Quantitative Trait Loci Mapping for Powdery Mildew Resistance in Wheat Genetic Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Identification of Pm Resistance
2.3. Microscopic Observation of Pm Spore Hyphae
2.4. Genotyping and Linkage Map Construction
2.5. QTL Analysis
2.6. Marker Development and Evaluation of Markers for MAS
3. Results
3.1. Pm Resistance in DH Population
3.2. Construction of Genetic Map
3.3. Genetic Loci of Pm Resistance
3.4. Molecular Markers for MAS
3.5. The Additive Effect of the QTL on Pm Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, Y.J.; Han, T.H. China’s wheat yield increase potential and realization path during the “14th Five-Year Plan” Period. Issu. Agric. Econ. 2021, 7, 38–46. [Google Scholar]
- Cowger, C.; Miranda, L.R.; Griffey, C.; Hall, M. Wheat powdery mildew. In Disease Resistance in Wheat; Sharma, I., Ed.; CABI: Oxfordshire, UK, 2012; pp. 84–119. [Google Scholar]
- Everts, K.L.; Leath, S.; Finney, P.L. Impact of powdery mildew and leaf rust on milling and baking quality of soft red winter wheat. Plant Dis. 2001, 85, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhou, Y.l.; Duan, X.Y.; Cao, X.R.; Zhou, Y.F. Sensitivity of Blumeria graminis f. sp. tritici isolates to triadimefon and fenpropidin in China in 2011. Acta Phytopathol. Sin. 2013, 43, 430–434. [Google Scholar]
- Li, M.; Dong, L.; Li, B.; Wang, Z.; Xie, J.; Qiu, D.; Li, Y.; Shi, W.; Yang, L.; Wu, Q.; et al. A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 2020, 228, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Yu, Z.Y.; Wang, D.M.; Xiao, L.N.; Su, F.Y.; Mu, Y.J.; Zheng, J.P.; Li, L.Z.; Yin, Y.; Yu, T.Y.; et al. Characterization and identification of the powdery mildew resistance gene in wheat breeding line ShiCG15-009. BMC Plant Biol. 2023, 23, 113. [Google Scholar] [CrossRef]
- He, H.G.; Liu, R.K.; Ma, P.T.; Du, H.N.; Zhang, H.H.; Wu, Q.H.; Yang, L.J.; Gong, S.J.; Liu, T.L.; Huo, N.X.; et al. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of greek durum wheat TRI 1796. Theor. Appl. Genet. 2021, 134, 53–62. [Google Scholar] [CrossRef]
- Parks, R.; Carbone, I.; Murphy, J.P.; Marshall, D.; Cowger, C. Virulence structure of the eastern U.S. wheat powdery mildew population. Plant Dis. 2008, 92, 1074–1082. [Google Scholar] [CrossRef]
- Cowger, C.; Mehra, L.; Arellano, C.; Meyers, E.; Murphy, J.P. Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern United States. Phytopathology 2018, 108, 402–411. [Google Scholar]
- Wang, B.; Meng, T.; Xiao, B.; Yu, T.; Yue, T.; Jin, Y.; Ma, P. Fighting wheat powdery mildew: From genes to fields. Theor. Appl. Genet. 2023, 136, 196. [Google Scholar] [CrossRef]
- Jin, Y.; Shi, F.; Liu, W.; Fu, X.; Gu, T.; Han, G.; Shi, Z.; Sheng, Y.; Xu, H.; Li, L.; et al. Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2978 wheat accessions. Plant Dis. 2021, 105, 3900–3908. [Google Scholar] [CrossRef]
- Jia, J.Z.; Miller, T.E.; Reader, S.M.; Gale, M.D. RFLP markers of powdery mildew resistance gene Pm12 in Wheat. Sci. China (Ser. B) 1993, 6, 589–594. [Google Scholar]
- Liu, Z.Y.; Sun, Q.X.; Li, H.J.; Ni, Z.F.; Yang, Z.M.; Tang, B.R.; Yang, A.D.; Jia, X. Molecular identification and marker-assisted selection of Pm21 gene conferring resistance to powdery mildew in wheat. Acta Genet. Sin. 1999, 26, 673–682+743. [Google Scholar] [PubMed]
- Cao, A.; Xing, L.; Wang, X.; Yang, X.; Wang, W.; Sun, Y.; Qian, C.; Ni, J.; Chen, Y.; Liu, D.; et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhu, S.; Zhao, R.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 2018, 11, 879–882. [Google Scholar] [CrossRef]
- Fu, B.S.; Liu, Y.; Zhang, Q.F.; Wu, X.Y.; Gao, H.D.; Cai, S.B.; Dai, T.B.; Wu, J.Z. Development of markers closely linked with wheat powdery mildew resistance gene Pm48. Acta Agron. Sin. 2017, 43, 307–312. [Google Scholar] [CrossRef]
- Chen, F.; Qiao, L.Y.; Li, R.; Liu, C.; Guo, H.J.; Zhang, S.W.; Chang, L.F.; Li, D.F.; Yan, X.T. Genetic analysis and chromosomal localization of powdery mildew resistance gene in wheat germplasm CH1357. Acta Agron. Sin. 2019, 45, 1503–1510. [Google Scholar]
- Zhang, D.Y.; Zhu, K.Y.; Dong, L.L.; Liang, Y.; Li, G.Q.; Fang, T.L.; Guo, G.H.; Wu, Q.H.; Xie, J.Z.; Chen, Y.X.; et al. Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J. 2019, 7, 761–770. [Google Scholar]
- Dong, Y.; Xu, D.; Xu, X.; Ren, Y.; Gao, F.; Song, J.; Jia, A.; Hao, Y.; He, Z.; Xia, X. Fine mapping of QPm.caas-3BS, a stable QTL for adult-plant resistance to powdery mildew in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2022, 135, 1083–1099. [Google Scholar] [CrossRef]
- Briggle, L.W. Near-isogenic lines of wheat with genes for resistance to Erysiphe graminis f. sp. tritici. Crop Sci. 1969, 9, 70–72. [Google Scholar] [CrossRef]
- Ma, P.T.; Xu, H.X.; Xu, Y.F.; Li, L.H.; Qie, Y.M.; Luo, Q.L.; Zhang, X.T.; Li, X.Q.; Zhou, Y.L.; An, D.G. Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 2015, 128, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yi, Y.; Ma, P.; Qie, Y.; Fu, X.; Xu, Y.; Zhang, X.; An, D. Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 2015, 128, 2077–2084. [Google Scholar] [CrossRef]
- Yao, M. Genetic Analysis of Powdery Mildew Resistance and Dwarfism in Pubing7-4, a Germplasm Derived from Wheat-Agropyron Cristatum Hybridization; CAAS: Beijing, China, 2015. [Google Scholar]
- Wang, Z.L.; Li, L.H.; He, Z.H.; Duan, X.Y.; Zhou, Y.L.; Chen, X.M.; Lillemo, M.; Singh, R.P.; Wang, H.; Xia, X.C. Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 2005, 89, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.H.; Zhang, L.Y.; Wang, J.K. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap® 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Population; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Winfield, M.O.; Allen, A.M.; Burridge, A.J.; Barker, G.L.; Benbow, H.R.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; Scopes, G.; et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 2016, 14, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Prasanna, R.B.; Timothy, J.C.; Lonardi, S. Efficient and Accurate Construction of Genetic Linkage Maps from the Minimum Spanning Tree of a Graph. PLOS Genet. 2008, 4, e1000212. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, S.C.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC, USA. Available online: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 1 August 2012).
- Zhai, H.J.; Feng, Z.Y.; Li, J.; Liu, X.Y.; Xiao, S.H.; Ni, Z.F.; Sun, Q.X. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSRbased linkage map. Front. Plant Sci. 2016, 7, 1617. [Google Scholar] [CrossRef]
- Summers, R.W.; Brown, J.K.M. Constraints on breeding for disease resistance in commercially competitive wheat cultivars. Plant Pathol. 2013, 62, 115–121. [Google Scholar] [CrossRef]
- Rabinovich, S. Importance of wheat-rye translocation for breeding modern cultivars of Triticum aestivum L. Euphytica 1998, 100, 323–340. [Google Scholar] [CrossRef]
- Chai, J.F.; Liu, X.; Jia, J.Z. Homoeologous cloning of ω-secalin gene family in a wheat 1BL/1RS translocation. Cell Res. 2005, 15, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Duan, X.; Wang, P.; Li, X.; Yuan, X.; Wang, Z.; Wan, L.; Yang, G.; Hong, D. Comprehensive speed breeding: A high-throughput and rapid generation system for long-day crops. Plant Biotechnol. J. 2022, 20, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022, 602, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.S. Chinese Wheat Improvement and Pedigree Analysis; China Agriculture Press: Beijing, China, 2003. [Google Scholar]
- Wu, J.P.; Xu, G.Y.; Li, G.Q.; Qiu, S.Y. Analysis of genetic basis and breeding model of Jinmai 33. J. Triticeae Crops 1996, 3, 8–9+26. [Google Scholar]
- Zhao, J.J.; Zheng, X.W.; Qiao, L.; Yang, C.K.; Wu, B.B.; He, Z.M.; Tang, Y.Q.; Li, G.R.; Yang, Z.J.; Zheng, J.; et al. Genome-wide association study reveals structural chromosome variations with phenotypic effects in wheat (Triticum aestivum L.). Plant J. 2022, 112, 1447–1461. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Li, G.; Zhang, X.; Li, X.; Guo, H.; Gong, W.; Jia, J.; Qiao, L.; Ren, Y.; Yang, Z.; et al. Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS ONE 2014, 9, e113455. [Google Scholar] [CrossRef]
- Feng, D.Q. Characteristics and high-yield cultivation techniques of Yannong19. Agric. Technol. Ser. 2011, 28, 585. [Google Scholar]
- Rong, J.K.; Millet, E.; Manisterski, J.; Feldman, M. A new powdery mildew resistance gene: Introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 2000, 115, 121–126. [Google Scholar] [CrossRef]
- Sun, H.; Hu, J.; Song, W.; Qiu, D.; Cui, L.; Wu, P.; Zhang, H.; Liu, H.; Yang, L.; Qu, Y.; et al. Pm61: A recessive gene for resistance to powdery mildew in wheat landrace Xuxusanyuehuang identified by comparative genomics analysis. Theor. Appl. Genet. 2018, 131, 2085–2097. [Google Scholar] [CrossRef]
- Hua, W.; Liu, Z.; Zhu, J.; Xie, C.; Yang, T.; Zhou, Y.; Duan, X.; Sun, Q.; Liu, Z. Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 223–230. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X.; Hu, J.; Wu, P.; Qiu, D.; Qu, Y.; Xie, J.; Wu, Q.; Zhang, H.; Yang, L.; et al. Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis. 2020, 104, 743–751. [Google Scholar] [CrossRef] [PubMed]
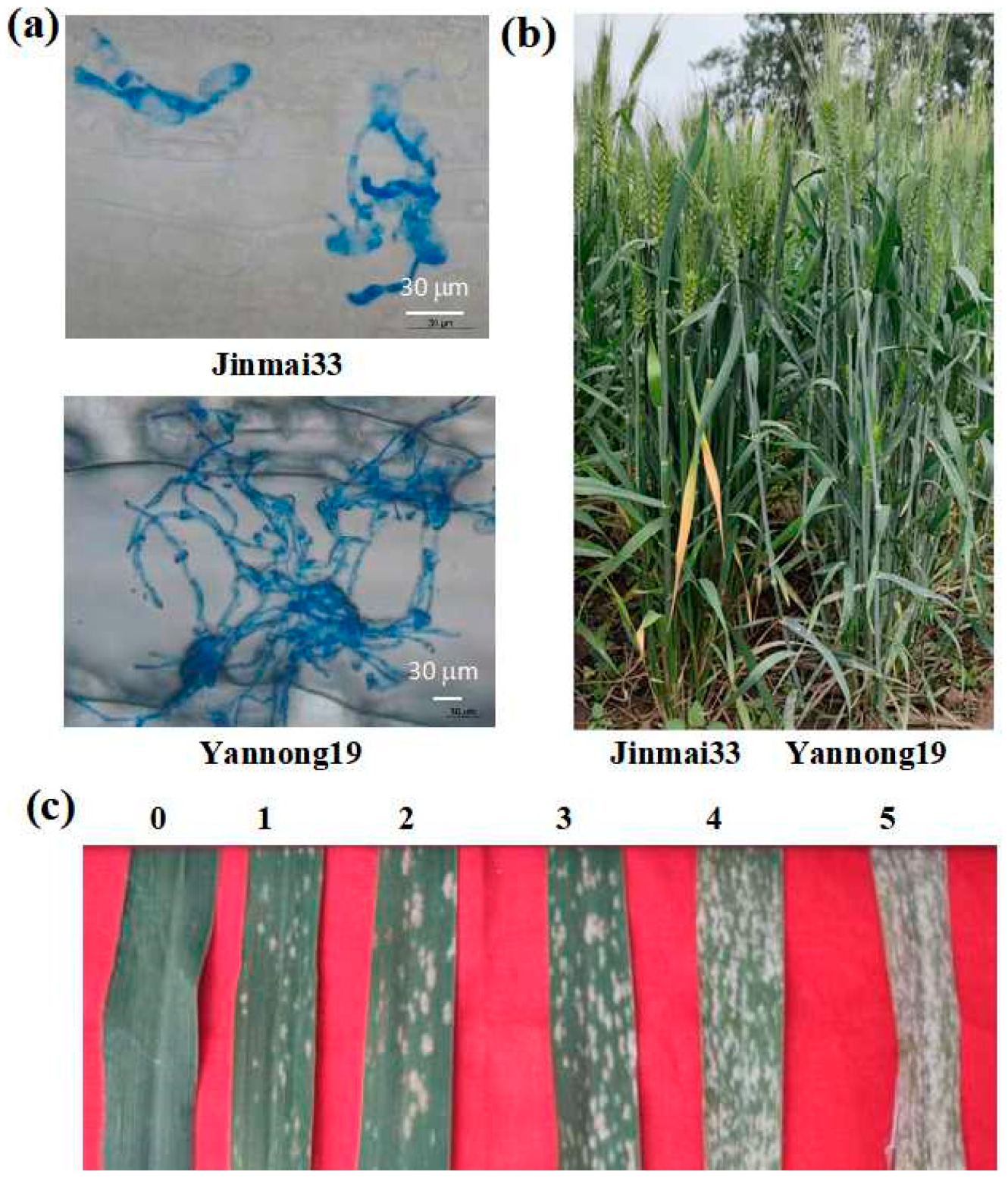
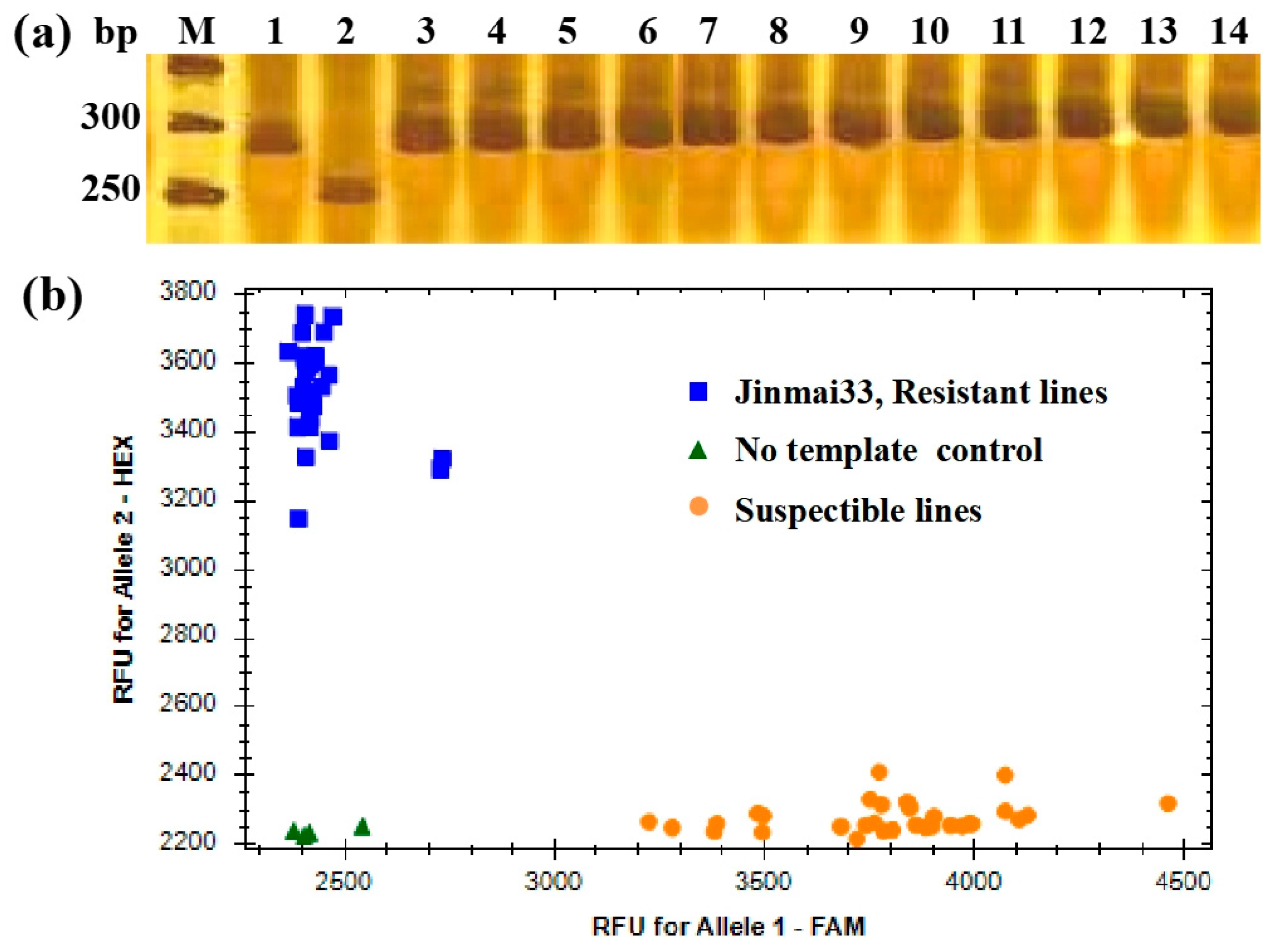
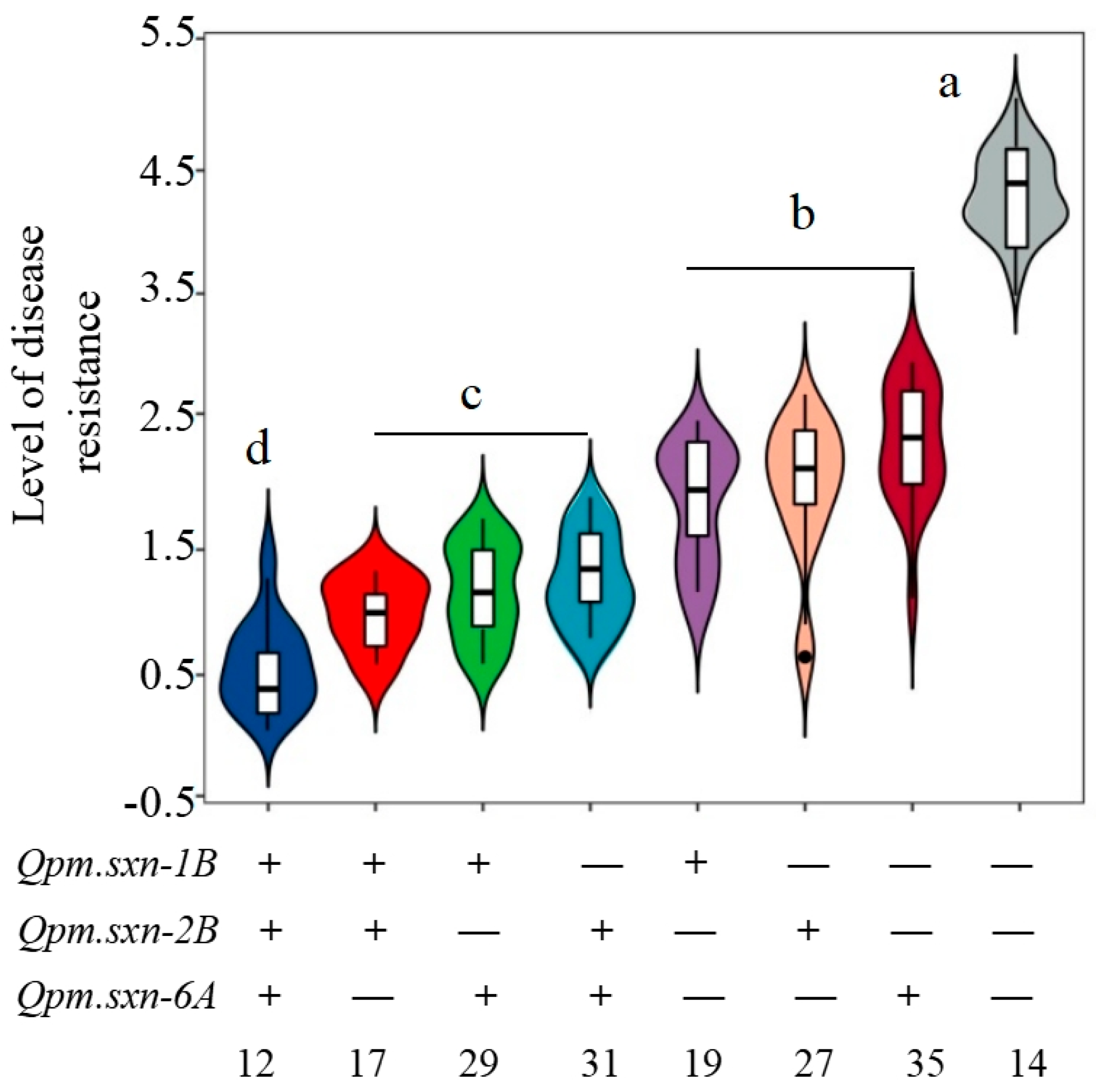
| Parent/Population | Total Number of Plants | Disease Resistant | Disease Susceptibility | Expected Separation Ratio | χ2 |
|---|---|---|---|---|---|
| Jinmai 33 | 20 | 20 | 0 | ||
| Yannong 19 | 20 | 0 | 20 | ||
| Jinmai 33 × Yannong 19 (E1) | 184 | 157 | 27 | 7:1 | 0.80 |
| Jinmai 33 × Yannong 19 (E2) | 184 | 162 | 22 | 7:1 | 0.05 |
| Jinmai 33 × Yannong 19 (E3) | 184 | 158 | 26 | 7:1 | 0.45 |
| Chromosomes | Number of Loci | Length (cM) | Density (cM)/Marker |
|---|---|---|---|
| 1A | 160 | 228.02 | 1.43 |
| 2A | 302 | 401.25 | 1.33 |
| 3A | 206 | 355.42 | 1.73 |
| 4A | 231 | 377.97 | 1.64 |
| 5A | 70 | 104.91 | 1.50 |
| 6A | 143 | 260.59 | 1.82 |
| 7A | 352 | 454.54 | 1.29 |
| 1B | 347 | 413.53 | 1.19 |
| 2B | 328 | 406.86 | 1.24 |
| 3B | 137 | 259.43 | 1.89 |
| 4B | 80 | 170.39 | 2.13 |
| 5B | 202 | 308.98 | 1.53 |
| 6B | 219 | 364.51 | 1.66 |
| 7B | 359 | 448.68 | 1.25 |
| 1D | 118 | 211.22 | 1.79 |
| 2D | 104 | 208.70 | 2.01 |
| 3D | 122 | 224.07 | 1.84 |
| 4D | 45 | 102.23 | 2.27 |
| 5D | 104 | 261.79 | 2.52 |
| 6D | 94 | 156.55 | 1.67 |
| 7D | 144 | 201.29 | 1.40 |
| A genome | 1464 | 2182.70 | 1.49 |
| B genome | 1672 | 2372.38 | 1.42 |
| D genome | 731 | 1365.85 | 1.87 |
| Total | 3867 | 5920.93 | 1.53 |
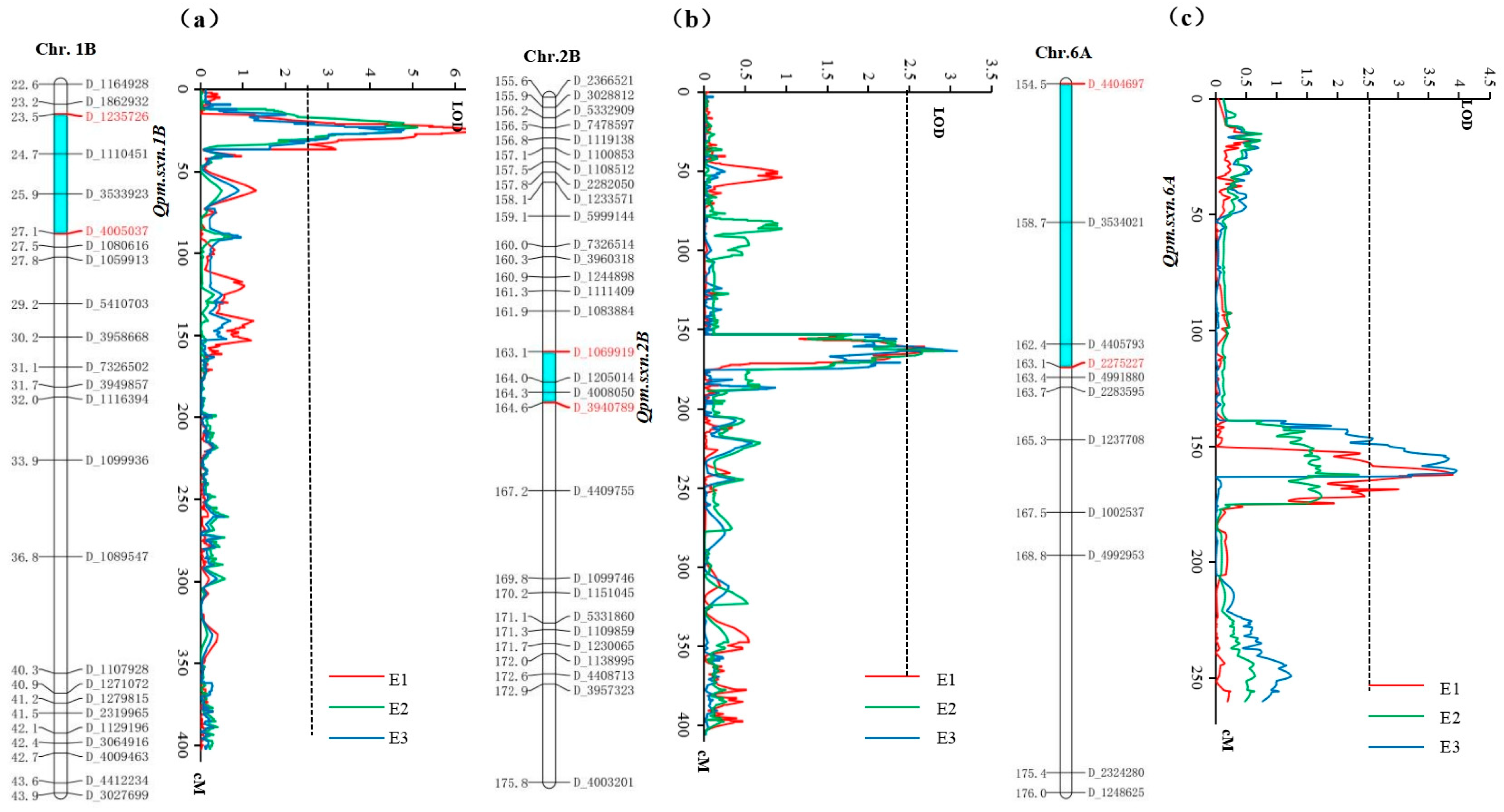
| QTL | Env. | Chr. | Flanking Markers | Genetic Distance (cM) | LOD Value | Phenotypic Variance (PVE) (%) | Additive Effect |
|---|---|---|---|---|---|---|---|
| Qpm.sxn-1B | E1 | 1B | D_4005037-D_1235726 | 23.48–27.14 | 6.32 | 11.56 | −0.40 |
| E2 | 1B | D_4005037-D_1235726 | 23.48–27.14 | 5.03 | 9.37 | −0.37 | |
| E3 | 1B | D_4005037-D_1235726 | 23.48–27.14 | 7.56 | 13.25 | −0.56 | |
| Qpm.sxn-2B | E1 | 2B | D_1069919-D_3940789 | 163.06–164.60 | 2.74 | 5.18 | −0.27 |
| E2 | 2B | D_1069919-D_3940789 | 163.06–164.60 | 3.16 | 5.23 | −0.29 | |
| E3 | 2B | D_1069919-D_3940789 | 163.06–164.60 | 3.02 | 4.98 | −0.21 | |
| Qpm.sxn-4B | E2 | 4B | D_1395268-D_1234524 | 39.83–41.39 | 2.69 | 4.59 | 0.27 |
| Qpm.sxn-4D | E1 | 4D | D_1121275-D_4991578 | 106.35–117.92 | 4.17 | 7.89 | 0.33 |
| Qpm.sxn-5A.1 | E2 | 5A | D_1320425-D_1013062 | 123.58–114.62 | 4.47 | 7.92 | 0.34 |
| Qpm.sxn-6A | E1 | 6A | D_4404697-D_2275227 | 154.54–163.06 | 3.68 | 7.13 | 0.32 |
| E2 | 6A | D_4404697-D_2275227 | 154.54–163.06 | 3.92 | 7.65 | 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Qiu, Y.; Cao, M.; Bi, H.; Si, G.; Meng, X. Quantitative Trait Loci Mapping for Powdery Mildew Resistance in Wheat Genetic Population. Genes 2024, 15, 1438. https://doi.org/10.3390/genes15111438
Zhao Z, Qiu Y, Cao M, Bi H, Si G, Meng X. Quantitative Trait Loci Mapping for Powdery Mildew Resistance in Wheat Genetic Population. Genes. 2024; 15(11):1438. https://doi.org/10.3390/genes15111438
Chicago/Turabian StyleZhao, Zhiyong, Yuliang Qiu, Menglin Cao, Hongyuan Bi, Guan Si, and Xianghai Meng. 2024. "Quantitative Trait Loci Mapping for Powdery Mildew Resistance in Wheat Genetic Population" Genes 15, no. 11: 1438. https://doi.org/10.3390/genes15111438
APA StyleZhao, Z., Qiu, Y., Cao, M., Bi, H., Si, G., & Meng, X. (2024). Quantitative Trait Loci Mapping for Powdery Mildew Resistance in Wheat Genetic Population. Genes, 15(11), 1438. https://doi.org/10.3390/genes15111438






