Genome-Wide Association Study of Body Weight Traits in Texel and Kazakh Crossbred Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Samples
2.3. Genotyping Analysis and Data Quality Control
2.4. Estimation of Genetic Parameters
2.5. GWAS Model Analysis Methods
2.6. Integrating Multiple GWAS Models: The E-GWAS Strategy
2.7. Annotation Using Multiple Databases
3. Results
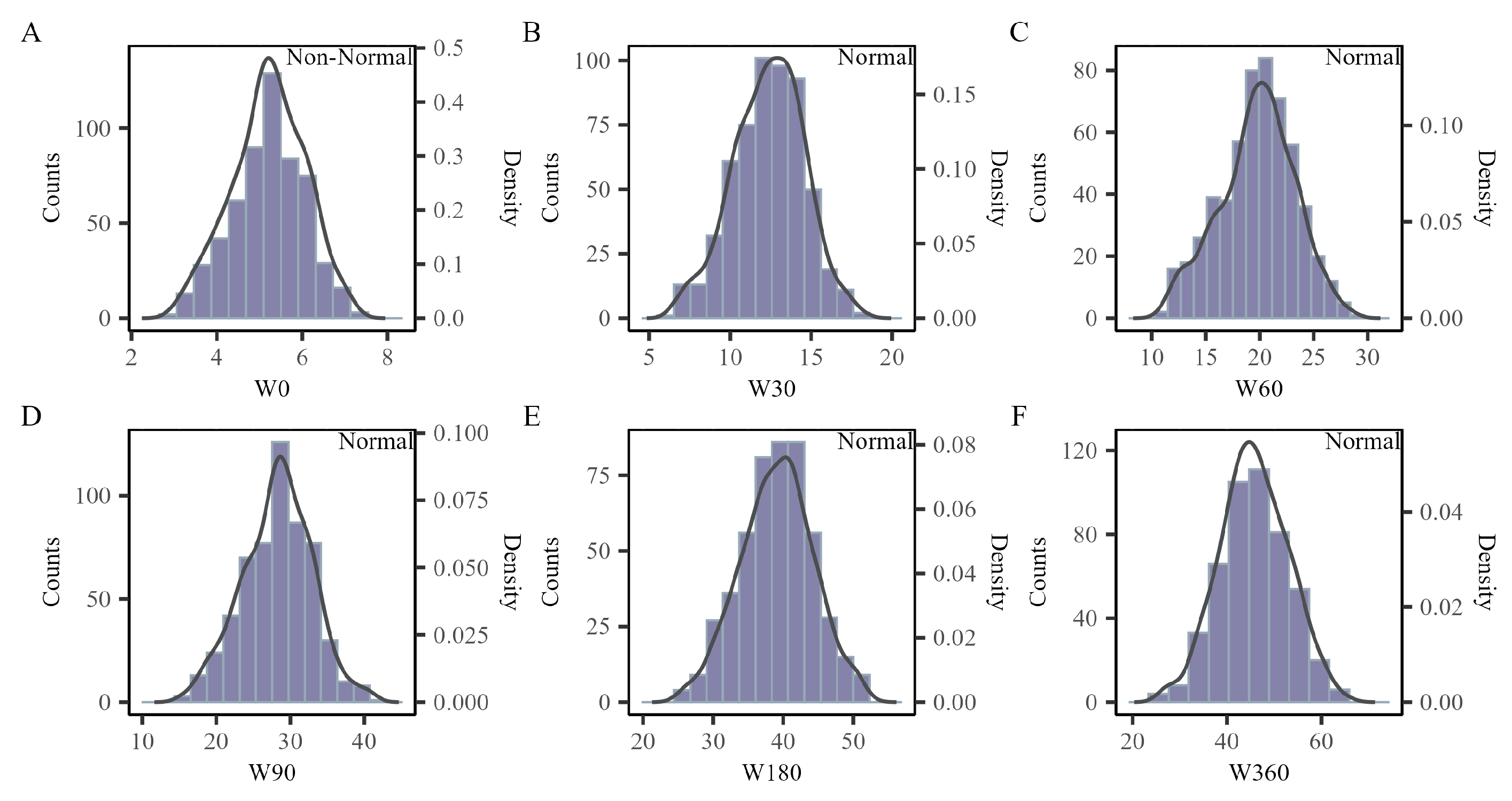
3.1. Phenotypic Descriptive Statistics
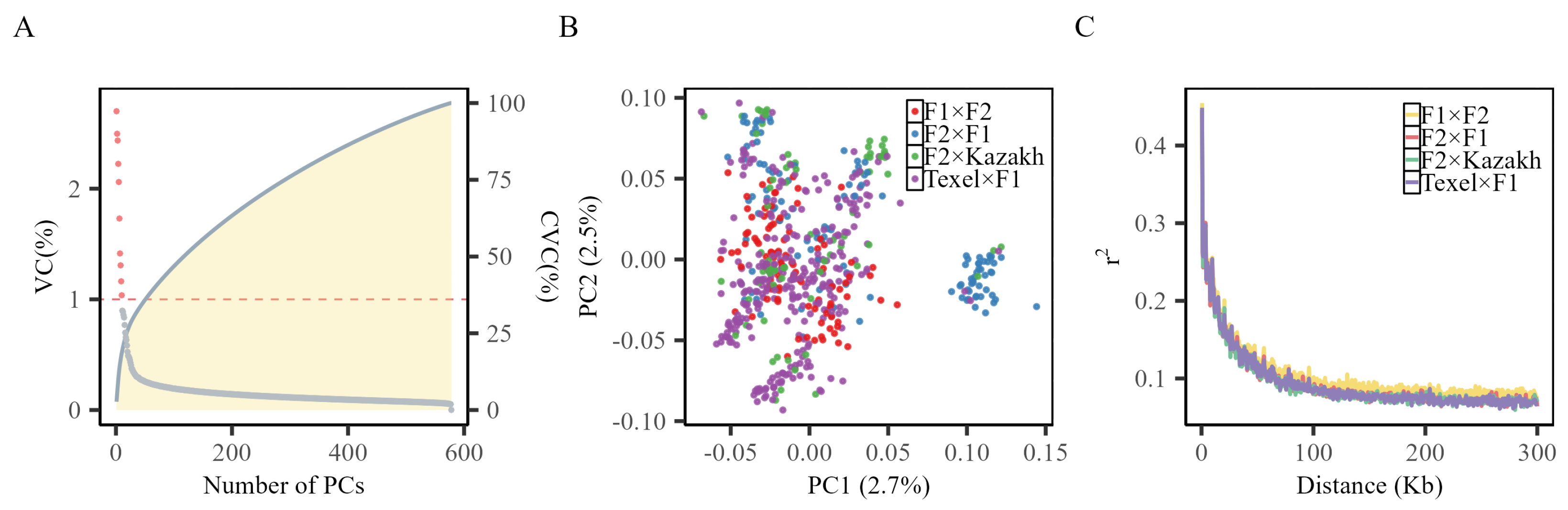
3.2. Population Structure Analysis and Linkage Disequilibrium Analysis
3.3. Estimation of Genetic Parameters
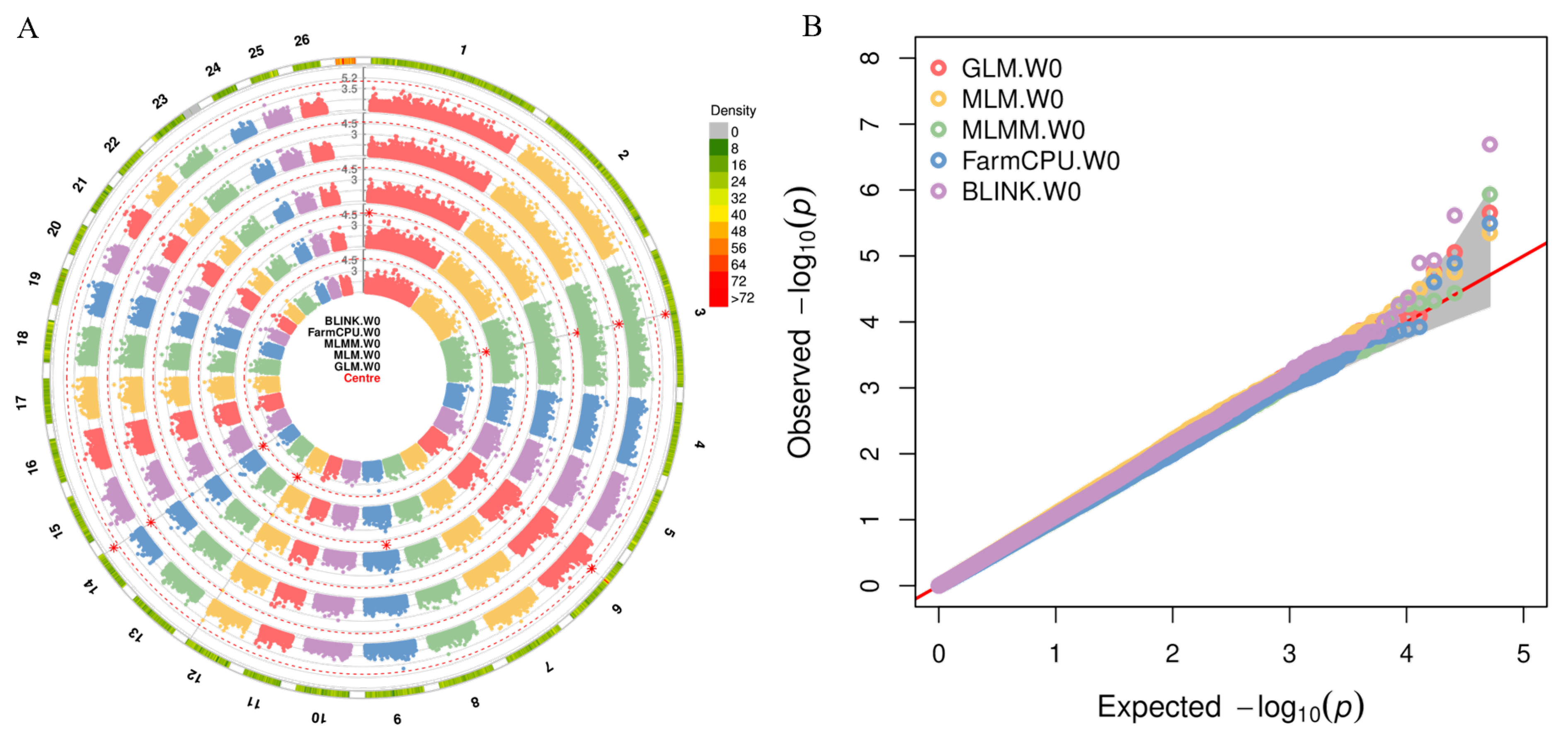
3.4. Genome-Wide Association Studies
3.5. E-GWAS Strategy Integrated Multiple GWAS Models
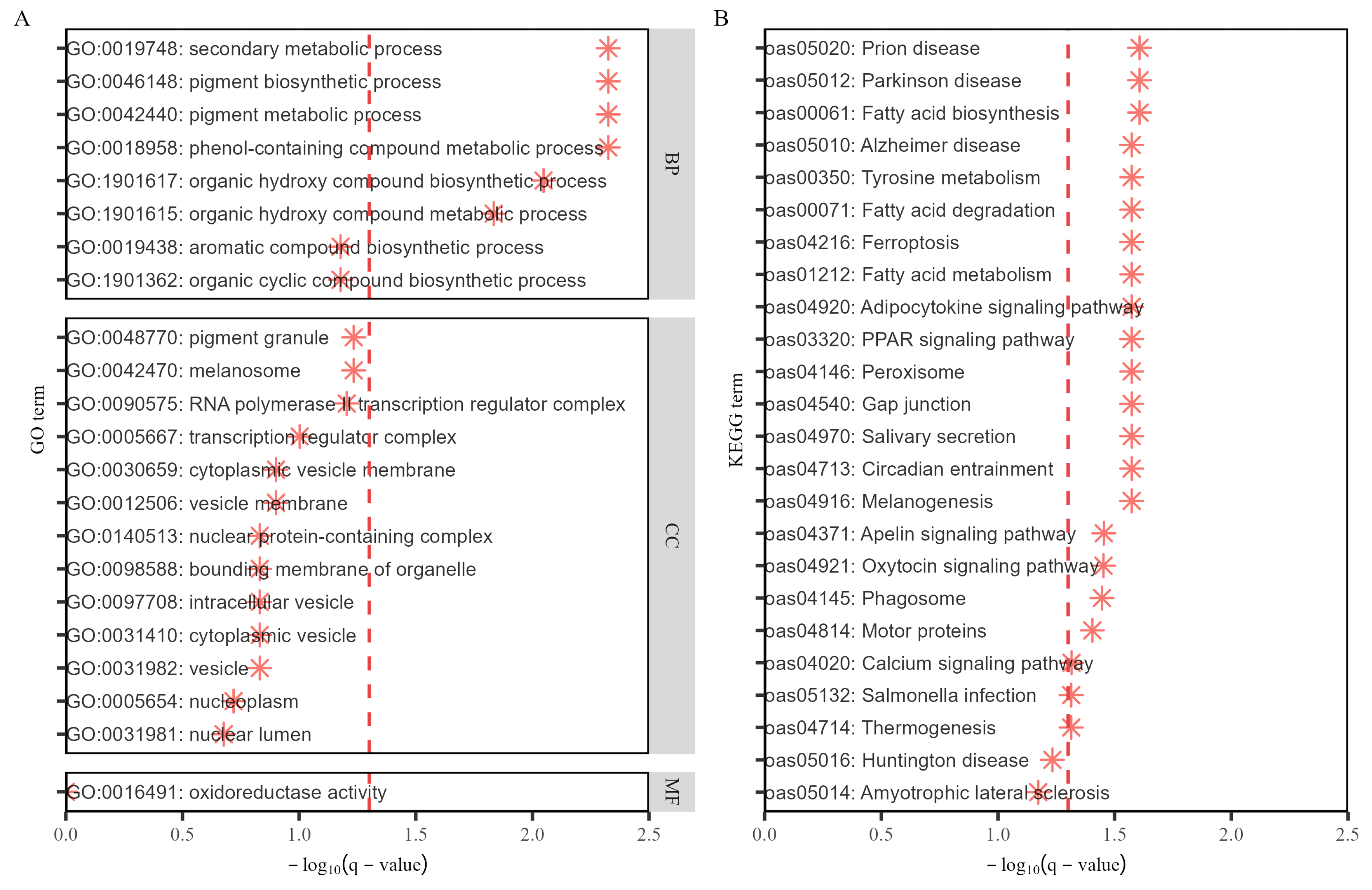
3.6. Post-GWAS Analysis Using Various Databases
4. Discussion
4.1. Influencing Factors
4.2. Genetic Parameters
4.3. Model Comparison
4.4. Post-GWAS and Gene Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohammadi, H.; Raffat, A.; Moradi, H.; Shoja, J.; Moradi, M.H. Estimation of linkage disequilibrium and whole-genome scan for detection of loci under selection associated with body weight in Zandi sheep breed. Agric. Biotechnol. J. 2018, 9, 151–172. [Google Scholar]
- Jumayi, X.; Liu, J.; Jiabaili, G. Discussion on the development trend of Altay sheep breeds. Xinjiang Livest. 2011, 3, 43–44. (In Chinese) [Google Scholar]
- Li, Z.; Li, B.; Liu, Y.; Hamutai, T.; Liu, M.; He, S.; Sulaiman, Y. Correlation analysis of body weight and body size of Texel × Kazakh sheep F1 generation. Herbiv. Livest. 2020, 2, 26–30. (In Chinese) [Google Scholar]
- Zhang, M.; Li, T.; Chu, H.; Tuan, Y.; Liu, J.; Chen, C.; Liu, Y.; Hamutai, T. Application of comprehensive analysis method in selection breeding of Texel–Kazakh sheep hybrids. Mod. Livest. Vet. Med. 2020, 49, 14–18. (In Chinese) [Google Scholar]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; Vries, J.d.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome–wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Su, P.; Huang, Y.M.; Xu, T.Q.; Lin, C.J.; Pan, C.Y. Application of Genome–Wide Association Study in Sheep Genetics and Breeding. Chin. J. Anim. Sci. 2023, 59, 74–85. [Google Scholar]
- Li, B.; Li, Z.Q.; Han, B.; Aishan, B.; Zhang, N.; Liu, M.J.; He, S.G.; Suleiman, Y.M. Correlation between Polymorphism and Productive Traits of Texel × Kazakh Sheep Analyzed Using Microsatellite Markers. Fujian J. Agric. Sci. 2020, 35, 243–253. (In Chinese) [Google Scholar]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second–generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome–wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinf. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome–wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed–model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, F.; Pan, Y.; Buckler, E.S.; Zhang, Z. A SUPER Powerful Method for Genome Wide Association Study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. Blink: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Zhou, G.L.; Xu, F.J.; Qiao, J.K.; Che, Z.X.; Xiang, T.; Liu, X.L.; Li, X.Y.; Zhao, S.H.; Zhu, M.J. E–GWAS: An ensemble-like GWAS strategy that provides effective control over false positive rates without decreasing true positives. Genet. Sel. Evol. 2023, 55, 46. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.T.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. Beagle: An application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 2012, 61, 170–173. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2023. Available online: http://www.R-project.org (accessed on 31 October 2023).
- Yin, L.; Zhang, H.; Tang, Z.; Yin, D.; Fu, Y.; Yuan, X.; Li, X.; Liu, X.; Zhao, S. Hiblup: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010, 11, 459–463. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. Bedtools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Bahreini Behzadi, M.R.; Shahroudi, F.E.; Van Vleck, L.D. Estimates of genetic parameters for growth traits in Kermani sheep. J. Anim. Breed. Genet. 2007, 124, 296–301. [Google Scholar] [CrossRef]
- Rahimi, S.M.; Rafat, S.A.; Jafari, S. Effects of environmental factors on growth traits in Makuie sheep. Biotechnol. Anim. Husb. 2014, 30, 185–192. [Google Scholar] [CrossRef]
- Benyi, K.; Norris, D.; Karbo, N.; Kgomo, K.A. Effects of genetic and environmental factors on pre-weaning and post-weaning growth in West African crossbred sheep. Trop. Anim. Health Prod. 2006, 38, 547–554. [Google Scholar] [CrossRef]
- Lupi, T.M.; Nogales, S.; León, J.M.; Barba, C.; Delgado, J.V. Analysis of the non-genetic factors affecting the growth of Segureño sheep. Ital. J. Anim. Sci. 2015, 14, 124. [Google Scholar] [CrossRef]
- Sharif, N.; Ali, A.; Dawood, M.; Khan, M.I.; Do, D.N. Environmental Effects and Genetic Parameters for Growth Traits of Lohi Sheep. Animals 2022, 12, 3590. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Latifi, M. Autosomal and sex-linked (co)variance components and genetic parameters for growth traits of Iranian Zandi sheep. Trop. Anim. Health. Prod. 2020, 52, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Qi, Y.; Liu, Y.; Rong, Y.; Ao, X.; Zhang, M.; Xia, Q.; Zhang, Y.; Wang, R. Study of the Influence of Non-Genetic Factors on the Growth and Development Traits and Cashmere Production Traits of Inner Mongolia White Cashmere Goats (Erlangshan Type). Vet. Sci. 2024, 11, 308. [Google Scholar] [CrossRef]
- Xiao, C.; Liu, Y.; Zhao, W.; Liang, Y.; Cui, C.; Yang, S.; Fang, W.; Miao, L.; Yuan, Z.; Lin, Z.; et al. The comparison of meat yield, quality, and flavor between small-tailed Han sheep and two crossbred sheep and the verification of related candidate genes. Front. Nutr. 2024, 11, 1399390. [Google Scholar] [CrossRef] [PubMed]
- Ellies-Oury, M.P.; Papillon, S.; Arranz, J.M.; Carpentier, D. Improvement of growth performance through crossbreeding in the Pyrenean suckling lamb protected geographical indication. Livest. Sci. 2022, 265, 105081. [Google Scholar] [CrossRef]
- Ba, Y.; Liang, L.; He, P.; Li, J.; He, X.; He, S.; Li, W. Comparison of early growth performance of offspring of kazakh sheep with different hybrid combinations. Xinjiang Agric. Sci. 2023, 60, 2331–2340. (In Chinese) [Google Scholar]
- Besufkad, S.; Goshme, S.; Abebe, A.; Bisrat, A.; Abebe, A.; Getachew, T.; Zewdie, T.; Lemma, S.; Areaya, A.; Gizaw, S. Estimates of genetic parameters for growth traits in dorper crossbred sheep population. Trop. Anim. Health Prod. 2024, 56, 264. [Google Scholar] [CrossRef]
- Dash, S.S.; Bangar, Y.C.; Magotra, A.; Patil, C.S.; Sharma, R.; Chauhan, A.; Dahiya, S.P. Bayesian estimates of genetic parameters for growth traits in Harnali sheep. J. Anim. Breed. Genet. 2024, early view.
- Oliveira, I.R.S.; Bastos, M.S.; Vesco, A.P.D.; Montalvan, Z.C.R.; Barreto Neto, A.D.; Barbosa, L.T. Genetic parameters for growth and reproductive traits in Santa Inês sheep. Small Rumin. Res. 2024, 239, 107327. [Google Scholar] [CrossRef]
- Krivoruchko, A.; Sermyagin, A.; Saprikina, T.; Golovanova, N.; Kvochko, A.; Yatsyk, O. Genome wide associations study of single nucleotide polymorphisms with productivity parameters in Jalgin merino for identification of new candidate genes. Gene Rep. 2021, 23, 101065. [Google Scholar] [CrossRef]
- Chen, Q.; Qu, K.; Ma, Z.; Zhan, J.; Zhang, F.; Shen, J.; Ning, Q.; Jia, P.; Zhang, J.; Chen, N.; et al. Genome-Wide Association Study Identifies Genomic Loci Associated with Neurotransmitter Concentration in Cattle. Front. Genet. 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Bao, A.; Hong, W.; Hou, C.; Zhang, Z.; Liang, X.; Aniwashi, J. Transcriptome profiling analysis reveals key genes of different coat color in sheep skin. PeerJ 2019, 7, e8077. [Google Scholar] [CrossRef] [PubMed]
- Dardente, H.; Lomet, D. Photoperiod and thyroid hormone regulate expression of l-dopachrome tautomerase (Dct), a melanocyte stem-cell marker, in tanycytes of the ovine hypothalamus. J. Neuroendocrinol. 2018, 30, e12640. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, J.; Wang, H.; Zhang, R.; An, X.; Yuan, C.; Guo, T.; Yue, Y. Genomic Selection for Live Weight in the 14th Month in Alpine Merino Sheep Combining GWAS Information. Animals 2023, 13, 3516. [Google Scholar] [CrossRef]
- Liu, Z.; Bai, C.; Shi, L.; He, Y.; Hu, M.; Sun, H.; Peng, H.; Lai, W.; Jiao, S.; Zhao, Z.; et al. Detection of selection signatures in South African Mutton Merino sheep using whole-genome sequencing data. Anim. Genet. 2022, 53, 224–229. [Google Scholar] [CrossRef]
- Luo, R.; Zhang, X.; Wang, L.; Zhang, L.; Li, G.; Zheng, Z. GLIS1, a potential candidate gene affect fat deposition in sheep tail. Mol. Biol. Rep. 2021, 48, 4925–4931. [Google Scholar] [CrossRef]
- Sweet-Jones, J.; Yurchenko, A.A.; Igoshin, A.V.; Yudin, N.S.; Swain, M.T.; Larkin, D.M. Resequencing and signatures of selection scan in two Siberian native sheep breeds point to candidate genetic variants for adaptation and economically important traits. Anim. Genet. 2021, 52, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Isaacs, K.; Becker, G.; Murdoch, B.M. A computational framework for improving genetic variants identification from 5061 sheep sequencing data. J. Anim. Sci. Biotechnol. 2023, 14, 127. [Google Scholar] [CrossRef]
- Seroussi, E.; Rosov, A.; Shirak, A.; Lam, A.; Gootwine, E. Unveiling genomic regions that underlie differences between Afec-Assaf sheep and its parental Awassi breed. Genet. Sel. Evol. 2017, 49, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, F.; Gao, G.; Yan, X.; Liu, H.; Liu, Z.; Wang, Z.; He, L.; Lv, Q.; Wang, Z.; et al. Genome-Wide Association Study of Body Weight Traits in Inner Mongolia Cashmere Goats. Front. Vet. Sci. 2021, 8, 752746. [Google Scholar] [CrossRef]
- Martins, R.; Machado, P.C.; Pinto, L.F.B.; Silva, M.R.; Schenkel, F.S.; Brito, L.F.; Pedrosa, V.B. Genome-wide association study and pathway analysis for fat deposition traits in nellore cattle raised in pasture-based systems. J. Anim. Breed. Genet. 2021, 138, 360–378. [Google Scholar] [CrossRef]
- Ramos, Z.; Garrick, D.J.; Blair, H.T.; Vera, B.; Ciappesoni, G.; Kenyon, P.R. Genomic Regions Associated with Wool, Growth and Reproduction Traits in Uruguayan Merino Sheep. Genes 2023, 14, 167. [Google Scholar] [CrossRef]
- Mohammadi, H.; Farahani, A.H.K.; Moradi, M.H.; Mastrangelo, S.; Di Gerlando, R.; Sardina, M.T.; Scatassa, M.L.; Portolano, B.; Tolone, M. Weighted Single-Step Genome-Wide Association Study Uncovers Known and Novel Candidate Genomic Regions for Milk Production Traits and Somatic Cell Score in Valle del Belice Dairy Sheep. Animals 2022, 12, 1155. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Bhushan, B.; Mishra, B.P. Genome-wide analysis of genetic diversity and selection signatures in three Indian sheep breeds. Livest. Sci. 2021, 243, 104367. [Google Scholar] [CrossRef]
- Estrada-Reyes, Z.M.; Rae, O.; Postley, C.; Jiménez Medrano, M.B.; Leal Gutiérrez, J.D.; Mateescu, R.G. Association study reveals Th17, Treg, and Th2 loci related to resistance to Haemonchus contortus in Florida Native sheep. J. Anim. Sci. 2019, 97, 4428–4444. [Google Scholar] [CrossRef]
- Lynch, R.M.; Naswa, S.; Rogers, G.L., Jr.; Kania, S.A.; Das, S.; Chesler, E.J.; Saxton, A.M.; Langston, M.A.; Voy, B.H. Identifying genetic loci and spleen gene coexpression networks underlying immunophenotypes in BXD recombinant inbred mice. Physiol. Genom. 2010, 41, 244–253. [Google Scholar] [CrossRef]
- Easa, A.A.; Selionova, M.; Aibazov, M.; Mamontova, T.; Sermyagin, A.; Belous, A.; Abdelmanova, A.; Deniskova, T.; Zinovieva, N. Identification of Genomic Regions and Candidate Genes Associated with Body Weight and Body Conformation Traits in Karachai Goats. Genes 2022, 13, 1773. [Google Scholar] [CrossRef]
- Yilmaz, O.; Kizilaslan, M.; Arzik, Y.; Behrem, S.; Ata, N.; Karaca, O.; Elmaci, C.; Cemal, I. Genome-wide association studies of preweaning growth and in vivo carcass composition traits in Esme sheep. J. Anim. Breed. Genet. 2022, 139, 26–39. [Google Scholar] [CrossRef]
- Xu, S.S.; Gao, L.; Shen, M.; Lyu, F. Whole-Genome Selective Scans Detect Genes Associated with Important Phenotypic Traits in Sheep (Ovis aries). Front. Genet. 2021, 12, 738879. [Google Scholar] [CrossRef]
- Antkowiak, M.; Szydlowski, M. Uncovering structural variants associated with body weight and obesity risk in labrador retrievers: A genome-wide study. Front. Genet. 2023, 14, 1235821. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zhao, Z.; Fang, X.; Yu, H.; Long, X.; Jiang, P.; Yang, R. Association of the ACSL5 gene G.33185918G>A and G.33186348C>T mutations with carcass and meat quality traits of Chinese Simmental-cross steers. J. Anim. Plant Sci. 2016, 26, 658–664. [Google Scholar]
- Pang, F.; Zhang, M.; Li, G.; Zhang, Z.; Huang, H.; Li, B.; Wang, C.; Yang, X.; Zheng, Y.; An, Q.; et al. Integrated mRNA and miRNA profiling in NIH/3T3 cells in response to bovine papillomavirus E6 gene expression. PeerJ 2019, 7, e7442. [Google Scholar] [CrossRef]
- Li, J.; Guan, M.; Qi, L.; Zhang, F.; Jia, C.; Meng, Q.; Han, J. Metalloproteins as risk factors for osteoarthritis: Improving and understanding causal estimates using Mendelian randomization. Clin. Rheumatol. 2024, 43, 2079–2091. [Google Scholar] [CrossRef]
- Zhu, M.; Jia, L.; Li, F.; Jia, J. Identification of KIAA0513 and Other Hub Genes Associated with Alzheimer Disease Using Weighted Gene Coexpression Network Analysis. Front. Genet. 2020, 11, 981. [Google Scholar] [CrossRef]
- Drew, K.; Lee, C.; Huizar, R.L.; Tu, F.; Borgeson, B.; McWhite, C.D.; Ma, Y.; Wallingford, J.B.; Marcotte, E.M. Integration of over 9000 mass spectrometry experiments builds a global map of human protein complexes. Mol. Syst. Biol. 2017, 13, 932. [Google Scholar] [CrossRef]
- Oddoux, S.; Randazzo, D.; Kenea, A.; Alonso, B.; Zaal, K.J.M.; Ralston, E. Misplaced Golgi Elements Produce Randomly Oriented Microtubules and Aberrant Cortical Arrays of Microtubules in Dystrophic Skeletal Muscle Fibers. Front. Cell Dev. Biol. 2019, 7, 176. [Google Scholar] [CrossRef]
- Maurin, J.; Morel, A.; Guérit, D.; Cau, J.; Urbach, S.; Blangy, A.; Bompard, G. The Beta-Tubulin Isotype TUBB6 Controls Microtubule and Actin Dynamics in Osteoclasts. Front. Cell Dev. Biol. 2021, 9, 778887. [Google Scholar] [CrossRef]
- Hsu, J.W.; Nien, C.Y.; Chen, H.W.; Tsai, F.Y.; Yeh, S.C.; Kao, Y.H.; Tsou, T.C. Di (2-ethylhexyl) phthalate exposure exacerbates metabolic disorders in diet-induced obese mice. Food Chem. Toxicol. 2021, 156, 112439. [Google Scholar] [CrossRef]
- Nagao, M.; Ogata, T.; Sawada, Y.; Gotoh, Y. Zbtb20 promotes astrocytogenesis during neocortical development. Nat. Commun. 2016, 7, 11102. [Google Scholar] [CrossRef] [PubMed]
- Rajawat, D.; Panigrahi, M.; Kumar, H.; Nayak, S.S.; Parida, S.; Bhushan, B.; Gaur, G.K.; Dutt, T.; Mishra, B.P. Identification of important genomic footprints using eight different selection signature statistics in domestic cattle breeds. Gene 2022, 816, 146165. [Google Scholar] [CrossRef]

| Factor | W0 | W30 | W60 | W90 | W180 | W360 |
|---|---|---|---|---|---|---|
| Hybrid Group | Y | Y | Y | Y | Y | Y |
| Rearing Group | N | Y | Y | Y | Y | Y |
| Birth Type | Y | Y | Y | Y | Y | Y |
| Gender | Y | Y | Y | Y | Y | Y |
| Age of Ewe | Y | Y | Y | Y | N | N |
| Trait | Number | Mean | SD | Median | Min | Max | Skew | Kurtosis | IQR |
|---|---|---|---|---|---|---|---|---|---|
| W0 | 573 | 5.21 | 0.86 | 5.20 | 2.90 | 7.30 | −0.18 | −0.34 | 1.20 |
| W30 | 569 | 12.37 | 2.19 | 12.47 | 6.52 | 18.29 | −0.18 | −0.15 | 2.96 |
| W60 | 561 | 19.76 | 3.47 | 19.97 | 10.76 | 28.70 | −0.22 | −0.28 | 4.41 |
| W90 | 568 | 28.25 | 4.71 | 28.44 | 15.21 | 41.12 | −0.11 | −0.07 | 6.45 |
| W180 | 493 | 39.08 | 5.15 | 39.19 | 25.24 | 52.15 | −0.08 | −0.24 | 6.83 |
| W360 | 488 | 45.73 | 7.24 | 45.49 | 25.99 | 65.67 | −0.01 | −0.20 | 9.77 |
| W0 | W30 | W60 | W90 | W180 | W360 | |
|---|---|---|---|---|---|---|
| W0 | ||||||
| W30 | ||||||
| W60 | ||||||
| W90 | ||||||
| W180 | ||||||
| W360 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Liu, M.; Zhang, H.; He, S.; Li, W.; Liang, L. Genome-Wide Association Study of Body Weight Traits in Texel and Kazakh Crossbred Sheep. Genes 2024, 15, 1521. https://doi.org/10.3390/genes15121521
Wang S, Liu M, Zhang H, He S, Li W, Liang L. Genome-Wide Association Study of Body Weight Traits in Texel and Kazakh Crossbred Sheep. Genes. 2024; 15(12):1521. https://doi.org/10.3390/genes15121521
Chicago/Turabian StyleWang, Sheng, Mingjun Liu, Huiguo Zhang, Sangang He, Wenrong Li, and Long Liang. 2024. "Genome-Wide Association Study of Body Weight Traits in Texel and Kazakh Crossbred Sheep" Genes 15, no. 12: 1521. https://doi.org/10.3390/genes15121521
APA StyleWang, S., Liu, M., Zhang, H., He, S., Li, W., & Liang, L. (2024). Genome-Wide Association Study of Body Weight Traits in Texel and Kazakh Crossbred Sheep. Genes, 15(12), 1521. https://doi.org/10.3390/genes15121521





