Genome-Wide Characterization of Solanum tuberosum CCoAOMT Gene Family and Identification of StCCoAOMT Genes Involved in Anthocyanin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Identification of S. tuberosum CCoAOMT Genes
2.3. Chromosomal Distribution and Collinearity Analysis
2.4. Phylogenetic Tree, Conserved Motif and Gene Structure Analysis
2.5. Cis-Regulatory Elements Analysis of the StCCoAOMT Promoters
2.6. Total RNA Extraction and qRT–PCR Analysis
2.7. Gene Cloning and Vector Construction
2.8. Subcellular Localization of StCCoAOMT10 in Tobacco
3. Results
3.1. Genome-Wide Identification and Physicochemical Analysis of the CCoAOMT Family in S. tuberosum
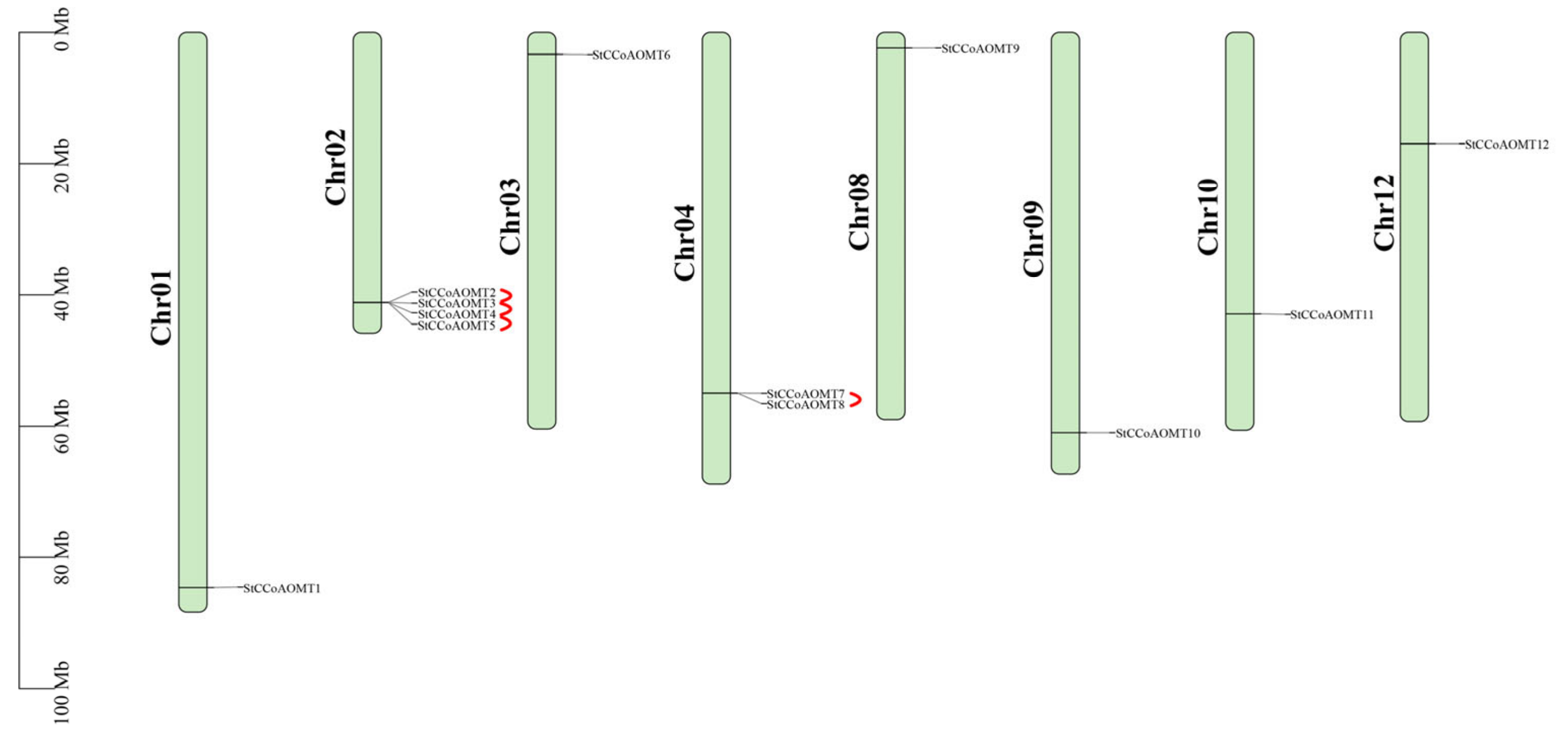
3.2. Chromosome Mapping of StCCoAOMT Genes
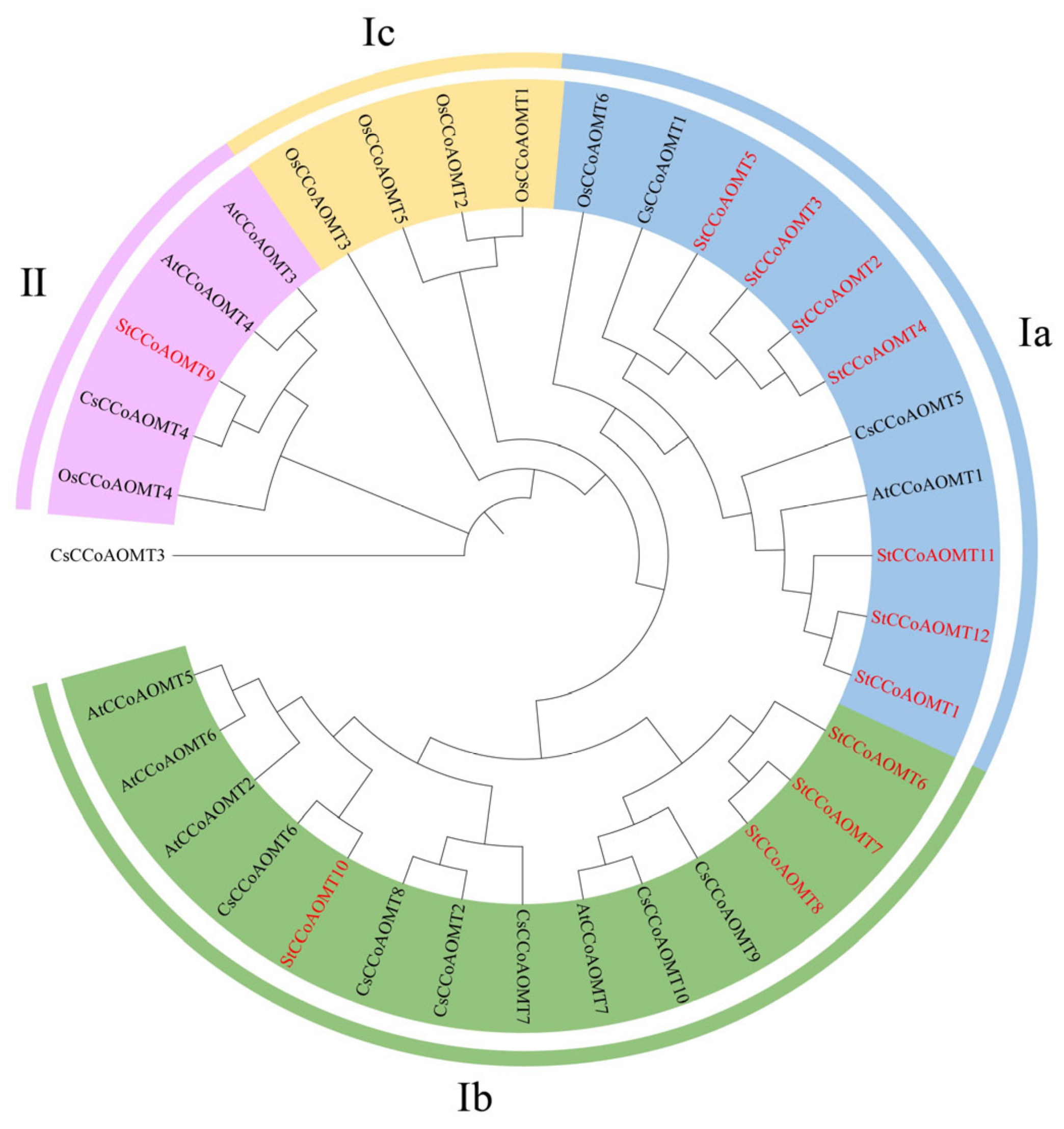
3.3. Evolutionary Analysis of StCCoAOMT Genes
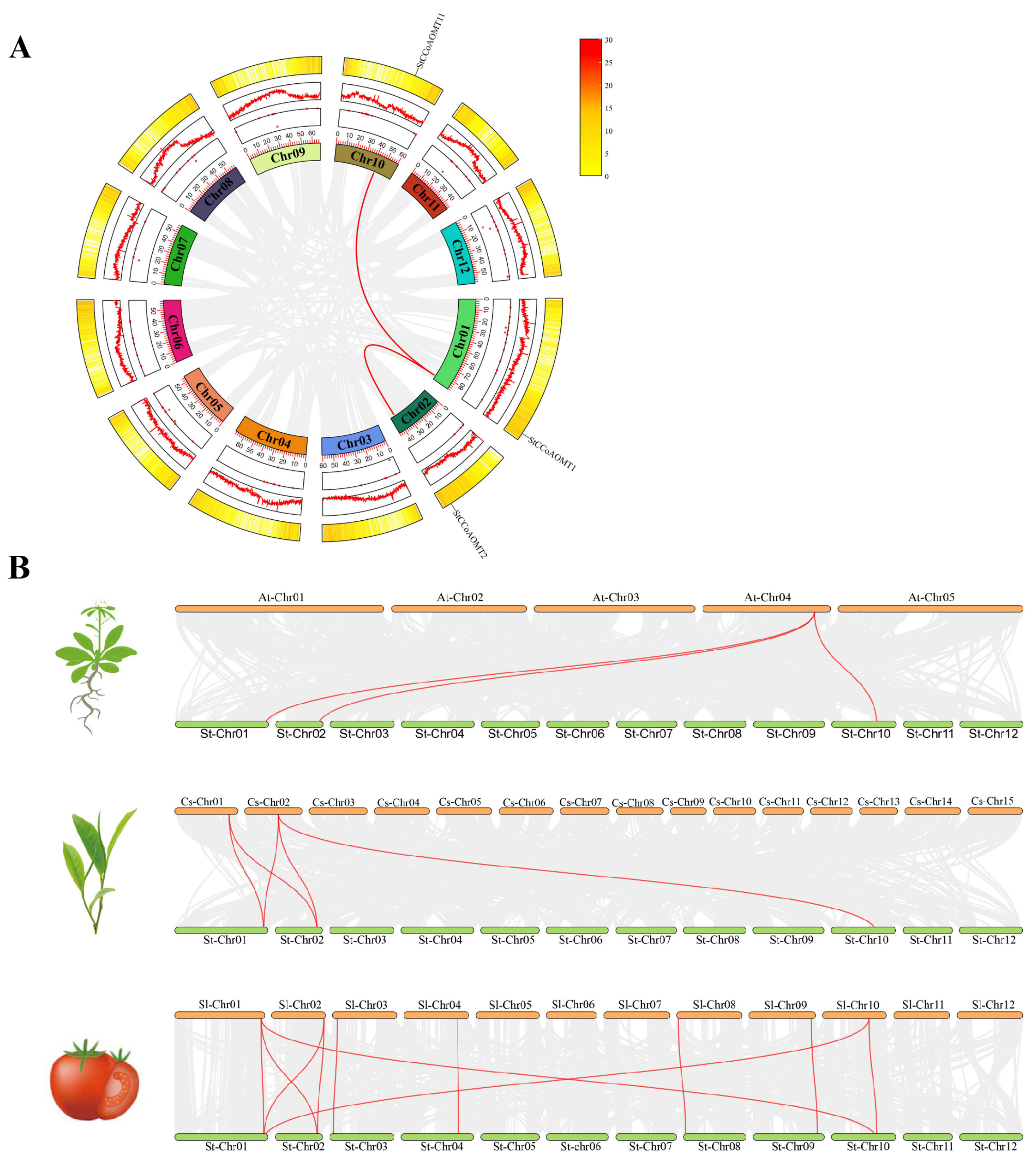
3.4. Intraspecific and Interspecific Collinearity Analysis of CCoAOMT Genes
3.5. Conserved Motif Composition and Gene Structure of StCCoAOMT Family Members
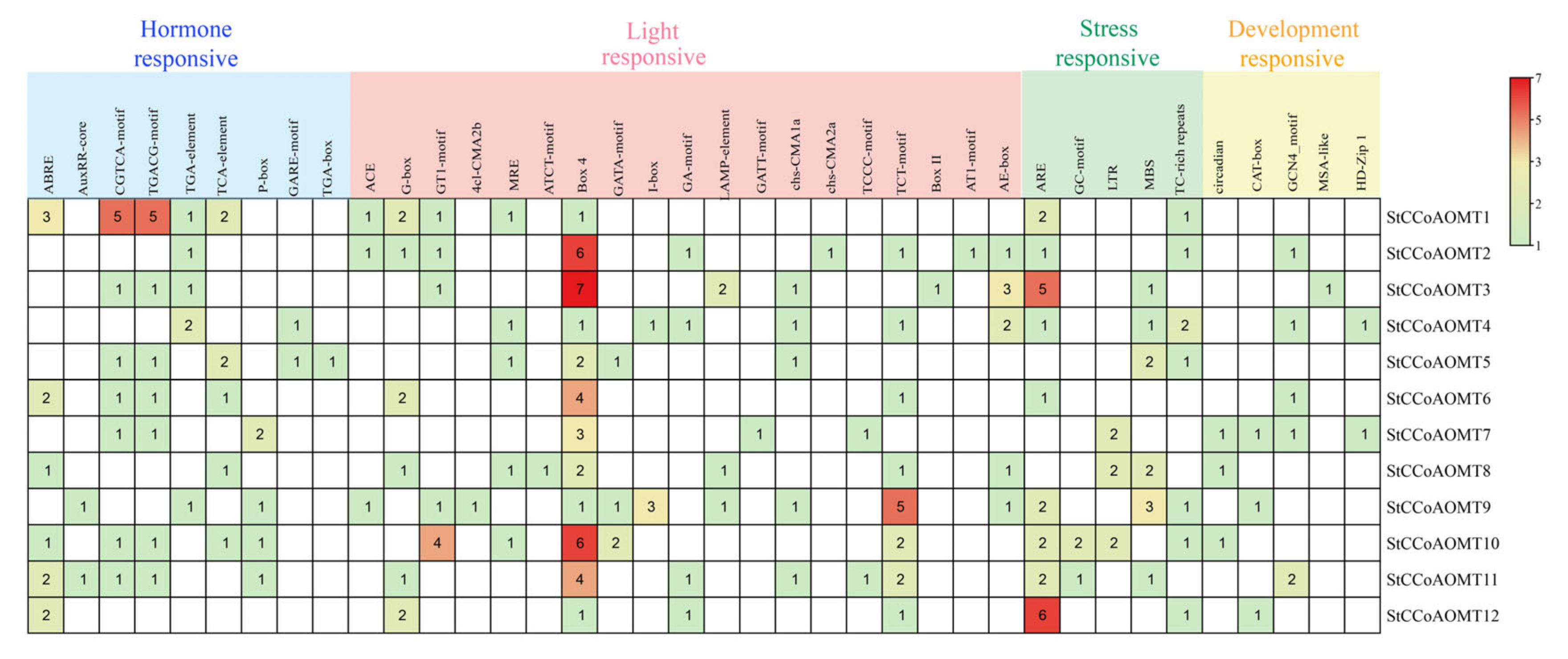
3.6. Cis-Regulatory Elements Analysis of StCCoAOMT Genes
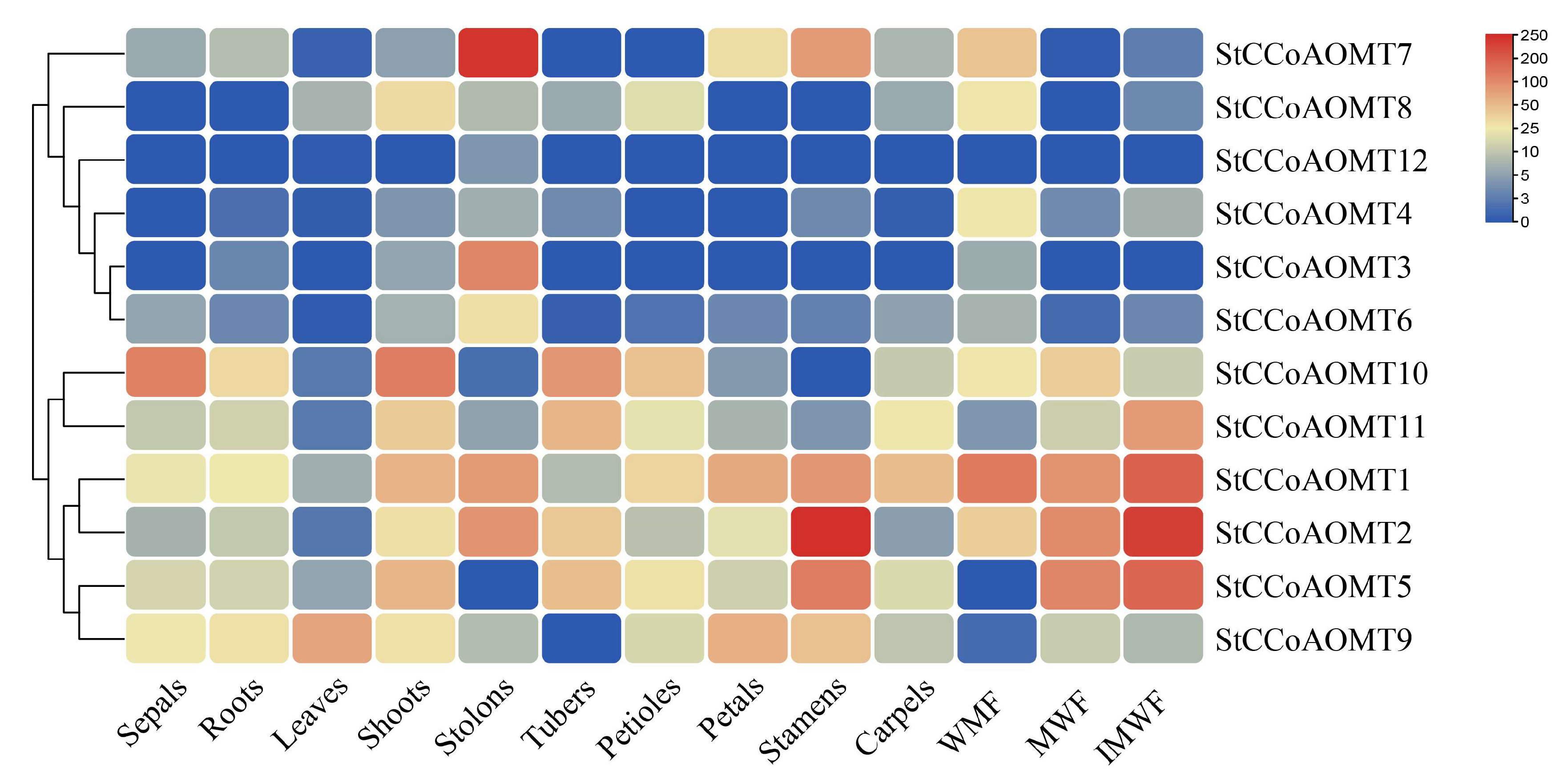
3.7. Expression Pattern of StCCoAOMT Genes in Various Tissues
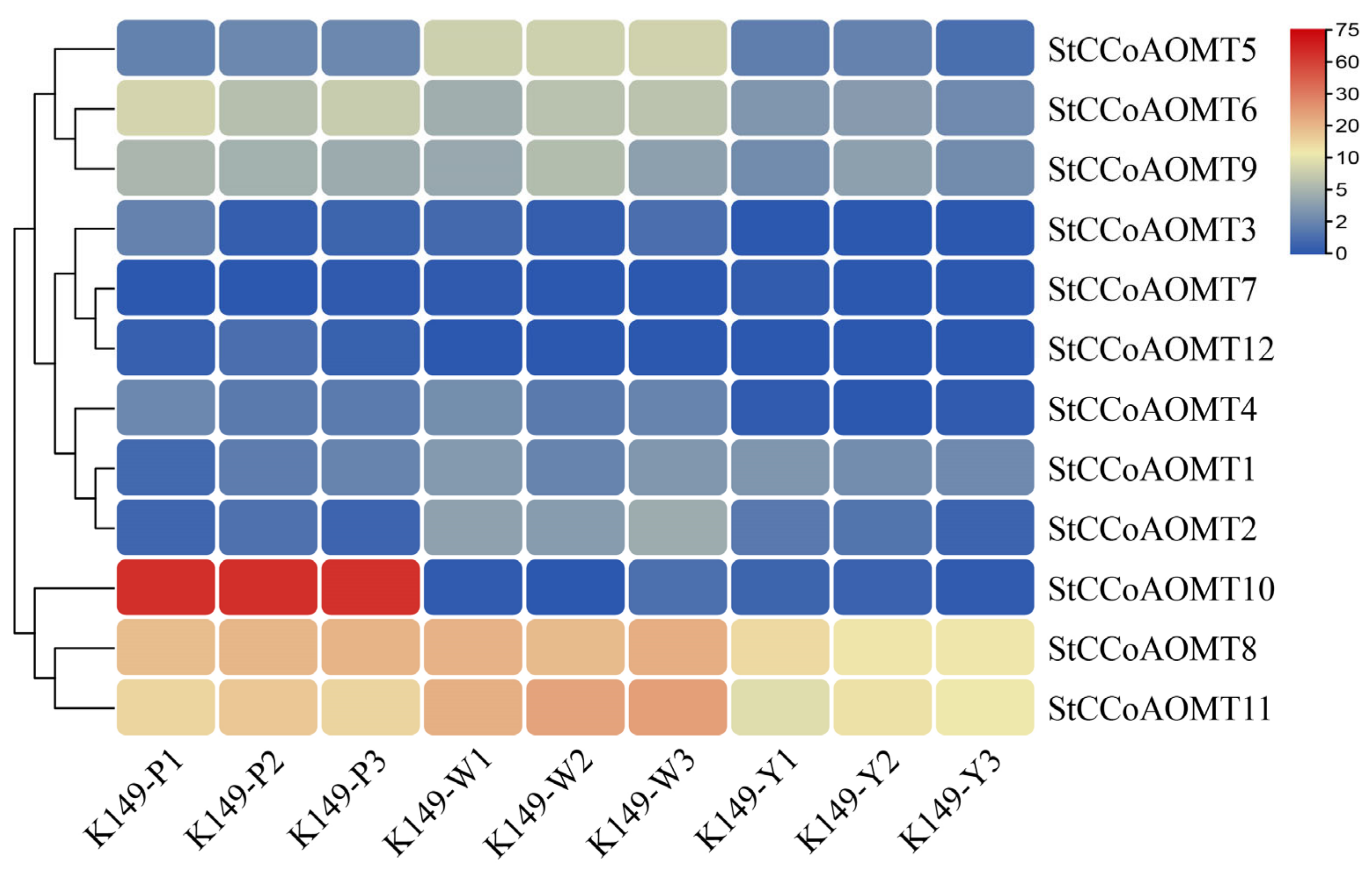
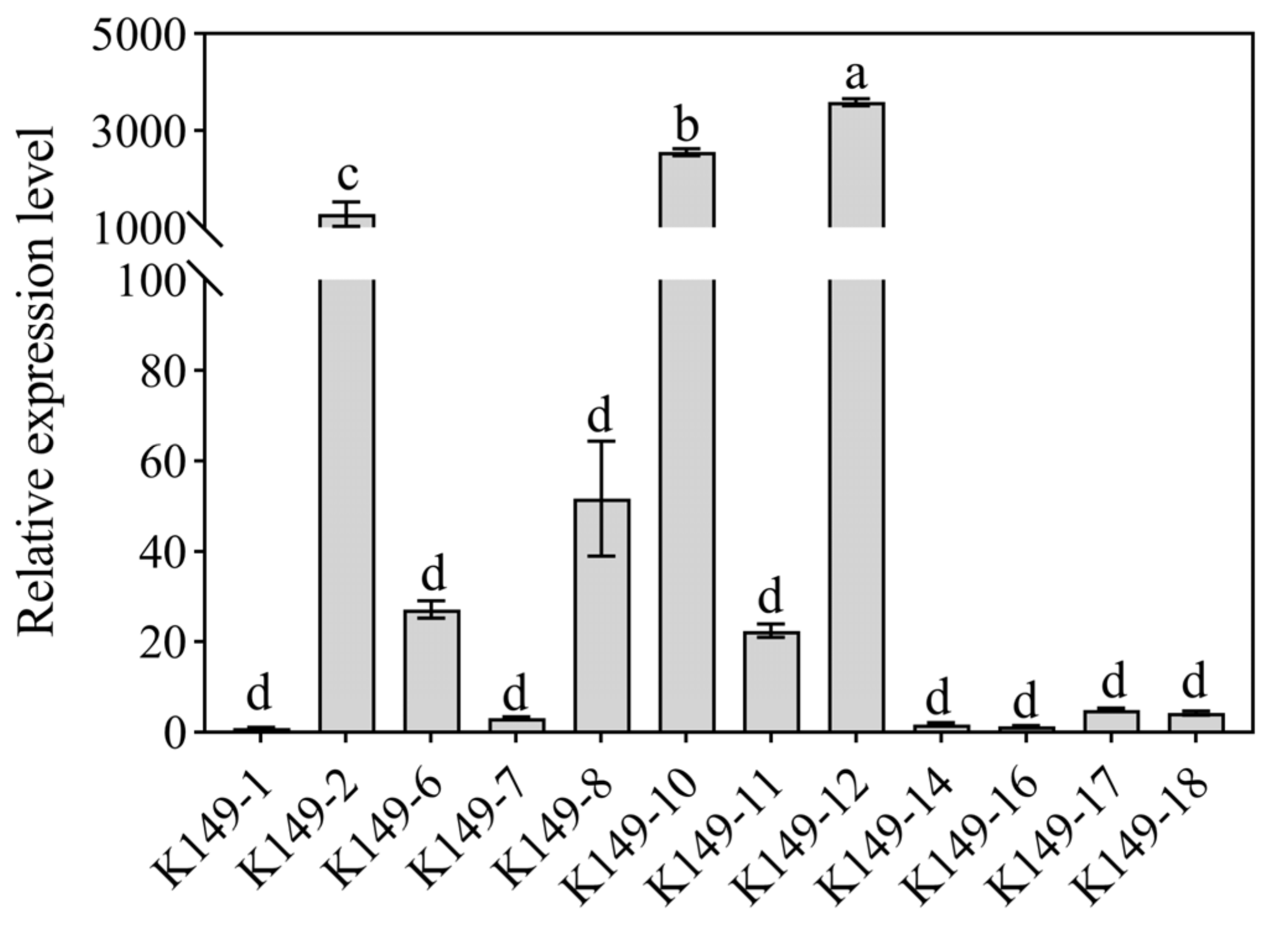
3.8. Identification of Candidate StCCoAOMT Gene Involved in Anthocyanin Biosynthesis
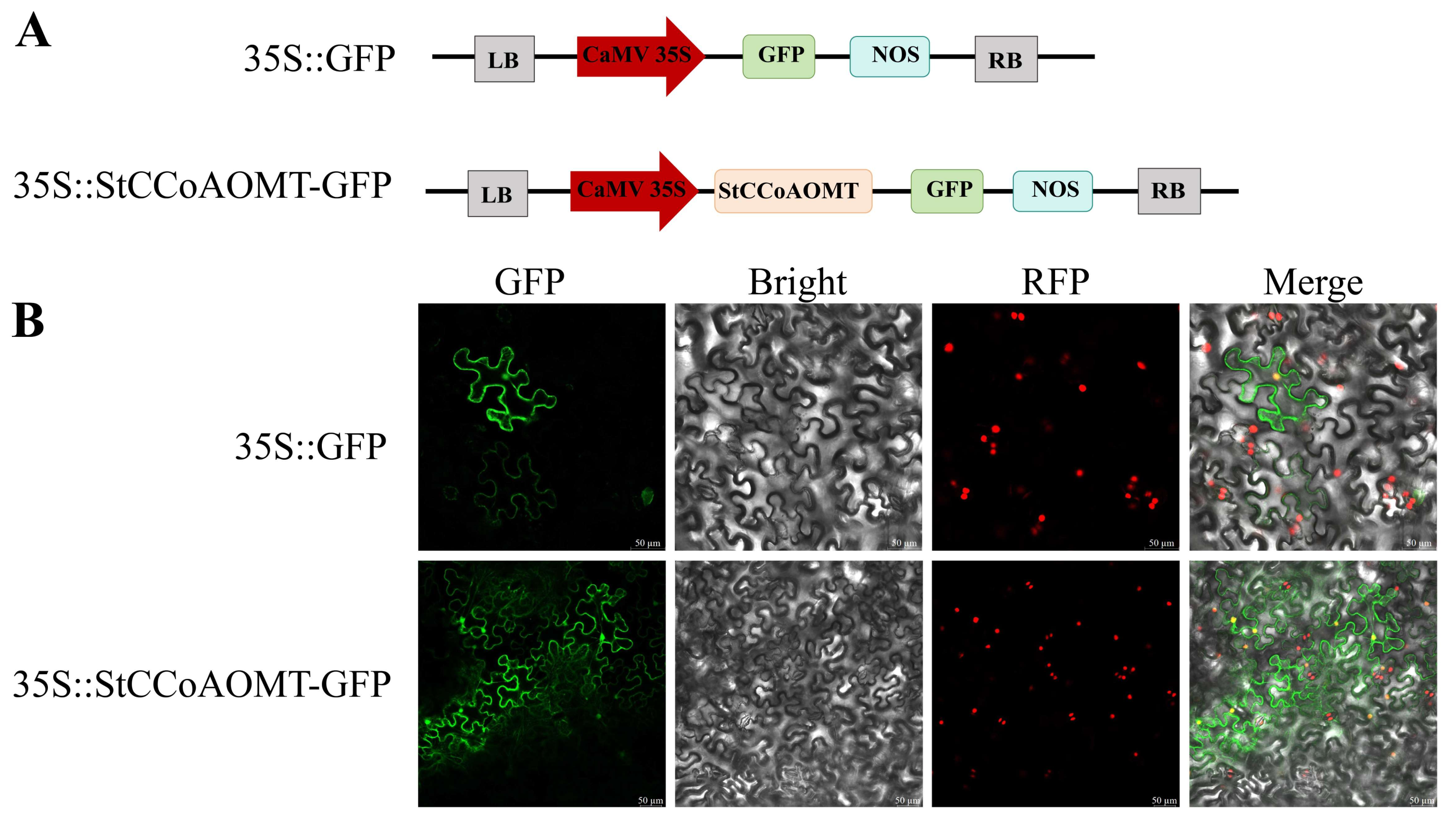
3.9. Subcellular Localization of the StCCoAOMT10 Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, M.; Li, Y.; Li, G.; Dong, T.; Liu, S.; Pei, L.; Wang, Q. Overexpression of StCYS1 gene enhances tolerance to salt stress in the transgenic potato (Solanum tuberosum L.) plant. J. Integr. Agric. 2020, 19, 2239–2246. [Google Scholar] [CrossRef]
- Birch, P.R.; Bryan, G.; Fenton, B.; Gilroy, E.M.; Hein, I.; Jones, J.T.; Prashar, A.; Taylor, M.A.; Torrance, L.; Toth, I.K. Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur. 2012, 4, 477–508. [Google Scholar] [CrossRef]
- Qu, G.; Peng, D.; Yu, Z.; Chen, X.; Cheng, X.; Yang, Y.; Ye, T.; Lv, Q.; Ji, W.; Deng, X. Advances in the role of auxin for transcriptional regulation of lignin biosynthesis. Funct. Plant Biol. 2021, 48, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Albert, N.W.; Zhou, Y.; Schwinn, K.E. Functions of flavonoid and betalain pigments in abiotic stress tolerance in plants. Ann. Plant Rev. 2018, 1, 21–62. [Google Scholar]
- Brunetti, C.; Fini, A.; Sebastiani, F.; Gori, A.; Tattini, M. Modulation of phytohormone signaling: A primary function of flavonoids in plant–environment interactions. Front. Plant Sci. 2018, 9, 1042. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.A. Ultraviolet-B radiation effects on plants: Induction of morphogenic responses. Physiol. Plantarum. 2002, 116, 423–429. [Google Scholar] [CrossRef]
- Erlejman, A.; Verstraeten, S.; Fraga, C.; Oteiza, P. The interaction of flavonoids with membranes: Potential determinant of flavonoid antioxidant effects. Free Radic. Res. 2004, 38, 1311–1320. [Google Scholar] [CrossRef]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES—Dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef]
- Emiliani, J.; Grotewold, E.; Ferreyra, M.L.F.; Casati, P. Flavonols protect Arabidopsis plants against UV-B deleterious effects. Mol. Plant. 2013, 6, 1376–1379. [Google Scholar] [CrossRef]
- Mutha, R.E.; Tatiya, A.U.; Surana, S.J. Flavonoids as natural phenolic compounds and their role in therapeutics: An overview. Futur. J. Pharm. Sci. 2021, 7, 25. [Google Scholar] [CrossRef]
- Braun, K.F.; Ehnert, S.; Freude, T.; Egaña, J.T.; Schenck, T.L.; Buchholz, A.; Schmitt, A.; Siebenlist, S.; Schyschka, L.; Neumaier, M. Quercetin protects primary human osteoblasts exposed to cigarette smoke through activation of the antioxidative enzymes HO-1 and SOD-1. Sci. World J. 2011, 11, 2348–2357. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, P.; Jagadeesan, R.; Sekaran, S.; Dhanasekaran, A.; Vimalraj, S. Flavonoids: Classification, function, and molecular mechanisms involved in bone remodelling. Front. Endocrinol. 2021, 12, 779638. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.L.; Guo, H.C.; Dong, Z.Y.; Zhao, Q. Pharmacological and nutritional activities of potato anthocyanins. Afr. J. Pharm. Pharmacol. 2009, 3, 463–468. [Google Scholar]
- Li, Z.; Ahammed, G.J. Plant stress response and adaptation via anthocyanins: A review. Plant Stress. 2023, 10, 100230. [Google Scholar] [CrossRef]
- Kaur, S.; Tiwari, V.; Kumari, A.; Chaudhary, E.; Sharma, A.; Ali, U.; Garg, M. Protective and defensive role of anthocyanins under plant abiotic and biotic stresses: An emerging application in sustainable agriculture. J. Biotechnol. 2023, 361, 12–29. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.; Visser, R.G.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Lewis, C.E.; Walker, J.R.; Lancaster, J.E.; Sutton, K.H. Determination of anthocyanins, flavonoids and phenolic acids in potatoes. I: Coloured cultivars of Solanum tuberosum L. J. Sci. Food Agric. 1998, 77, 45–57. [Google Scholar] [CrossRef]
- Naito, K.; Umemura, Y.; Mori, M.; Sumida, T.; Okada, T.; Takamatsu, N.; Okawa, Y.; Hayashi, K.; Saito, N.; Honda, T. Acylated pelargonidin glycosides from a red potato. Phytochemistry 1998, 47, 109–112. [Google Scholar] [CrossRef]
- Azuma, A.; Ban, Y.; Sato, A.; Kono, A.; Shiraishi, M.; Yakushiji, H.; Kobayashi, S. MYB diplotypes at the color locus affect the ratios of tri/di-hydroxylated and methylated/non-methylated anthocyanins in grape berry skin. Genomes 2015, 11, 31. [Google Scholar] [CrossRef]
- Hugueney, P.; Provenzano, S.; Verries, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 2009, 150, 2057–2070. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Hugueney, P.; Verriès, C.; This, P.; Ageorges, A. Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biol. 2011, 11, 179. [Google Scholar] [CrossRef] [PubMed]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Wei, L.; Zhao, X.; Gu, X.; Peng, J.; Song, W.; Deng, B.; Cao, Y.; Hu, S. Genome-wide identification and expression analysis of Dendrocalamus farinosus CCoAOMT gene family and the role of DfCCoAOMT14 involved in lignin synthesis. Int. J. Mol. Sci. 2023, 24, 8965. [Google Scholar] [CrossRef]
- Lu, S.; Zhuge, Y.; Hao, T.; Liu, Z.; Zhang, M.; Fang, J. Systematic analysis reveals O-methyltransferase gene family members involved in flavonoid biosynthesis in grape. Plant Physiol. Biochem. 2022, 173, 33–45. [Google Scholar] [CrossRef]
- Joshi, C.P.; Chiang, V.L. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol. 1998, 37, 663–674. [Google Scholar] [CrossRef]
- Liao, Z.; Liu, X.; Zheng, J.; Zhao, C.; Wang, D.; Xu, Y.; Sun, C. A multifunctional true caffeoyl coenzyme AO-methyltransferase enzyme participates in the biosynthesis of polymethoxylated flavones in citrus. Plant Mol. Biol. 2023, 192, 2049–2066. [Google Scholar]
- Gauthier, A.; Gulick, P.J.; Ibrahim, R.K. Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds. Arch. Biochem. Biophys. 1998, 351, 243–249. [Google Scholar] [CrossRef]
- Berim, A.; Hyatt, D.C.; Gang, D.R. A set of regioselective O-methyltransferases gives rise to the complex pattern of methoxylated flavones in sweet basil. Plant Physiol. 2012, 160, 1052–1069. [Google Scholar] [CrossRef]
- Zhou, J.M.; Seo, Y.W.; Ibrahim, R.K.; Biochemistry. Biochemical characterization of a putative wheat caffeic acid O-methyltransferase. Plant Physiol. Biochem. 2009, 47, 322–326. [Google Scholar] [CrossRef]
- Kim, B.; Lee, H.; Park, Y.; Lim, Y.; Ahn, J.H. Characterization of an O-methyltransferase from soybean. Plant Physiol. Biochem. 2006, 44, 236–241. [Google Scholar] [CrossRef]
- Cacace, S.; Schröder, G.; Wehinger, E.; Strack, D.; Schmidt, J.; Schröder, J. A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry 2003, 62, 127–137. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; Hou, X.; Chen, J. Evolution and Analysis of Caffeic Acid Transferase (COMT) in Seed Plants. Biochem. Genet. 2024, 62, 1953–1976. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, L.; Seoka, M.; Nakata, A.; Yahata, M.; Shimada, T.; Fujii, H.; Endo, T.; Yoshioka, T.; Kan, T.J. Characterization of a caffeic acid 8-O-methyltransferase from citrus and its function in nobiletin biosynthesis. Agric. Food Chem. 2021, 70, 543–553. [Google Scholar] [CrossRef]
- Yan, Y.; Sun, S.; Zhao, N.; Yang, W.; Shi, Q.; Gong, B. COMT1 overexpression resulting in increased melatonin biosynthesis contributes to the alleviation of carbendazim phytotoxicity and residues in tomato plants. Environ. Pollut. 2019, 252, 51–61. [Google Scholar] [CrossRef]
- Do, C.-T.; Pollet, B.; Thévenin, J.; Sibout, R.; Denoue, D.; Barrière, Y.; Lapierre, C.; Jouanin, L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 2007, 226, 1117–1129. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Y.H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Mol. Genet. Genom. 2010, 51, 144–163. [Google Scholar] [CrossRef]
- Rakoczy, M.; Femiak, I.; Alejska, M.; Figlerowicz, M.; Podkowinski, J. Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol. Genet. Genom. 2018, 293, 1077–1089. [Google Scholar] [CrossRef]
- Yang, G.; Pan, W.; Zhang, R.; Pan, Y.; Guo, Q.; Song, W.; Zheng, W.; Nie, X. Genome-wide identification and characterization of caffeoyl-coenzyme A O-methyltransferase genes related to the Fusarium head blight response in wheat. BMC Genom. 2021, 22, 504. [Google Scholar] [CrossRef] [PubMed]
- Ibdah, M.; Zhang, X.H.; Schmidt, J.; Vogt, T. A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. Biol. Chem. 2003, 278, 43961–43972. [Google Scholar] [CrossRef]
- Sung, S.H.; Kim, B.-G.; Chong, Y.; Ahn, J.H. Characterization of phenylpropanoid O-methyltransferase from rice: Molecular basis for the different reactivity toward different substrates. Plant Biol. 2011, 54, 314–320. [Google Scholar] [CrossRef]
- Zhong, R.; Morrison, W.H., II.; Himmelsbach, D.S.; Poole, F.L.; Ye, Z.H. Essential role of caffeoyl coenzyme AO-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 2000, 124, 563–578. [Google Scholar] [CrossRef]
- Zhong, R.; Morrison, W.H., III.; Negrel, J.; Ye, Z.H. Dual methylation pathways in lignin biosynthesis. Plant Cell 1998, 10, 2033–2045. [Google Scholar] [CrossRef]
- Gomez Roldan, M.V.; Outchkourov, N.; van Houwelingen, A.; Lammers, M.; Romero de la Fuente, I.; Ziklo, N.; Aharoni, A.; Hall, R.D.; Beekwilder, J. An O-methyltransferase modifies accumulation of methylated anthocyanins in seedlings of tomato. Plant J. 2014, 80, 695–708. [Google Scholar] [CrossRef]
- Cheng, J.; Wei, G.; Zhou, H.; Gu, C.; Vimolmangkang, S.; Liao, L.; Han, Y. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 2014, 166, 1044–1058. [Google Scholar] [CrossRef]
- Itoh, N.; Iwata, C.; Toda, H. Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa. BMC Plant Biol. 2016, 16, 180. [Google Scholar] [CrossRef]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.e.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wei, K.; Chen, H. Global identification, structural analysis and expression characterization of cytochrome P450 monooxygenase superfamily in rice. BMC Genom. 2018, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, T.; Ma, Y.; Wang, N.; Wang, W.; Tang, J.; Zhang, C.; Hou, X.; Hou, H. Ectopic Expression of BcCUC2 Involved in Sculpting the Leaf Margin Serration in Arabidopsis thaliana. Genes 2023, 14, 1272. [Google Scholar] [CrossRef]
- Lam, K.C.; Ibrahim, R.K.; Behdad, B.; Dayanandan, S. Structure, function, and evolution of plant O-methyltransferases. Genome 2007, 50, 1001–1013. [Google Scholar] [CrossRef]
- Struck, A.W.; Thompson, M.L.; Wong, L.S.; Micklefield, J. S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemInform 2012, 13, 2642–2655. [Google Scholar]
- Provenzano, S.; Spelt, C.; Hosokawa, S.; Nakamura, N.; Brugliera, F.; Demelis, L.; Geerke, D.P.; Schubert, A.; Tanaka, Y.; Quattrocchio, F. Genetic control and evolution of anthocyanin methylation. Plant Physiol. 2014, 165, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Koirala, N.; Thuan, N.H.; Ghimire, G.P.; Van Thang, D.; Sohng, J.K. Methylation of flavonoids: Chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzym. Microb. Technol. 2016, 86, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Widiez, T.; Hartman, T.G.; Dudai, N.; Yan, Q.; Lawton, M.; Havkin-Frenkel, D.; Belanger, F.C. Functional characterization of two new members of the caffeoyl CoA O-methyltransferase-like gene family from Vanilla planifolia reveals a new class of plastid-localized O-methyltransferases. Plant Mol. Biol. 2011, 76, 475–488. [Google Scholar] [CrossRef]
- Pakusch, A.E.; Kneusel, R.E.; Matern, U. S-adenosyl-L-methionine: Trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch. Biochem. Biophys. 1989, 271, 488–494. [Google Scholar] [CrossRef]
- Chen, C.; Meyermans, H.; Burggraeve, B.; De Rycke, R.M.; Inoue, K.; De Vleesschauwer, V.; Steenackers, M.; Van Montagu, M.C.; Engler, G.J.; Boerjan, W.A. Cell-specific and conditional expression of caffeoyl-coenzyme A-3-O-methyltransferase in poplar. Plant Physiol. 2000, 123, 853–868. [Google Scholar] [CrossRef]
- Chun, H.J.; Lim, L.H.; Cheong, M.S.; Baek, D.; Park, M.S.; Cho, H.M.; Lee, S.H.; Jin, B.J.; No, D.H.; Cha, Y.J. Arabidopsis CCoAOMT1 plays a role in drought stress response via ROS-and ABA-dependent manners. Plants 2021, 10, 831. [Google Scholar] [CrossRef]
- Wang, P.; Guo, L.; Morgan, J.; Dudareva, N.; Chapple, C. Transcript and metabolite network perturbations in lignin biosynthetic mutants of Arabidopsis. Plant Physiol. 2022, 190, 2828–2846. [Google Scholar] [CrossRef] [PubMed]
- Fellenberg, C.; Böttcher, C.; Vogt, T. Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry 2009, 70, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.X.; Wu, T.T.; Ding, C.H.; Zhou, P.C.; Chen, Z.Z.; Gou, J.Y. SAHH and SAMS form a methyl donor complex with CCoAOMT7 for methylation of phenolic compounds. Biochem. Biophys. Res. Commun. 2019, 520, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Wong, D.C.J.; Lopez Gutierrez, R.; Gambetta, G.A.; Castellarin, S.D. Genome-wide analysis of cis-regulatory element structure and discovery of motif-driven gene co-expression networks in grapevine. DNA Res. 2017, 24, 311–326. [Google Scholar] [CrossRef]
- Leyva, A.; Liang, X.; Pintor-Toro, J.A.; Dixon, R.A.; Lamb, C.J. cis-Element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. Plant Cell 1992, 4, 263–271. [Google Scholar]
- Lacombe, E.; Van Doorsselaere, J.; Boerjan, W.; Boudet, A.M.; Grima-Pettenati, J. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein–DNA complex formation. Plant Cell 1992, 4, 263–271. [Google Scholar] [CrossRef]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef]
- Patzlaff, A.; McInnis, S.; Courtenay, A.; Surman, C.; Newman, L.J.; Smith, C.; Bevan, M.W.; Mansfield, S.; Whetten, R.W.; Sederoff, R.R. Characterisation of a pine MYB that regulates lignification. Plant J. 2003, 36, 743–754. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Ageorges, A.; Fernandez, L.; Vialet, S.; Merdinoglu, D.; Terrier, N.; Romieu, C. Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 2006, 170, 372–383. [Google Scholar] [CrossRef]
- Giordano, D.; Provenzano, S.; Ferrandino, A.; Vitali, M.; Pagliarani, C.; Roman, F.; Cardinale, F.; Castellarin, S.D.; Schubert, A. Characterization of a multifunctional caffeoyl-CoA O-methyltransferase activated in grape berries upon drought stress. Plant Physiol. Biochem. 2016, 101, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, L.; Xu, L.; Zeng, L.; Xu, J. Genome-wide identification of the AOMT gene family in wax apple and functional characterization of SsAOMTs to anthocyanin methylation. Plant Sci. 2023, 14, 1213642. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Number of Amino Acids | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| StCCoAOMT1 | Soltu.DM.01G047320 | 248 | 27.99 | 5.53 | 37.62 | 101.45 | −0.227 | Cytoplasm |
| StCCoAOMT2 | Soltu.DM.02G028520 | 242 | 27.33 | 5.29 | 42.1 | 97.15 | −0.251 | Cytoplasm |
| StCCoAOMT3 | Soltu.DM.02G028530 | 175 | 19.69 | 5.18 | 40.75 | 109.26 | −0.045 | Cytoplasm |
| StCCoAOMT4 | Soltu.DM.02G028540 | 242 | 27.25 | 5.29 | 39.05 | 97.15 | −0.244 | Cytoskeleton |
| StCCoAOMT5 | Soltu.DM.02G028550 | 242 | 27.28 | 5.3 | 39.05 | 97.56 | −0.245 | Cytoskeleton |
| StCCoAOMT6 | Soltu.DM.03G003400 | 441 | 49.72 | 5.13 | 42.69 | 106.37 | −0.126 | Cytoplasm |
| StCCoAOMT7 | Soltu.DM.04G025040 | 244 | 27.57 | 5.3 | 37.84 | 98.73 | −0.205 | Cytoplasm |
| StCCoAOMT8 | Soltu.DM.04G025050 | 282 | 31.76 | 5.32 | 46.16 | 95.11 | −0.193 | Chloroplast |
| StCCoAOMT9 | Soltu.DM.08G001680 | 288 | 32.22 | 7.08 | 36.53 | 97.74 | −0.106 | Chloroplast |
| StCCoAOMT10 | Soltu.DM.09G025040 | 234 | 26.33 | 5.51 | 35.93 | 103.33 | −0.118 | Cytoplasm |
| StCCoAOMT11 | Soltu.DM.10G014940 | 245 | 27.85 | 5.14 | 34.38 | 100.73 | −0.271 | Cytoplasm |
| StCCoAOMT12 | Soltu.DM.12G013090 | 123 | 13.72 | 4.93 | 55.56 | 111.71 | −0.031 | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Sheng, S.; Wang, T.; Song, J.; Wang, D.; Zhang, Y.; Cheng, J.; Zheng, T.; Lv, Z.; Zhu, X.; et al. Genome-Wide Characterization of Solanum tuberosum CCoAOMT Gene Family and Identification of StCCoAOMT Genes Involved in Anthocyanin Biosynthesis. Genes 2024, 15, 1466. https://doi.org/10.3390/genes15111466
Peng Y, Sheng S, Wang T, Song J, Wang D, Zhang Y, Cheng J, Zheng T, Lv Z, Zhu X, et al. Genome-Wide Characterization of Solanum tuberosum CCoAOMT Gene Family and Identification of StCCoAOMT Genes Involved in Anthocyanin Biosynthesis. Genes. 2024; 15(11):1466. https://doi.org/10.3390/genes15111466
Chicago/Turabian StylePeng, Yaxuan, Suao Sheng, Tongtong Wang, Jiafeng Song, Daijuan Wang, Yixuan Zhang, Jielan Cheng, Tingting Zheng, Zhaoyan Lv, Xiaobiao Zhu, and et al. 2024. "Genome-Wide Characterization of Solanum tuberosum CCoAOMT Gene Family and Identification of StCCoAOMT Genes Involved in Anthocyanin Biosynthesis" Genes 15, no. 11: 1466. https://doi.org/10.3390/genes15111466
APA StylePeng, Y., Sheng, S., Wang, T., Song, J., Wang, D., Zhang, Y., Cheng, J., Zheng, T., Lv, Z., Zhu, X., & Hou, H. (2024). Genome-Wide Characterization of Solanum tuberosum CCoAOMT Gene Family and Identification of StCCoAOMT Genes Involved in Anthocyanin Biosynthesis. Genes, 15(11), 1466. https://doi.org/10.3390/genes15111466





