Scholar] [CrossRef]
- Kiba, T.; Kudo, T.; Kojima, M.; Sakakibara, H. Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 2011, 62, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Yue, X.L.; Fang, S.Z.; Qian, M.Y.; Zhou, S.T.; Shang, X.L.; Yang, W.X. Responses of nitrogen metabolism, photosynthetic parameter and growth to nitrogen fertilization in Cyclocarya paliurus. For. Ecol. Manag. 2021, 502, 119715. [Google Scholar] [CrossRef]
- Lezhneva, L.; Kiba, T.; Feriabourrelier, A.B.; Lafouge, F.; Boutet-Mercey, S.; Zoufan, P.; Sakakibara, H.; Daniel-Vedele, F.; Krapp, A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2015, 80, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cui, Y.N.; Yu, M.; Su, B.D.; Gong, W.; Baluška, F.; Komis, G.; Šamaj, J.; Shan, X.Y.; Li, J.X. Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol. 2019, 181, 480–498. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Warzybok, A.; Kłobus, G. The genomic organization and transcriptional pattern of genes encoding nitrate transporters 1 (NRT1) in cucumber. Plant Soil 2012, 364, 254–260. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
| Nutrient Elements | N1 (mg/L) | N2 (mg/L) | N3 (mg/L) | N4 (mg/L) | N5 (mg/L) |
|---|---|---|---|---|---|
| N | 29.75 | 59.50 | 119.00 | 178.50 | 238.00 |
| P | 15.50 | 15.50 | 15.50 | 15.50 | 15.50 |
| K | 298.00 | 298.00 | 298.00 | 298.00 | 298.00 |
| Ca | 210.00 | 210.00 | 210.00 | 210.00 | 210.00 |
| Mg | 48.10 | 48.10 | 48.10 | 48.10 | 48.10 |
| B | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| Mn | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| Zn | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| Cu | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| Mo | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 |
| Fe | 5.60 | 5.60 | 5.60 | 5.60 | 5.60 |
| Cl | 553.45 | 519.72 | 395.47 | 271.22 | 146.74 |
| Na | 4.60 | 4.60 | 4.60 | 4.60 | 4.60 |
| Gene Name | Gene ID | Chromosome | Amino Acid Number (aa) | Isoelectric Point (pI) | Molecular Weight (Mw, kDa) | Instability Index (II) | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|
| PaNRT1.1 | KAJ8648131.1 | Chr1 | 582 | 8.55 | 65.66 | 28.62 | 0.071 |
| PaNRT1.2 | KAJ8648506.1 | Chr1 | 591 | 6.90 | 65.80 | 34.01 | 0.163 |
| PaNRT1.3 | KAJ8649386.1 | Chr1 | 570 | 8.26 | 62.41 | 47.64 | 0.451 |
| PaNRT1.4 | KAJ8650603.1 | Chr1 | 673 | 8.84 | 73.85 | 42.48 | 0.237 |
| PaNRT1.5 | KAJ8650604.1 | Chr1 | 641 | 8.99 | 70.60 | 42.03 | 0.262 |
| PaNRT1.6 | KAJ8650605.1 | Chr1 | 581 | 6.82 | 64.02 | 41.71 | 0.386 |
| PaNRT1.7 | KAJ8650614.1 | Chr1 | 591 | 6.66 | 65.06 | 41.51 | 0.351 |
| PaNRT1.8 | KAJ8650615.1 | Chr1 | 1003 | 8.83 | 109.84 | 39.17 | 0.059 |
| PaNRT1.9 | KAJ8650804.1 | Chr1 | 1028 | 8.82 | 115.40 | 38.84 | 0.065 |
| PaNRT1.10 | KAJ8643124.1 | Chr2 | 599 | 8.75 | 65.72 | 32.66 | 0.403 |
| PaNRT1.11 | KAJ8643206.1 | Chr2 | 476 | 8.98 | 52.64 | 35.40 | 0.190 |
| PaNRT1.12 | KAJ8644789.1 | Chr2 | 581 | 9.02 | 63.81 | 53.51 | 0.289 |
| PaNRT1.13 | KAJ8644791.1 | Chr2 | 507 | 8.76 | 55.89 | 40.64 | 0.363 |
| PaNRT1.14 | KAJ8645062.1 | Chr2 | 680 | 8.82 | 75.05 | 36.46 | 0.185 |
| PaNRT1.15 | KAJ8645064.1 | Chr2 | 574 | 8.86 | 63.43 | 33.50 | 0.369 |
| PaNRT1.16 | KAJ8645606.1 | Chr2 | 589 | 8.56 | 64.59 | 33.15 | 0.351 |
| PaNRT1.17 | KAJ8645953.1 | Chr2 | 539 | 7.14 | 59.38 | 35.94 | 0.178 |
| PaNRT1.18 | KAJ8646339.1 | Chr2 | 628 | 8.58 | 69.00 | 36.58 | 0.164 |
| PaNRT1.19 | KAJ8646648.1 | Chr2 | 1130 | 8.73 | 125.09 | 37.08 | 0.283 |
| PaNRT1.20 | KAJ8646649.1 | Chr2 | 578 | 8.82 | 63.82 | 36.49 | 0.265 |
| PaNRT1.21 | KAJ8646894.1 | Chr2 | 585 | 5.55 | 64.36 | 27.09 | 0.333 |
| PaNRT1.22 | KAJ8634464.1 | Chr3 | 534 | 8.83 | 59.55 | 38.84 | 0.336 |
| PaNRT1.23 | KAJ8635043.1 | Chr3 | 534 | 8.93 | 59.65 | 41.24 | 0.306 |
| PaNRT1.24 | KAJ8635775.1 | Chr3 | 586 | 8.92 | 64.87 | 25.21 | 0.293 |
| PaNRT1.25 | KAJ8637006.1 | Chr3 | 532 | 9.06 | 57.82 | 34.75 | 0.425 |
| PaNRT1.26 | KAJ8637010.1 | Chr3 | 564 | 9.11 | 61.16 | 37.97 | 0.436 |
| PaNRT1.27 | KAJ8637866.1 | Chr3 | 625 | 8.64 | 69.15 | 40.26 | 0.153 |
| PaNRT1.28 | KAJ8637867.1 | Chr3 | 611 | 9.10 | 67.60 | 40.54 | 0.233 |
| PaNRT1.29 | KAJ8638117.1 | Chr3 | 604 | 5.33 | 66.19 | 23.04 | 0.376 |
| PaNRT1.30 | KAJ8618733.1 | Chr4 | 602 | 9.01 | 66.20 | 44.95 | 0.320 |
| PaNRT1.31 | KAJ8639158.1 | Chr5 | 543 | 9.47 | 60.90 | 33.41 | 0.294 |
| PaNRT1.32 | KAJ8639160.1 | Chr5 | 550 | 9.13 | 61.89 | 36.91 | 0.305 |
| PaNRT1.33 | KAJ8639166.1 | Chr5 | 548 | 9.22 | 61.49 | 36.79 | 0.335 |
| PaNRT1.34 | KAJ8639169.1 | Chr5 | 551 | 9.32 | 61.53 | 30.50 | 0.305 |
| PaNRT3 | KAJ8639622.1 | Chr5 | 389 | 6.48 | 43.81 | 49.26 | -0.398 |
| PaNRT1.35 | KAJ8641744.1 | Chr5 | 552 | 8.98 | 61.36 | 35.84 | 0.292 |
| PaNRT1.36 | KAJ8641745.1 | Chr5 | 631 | 8.90 | 69.79 | 35.37 | 0.178 |
| PaNRT1.37 | KAJ8642414.1 | Chr5 | 587 | 9.21 | 66.10 | 43.17 | 0.224 |
| PaNRT1.38 | KAJ8625967.1 | Chr6 | 613 | 6.76 | 68.27 | 28.61 | 0.165 |
| PaNRT1.39 | KAJ8626045.1 | Chr6 | 679 | 6.69 | 75.60 | 33.93 | 0.199 |
| PaNRT1.40 | KAJ8629498.1 | Chr7 | 615 | 9.05 | 68.12 | 39.34 | 0.276 |
| PaNRT2.1 | KAJ8630468.1 | Chr7 | 506 | 9.05 | 54.10 | 39.77 | 0.381 |
| PaNRT1.41 | KAJ8630599.1 | Chr7 | 613 | 8.30 | 67.57 | 48.46 | 0.335 |
| PaNRT1.42 | KAJ8630600.1 | Chr7 | 531 | 8.90 | 58.82 | 41.00 | 0.460 |
| PaNRT1.43 | KAJ8631817.1 | Chr7 | 582 | 8.50 | 65.23 | 41.23 | 0.161 |
| PaNRT1.44 | KAJ8632061.1 | Chr8 | 546 | 6.57 | 60.94 | 32.89 | 0.139 |
| PaNRT1.45 | KAJ8632063.1 | Chr8 | 659 | 6.79 | 73.01 | 28.70 | 0.213 |
| PaNRT1.46 | KAJ8632194.1 | Chr8 | 586 | 9.04 | 64.78 | 34.61 | 0.288 |
| PaNRT1.47 | KAJ8633780.1 | Chr8 | 612 | 7.82 | 68.15 | 33.15 | 0.143 |
| PaNRT1.48 | KAJ8633886.1 | Chr8 | 590 | 6.37 | 65.42 | 33.31 | 0.301 |
| PaNRT1.49 | KAJ8634373.1 | Chr8 | 521 | 9.11 | 58.11 | 41.06 | 0.362 |
| PaNRT1.50 | KAJ8621555.1 | Chr10 | 575 | 9.25 | 63.17 | 49.25 | 0.410 |
| PaNRT2.2 | KAJ8623074.1 | Chr10 | 393 | 9.87 | 42.20 | 37.24 | 0.585 |
| PaNRT1.51 | KAJ8623589.1 | Chr10 | 605 | 8.54 | 67.25 | 24.56 | 0.285 |
| PaNRT2.3 | KAJ8623672.1 | Chr11 | 530 | 9.01 | 57.24 | 42.03 | 0.335 |
| PaNRT1.52 | KAJ8624254.1 | Chr11 | 599 | 9.04 | 67.00 | 41.35 | 0.208 |
| PaNRT1.53 | KAJ8624412.1 | Chr11 | 569 | 8.69 | 63.58 | 22.86 | 0.272 |
| PaNRT1.54 | KAJ8625368.1 | Chr11 | 552 | 8.89 | 61.29 | 42.34 | 0.326 |
| PaNRT1. 55 | KAJ8616190.1 | Chr11 | 597 | 9.39 | 67.05 | 38.00 | 0.252 |
| PaNRT1. 56 | KAJ8616201.1 | Chr11 | 607 | 8.42 | 68.21 | 27.41 | 0.159 |
| PaNRT1. 57 | KAJ8616588.1 | Chr11 | 640 | 9.12 | 71.34 | 30.74 | 0.236 |
| Treatment | Nitrate Reductase (NR) Activity (µg/(h2·g FW)) | Nitrite Reductases (NiR) Activity (µg/(h2·g FW)) | Glutamine Synthease (GS) Activity (µg/(h2·g FW)) | Glutamate Synthase (GOGAT) Activity (U/mg FW) | Superoxide Dismutase (SOD) Activity (U/g FW) | Peroxidase (POD) Activity (U/g FW) | Catalase (CAT) Activity (U/g FW) | Nitrogen Content (mg/g) |
|---|---|---|---|---|---|---|---|---|
| N1 | 11.47 ± 1.14 c | 48.13 ± 3.73 c | 16.43 ± 1.01 c | 0.35 ± 0.04 c | 34.90 ± 2.65 a | 3.94 ± 0.35 b | 0.39 ± 0.03 a | 6.78 ± 0.68 c |
| N2 | 23.00 ± 1.71 b | 90.97 ± 11.37 b | 30.60 ± 2.44 b | 0.73 ± 0.06 b | 20.61 ± 1.54 b | 2.37 ± 0.19 c | 0.23 ± 0.03 b | 8.46 ± 0.78 b |
| N3 | 34.03 ± 1.86 a | 171.33 ± 14.90 a | 42.93 ± 3.03 a | 1.19 ± 0.08 a | 9.97 ± 0.76 c | 1.00 ± 0.09 d | 0.11 ± 0.01 c | 11.25 ± 0.71 a |
| N4 | 19.37 ± 1.40 b | 80.43 ± 7.18 b | 28.67 ± 2.48 b | 0.66 ± 0.09 b | 23.19 ± 2.14 b | 2.80 ± 0.19 c | 0.29 ± 0.03 b | 7.58 ± 0.44 b |
| N5 | 12.13 ± 0.83 c | 37.93 ± 4.93 c | 13.43 ± 1.01 c | 0.26 ± 0.03 c | 41.25 ± 2.98 a | 5.02 ± 0.42 a | 0.46 ± 0.04 a | 5.93 ± 0.42 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Jiang, R.; Qin, J. Genome-Wide Identification and Expression Analysis of PaNRT Gene Family Under Various Nitrogen Conditions in Avocado (Persea americana Mill.). Genes 2024, 15, 1600. https://doi.org/10.3390/genes15121600
Tian Y, Jiang R, Qin J. Genome-Wide Identification and Expression Analysis of PaNRT Gene Family Under Various Nitrogen Conditions in Avocado (Persea americana Mill.). Genes. 2024; 15(12):1600. https://doi.org/10.3390/genes15121600
Chicago/Turabian StyleTian, Yuan, Ruiyuan Jiang, and Jian Qin. 2024. "Genome-Wide Identification and Expression Analysis of PaNRT Gene Family Under Various Nitrogen Conditions in Avocado (Persea americana Mill.)" Genes 15, no. 12: 1600. https://doi.org/10.3390/genes15121600
APA StyleTian, Y., Jiang, R., & Qin, J. (2024). Genome-Wide Identification and Expression Analysis of PaNRT Gene Family Under Various Nitrogen Conditions in Avocado (Persea americana Mill.). Genes, 15(12), 1600. https://doi.org/10.3390/genes15121600





