A 3D Bioprinting Approach to Studying Retinal Müller Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Post-Printing Viability
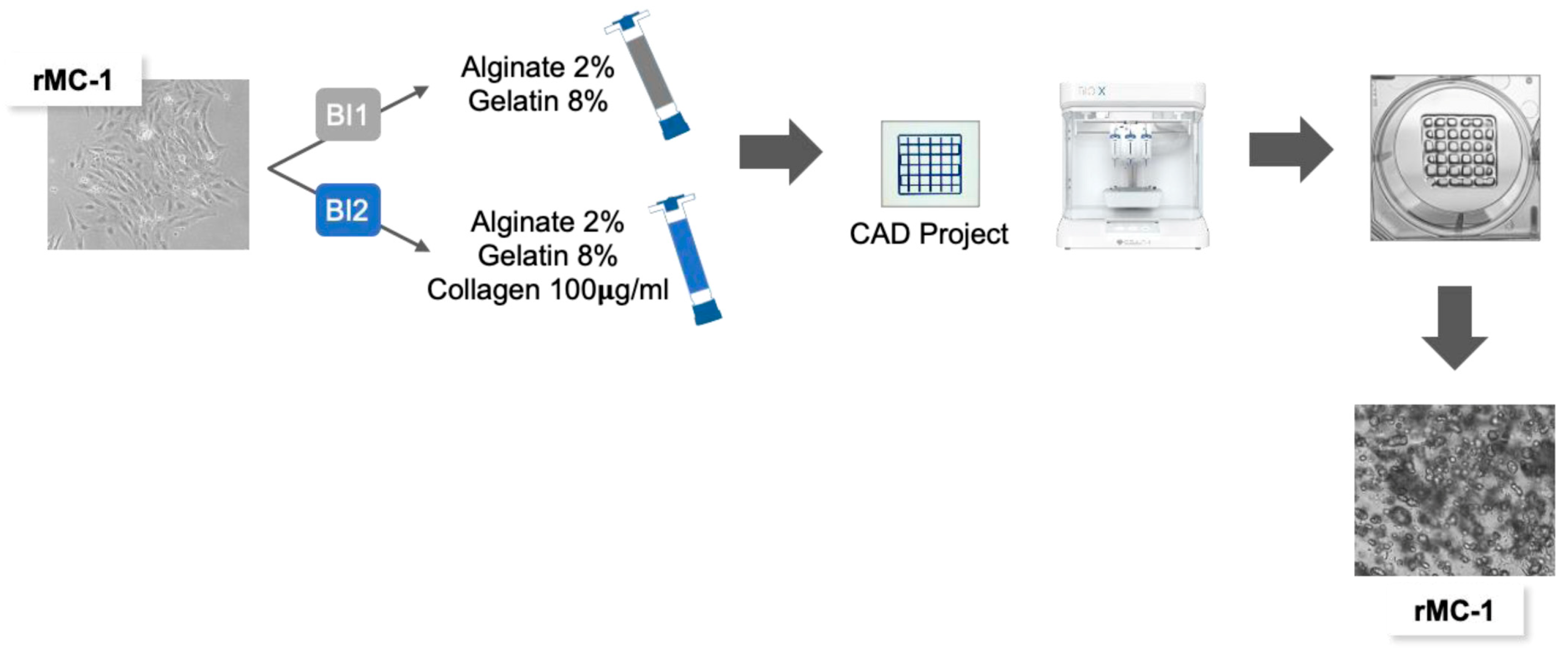
2.2. Sodium Alginate–Gelatin-Based Bioinks Preparation
2.3. Bioprinting Process
2.4. Morphological Analysis and Imaging
2.5. Protein Extraction and Western Blot Analysis
3. Results
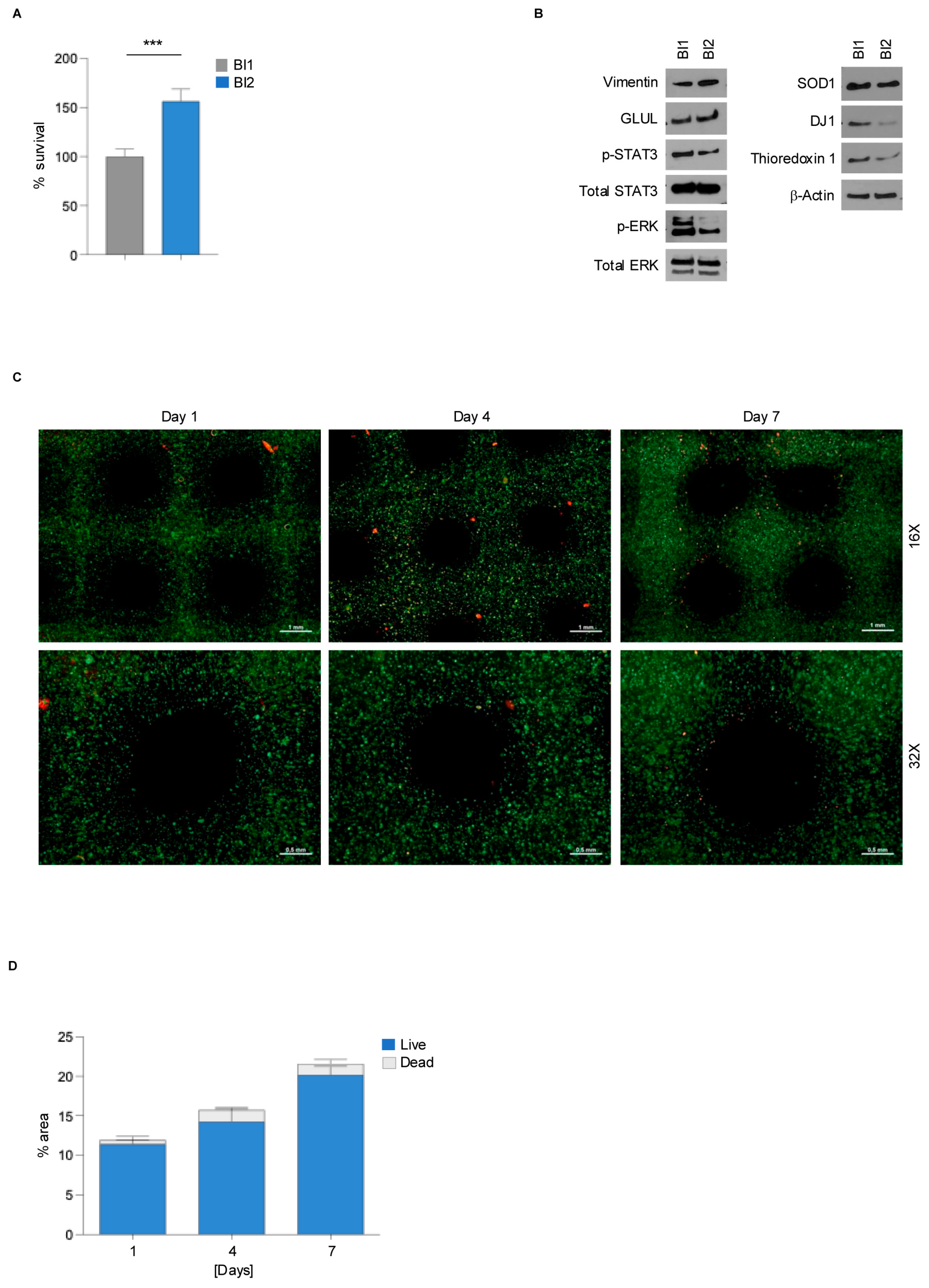
3.1. Collagen-Based Bioink Supports rMC-1 Cells Viability
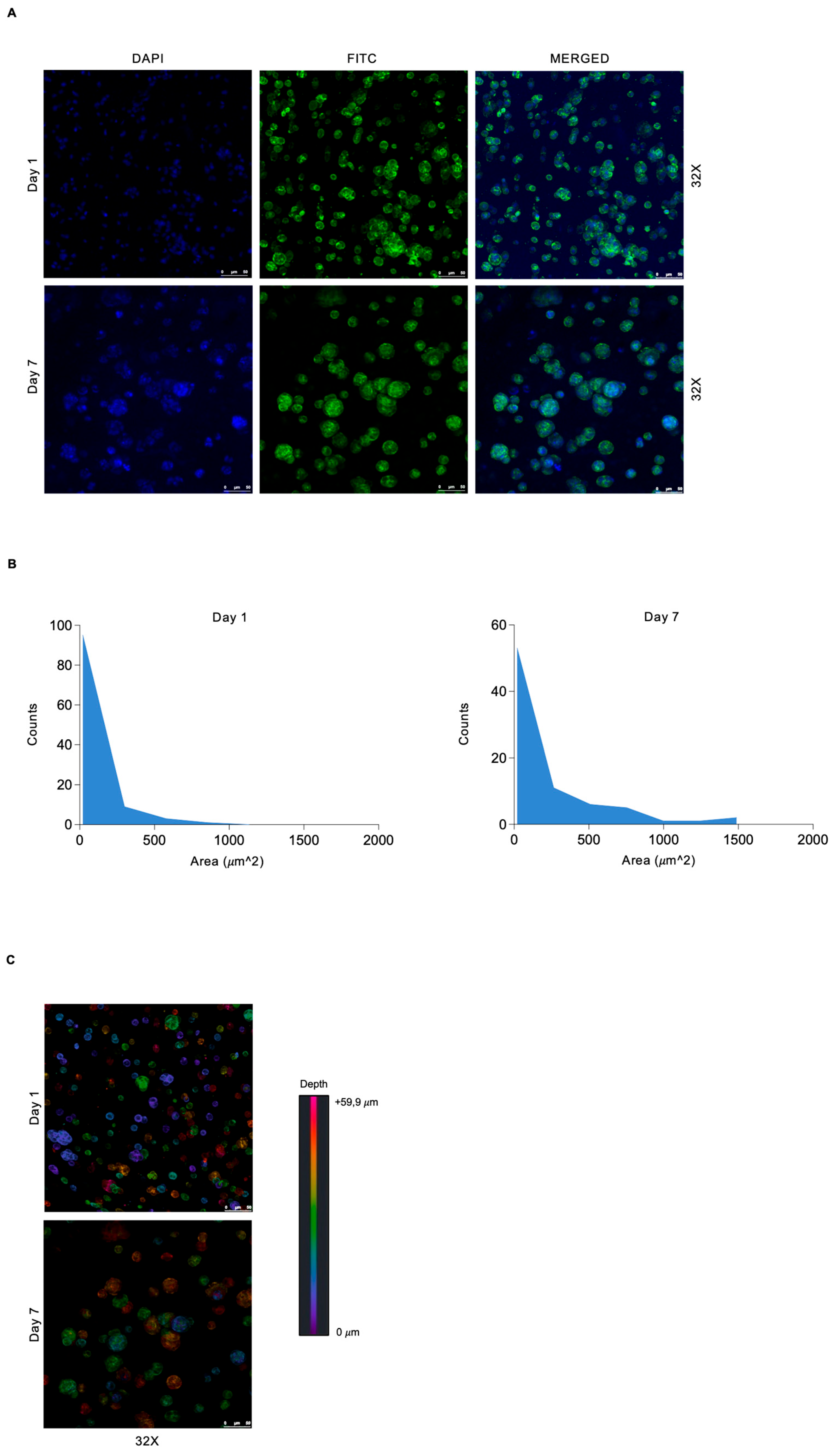
3.2. Spatially Defined Pattern of 3D Bioprinted rMC-1 Cells Shows Heterogeneous Spatial Distribution and Tendency to Form Spheroid Structures
3.3. 3D Bioprinted Müller Cells Show Reduced Gliosis and Oxidative Stress Compared to Conventional 2D Culture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Atawi, S. Three-Dimensional Bioprinting in Ophthalmic Care. Int. J. Ophthalmol. 2023, 16, 1702–1711. [Google Scholar] [CrossRef] [PubMed]
- Ozbolat, I.T.; Peng, W.; Ozbolat, V. Application Areas of 3D Bioprinting. Drug Discov. Today 2016, 21, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The Bioink: A Comprehensive Review on Bioprintable Materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, C.; Keenan, P.; Kirkwood, D.; Oji, S.; Webster, C.; Russell, K.A.; Koch, T.G. A Review of Recent Advances in 3D Bioprinting with an Eye on Future Regenerative Therapies in Veterinary Medicine. Front. Vet. Sci. 2020, 7, 584193. [Google Scholar] [CrossRef]
- Gu, Y.; Schwarz, B.; Forget, A.; Barbero, A.; Martin, I.; Shastri, V.P. Advanced Bioink for 3D Bioprinting of Complex Free-Standing Structures with High Stiffness. Bioengineering 2020, 7, 141. [Google Scholar] [CrossRef]
- Albanna, M.; Binder, K.W.; Murphy, S.V.; Kim, J.; Qasem, S.A.; Zhao, W.; Tan, J.; El-Amin, I.B.; Dice, D.D.; Marco, J.; et al. In Situ Bioprinting of Autologous Skin Cells Accelerates Wound Healing of Extensive Excisional Full-Thickness Wounds. Sci. Rep. 2019, 9, 1856. [Google Scholar] [CrossRef]
- Skytt, D.M.; Toft-Kehler, A.K.; Brændstrup, C.T.; Cejvanovic, S.; Gurubaran, I.S.; Bergersen, L.H.; Kolko, M. Glia-Neuron Interactions in the Retina Can Be Studied in Cocultures of Müller Cells and Retinal Ganglion Cells. Biomed Res. Int. 2016, 2016, 1087647. [Google Scholar] [CrossRef]
- Coughlin, B.A.; Trombley, B.T.; Mohr, S. Interleukin-6 (IL-6) Mediates Protection against Glucose Toxicity in Human Müller Cells via Activation of VEGF-A Signaling. Biochem. Biophys. Res. Commun. 2019, 517, 227–232. [Google Scholar] [CrossRef]
- Gao, H.; A, L.; Huang, X.; Chen, X.; Xu, H. Müller Glia-Mediated Retinal Regeneration. Mol. Neurobiol. 2021, 58, 2342–2361. [Google Scholar] [CrossRef]
- Reichenbach, A.; Bringmann, A. Glia of the Human Retina. Glia 2020, 68, 768–796. [Google Scholar] [CrossRef]
- Poitry, S.; Poitry-Yamate, C.; Ueberfeld, J.; MacLeish, P.R.; Tsacopoulos, M. Mechanisms of Glutamate Metabolic Signaling in Retinal Glial (Müller) Cells. J. Neurosci. Off. J. Soc. Neurosci. 2000, 20, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Grosche, A.; Pannicke, T.; Reichenbach, A. GABA and Glutamate Uptake and Metabolism in Retinal Glial (Müller) Cells. Front. Endocrinol. 2013, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Aponte, D.E.; Méndez-González, M.P.; Rivera-Pagán, A.F.; Kucheryavykh, Y.V.; Kucheryavykh, L.Y.; Skatchkov, S.N.; Eaton, M.J. Hyperglycemia Reduces Functional Expression of Astrocytic Kir4.1 Channels and Glial Glutamate Uptake. Neuroscience 2015, 310, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Labin, A.M.; Safuri, S.K.; Ribak, E.N.; Perlman, I. Müller Cells Separate between Wavelengths to Improve Day Vision with Minimal Effect upon Night Vision. Nat. Commun. 2014, 5, 4319. [Google Scholar] [CrossRef]
- Bringmann, A.; Wiedemann, P. Müller Glial Cells in Retinal Disease. Ophthalmologica 2012, 227, 1–19. [Google Scholar] [CrossRef]
- Puro, D.G. Diabetes-Induced Dysfunction of Retinal Müller Cells. Trans. Am. Ophthalmol. Soc. 2002, 100, 339–352. [Google Scholar]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-Culture Models as Drug-Testing Platforms in Breast Cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Jung, S.S.; Son, J.; Yi, S.J.; Kim, K.; Park, H.S.; Kang, H.-W.; Kim, H.K. Development of Müller Cell-Based 3D Biomimetic Model Using Bioprinting Technology. Biomed. Mater. 2022, 18, 015009. [Google Scholar] [CrossRef]
- Vecchiotti, D.; Verzella, D.; Capece, D.; Nolfi, M.D.V.; Di Francesco, B.; Cornice, J.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Biochemical Methods to Analyze the Subcellular Localization of NF-ΚB Proteins Using Cell Fractionation. Methods Mol. Biol. 2021, 2366, 19–25. [Google Scholar] [CrossRef]
- Fischietti, M.; Fratini, E.; Verzella, D.; Vecchiotti, D.; Capece, D.; Di Francesco, B.; Esposito, G.; Balata, M.; Ioannuci, L.; Sykes, P.; et al. Low Radiation Environment Switches the Overgrowth-Induced Cell Apoptosis Toward Autophagy. Front. Public Health 2021, 8, 594789. [Google Scholar] [CrossRef]
- Rosato, C.; Bettegazzi, B.; Intagliata, P.; Balbontin Arenas, M.; Zacchetti, D.; Lanati, A.; Zerbini, G.; Bandello, F.; Grohovaz, F.; Codazzi, F. Redox and Calcium Alterations of a Müller Cell Line Exposed to Diabetic Retinopathy-like Environment. Front. Cell. Neurosci. 2022, 16, 862325. [Google Scholar] [CrossRef] [PubMed]
- Sarthy, V.P.; Brodjian, S.J.; Dutt, K.; Kennedy, B.N.; French, R.P.; Crabb, J.W. Establishment and Characterization of a Retinal Müller Cell Line. Investig. Ophthalmol. Vis. Sci. 1998, 39, 212–216. [Google Scholar]
- Prieto-López, L.; Pereiro, X.; Vecino, E. The Mechanics of the Retina: Müller Glia Role on Retinal Extracellular Matrix and Modelling. Front. Med. 2024, 11, 1393057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wehrle, E.; Rubert, M.; Müller, R. 3D Bioprinting of Human Tissues: Biofabrication, Bioinks, and Bioreactors. Int. J. Mol. Sci. 2021, 22, 3971. [Google Scholar] [CrossRef] [PubMed]
- Toft-Kehler, A.K.; Gurubaran, I.S.; Desler, C.; Rasmussen, L.J.; Skytt, D.M.; Kolko, M. Oxidative Stress-Induced Dysfunction of Müller Cells During Starvation. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2721–2728. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Li, F.; Miao, Y.; Xu, L.-J.; Zhao, Y.; Li, Q.; Zhang, S.-H.; Wu, J.; Sun, X.-H.; Wang, Z. Involvement of the MEK-ERK/P38-CREB/c-Fos Signaling Pathway in Kir Channel Inhibition-Induced Rat Retinal Müller Cell Gliosis. Sci. Rep. 2017, 7, 1480. [Google Scholar] [CrossRef]
- Au, N.P.B.; Ma, C.H.E. Neuroinflammation, Microglia and Implications for Retinal Ganglion Cell Survival and Axon Regeneration in Traumatic Optic Neuropathy. Front. Immunol. 2022, 13, 860070. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Lu, R.; Soden, P.A.; Lee, E. Tissue-Engineered Models for Glaucoma Research. Micromachines 2020, 11, 612. [Google Scholar] [CrossRef]
- Larochelle, R.D.; Mann, S.E.; Ifantides, C. 3D Printing in Eye Care. Ophthalmol. Ther. 2021, 10, 733–752. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, E.J.; Hughes, J.M.; Van Geest, R.J.; Vogels, I.M.C.; Goldschmeding, R.; Van Noorden, C.J.F.; Schlingemann, R.O.; Klaassen, I. Effect of VEGF-A on Expression of Profibrotic Growth Factor and Extracellular Matrix Genes in the Retina. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4267–4276. [Google Scholar] [CrossRef] [PubMed]
- Ihanamäki, T.; Pelliniemi, L.J.; Vuorio, E. Collagens and Collagen-Related Matrix Components in the Human and Mouse Eye. Prog. Retin. Eye Res. 2004, 23, 403–434. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Niu, X.; Hou, S.; Li, Z.; Li, P.; Fan, Y. Apatite Minerals Derived from Collagen Phosphorylation Modification Induce the Hierarchical Intrafibrillar Mineralization of Collagen Fibers. J. Biomed. Mater. Res. A 2019, 107, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Sorushanova, A.; Delgado, L.M.; Wu, Z.; Shologu, N.; Kshirsagar, A.; Raghunath, R.; Mullen, A.M.; Bayon, Y.; Pandit, A.; Raghunath, M.; et al. The Collagen Suprafamily: From Biosynthesis to Advanced Biomaterial Development. Adv. Mater. 2019, 31, e1801651. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, L.; Sun, L.; She, Z.D.; Tan, R.W.; Niu, X.F. Biocompatibility and Immunotoxicology of the Preclinical Implantation of a Collagen-Based Artificial Dermal Regeneration Matrix. Biomed. Environ. Sci. 2018, 31, 829–842. [Google Scholar] [CrossRef]
- Heravi, M.; Rasoulinejad, S.A. Potential of Müller Glial Cells in Regeneration of Retina; Clinical and Molecular Approach. Int. J. Organ Transplant. Med. 2022, 13, 50–59. [Google Scholar]
- Das, A.V.; Mallya, K.B.; Zhao, X.; Ahmad, F.; Bhattacharya, S.; Thoreson, W.B.; Hegde, G.V.; Ahmad, I. Neural Stem Cell Properties of Müller Glia in the Mammalian Retina: Regulation by Notch and Wnt Signaling. Dev. Biol. 2006, 299, 283–302. [Google Scholar] [CrossRef]
- Fernández-Sánchez, L.; Lax, P.; Campello, L.; Pinilla, I.; Cuenca, N. Astrocytes and Müller Cell Alterations During Retinal Degeneration in a Transgenic Rat Model of Retinitis Pigmentosa. Front. Cell. Neurosci. 2015, 9, 484. [Google Scholar] [CrossRef]
- Li, L.; Sheedlo, H.J.; Turner, J.E. Muller Cell Expression of Glial Fibrillary Acidic Protein (GFAP) in RPE-Cell Transplanted Retinas of RCS Dystrophic Rats. Curr. Eye Res. 1993, 12, 841–849. [Google Scholar] [CrossRef]
- Hagemann, T.L.; Coyne, S.; Levin, A.; Wang, L.; Feany, M.B.; Messing, A. STAT3 Drives GFAP Accumulation and Astrocyte Pathology in a Mouse Model of Alexander Disease. Cells 2023, 12, 978. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.-K.; Park, Y.; Yoon, B.; Bae, J.-S.; Han, S.-W.; Heo, J.-E.; Kim, D.-E.; Ryu, K.-Y. Reduced Secretion of LCN2 (Lipocalin 2) from Reactive Astrocytes through Autophagic and Proteasomal Regulation Alleviates Inflammatory Stress and Neuronal Damage. Autophagy 2023, 19, 2296–2317. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, Q.; Wang, B. Effects of Apelin on the Fibrosis of Retinal Tissues and Müller Cells in Diabetes Retinopathy through the JAK2/STAT3 Signalling Pathway. Autoimmunity 2023, 56, 2259129. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Zhang, T.; Chen, Y.; Chu-Tan, J.; Jin, K.; Lee, S.-R.; Yam, M.X.; Madigan, M.C.; Fernando, N.; Cioanca, A.; et al. Inhibiting the Activation of MAPK (ERK1/2) in Stressed Müller Cells Prevents Photoreceptor Degeneration. Theranostics 2022, 12, 6705–6722. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Luo, L.-J.; Yang, C.-J.; Lai, J.-Y. Highly Retina-Permeating and Long-Acting Resveratrol/Metformin Nanotherapeutics for Enhanced Treatment of Macular Degeneration. ACS Nano 2023, 17, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Tezel, G.; Chauhan, B.C.; LeBlanc, R.P.; Wax, M.B. Immunohistochemical Assessment of the Glial Mitogen-Activated Protein Kinase Activation in Glaucoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 3025–3033. [Google Scholar] [CrossRef]
- Fischer, A.J.; Scott, M.A.; Tuten, W. Mitogen-Activated Protein Kinase-Signaling Stimulates Müller Glia to Proliferate in Acutely Damaged Chicken Retina. Glia 2009, 57, 166–181. [Google Scholar] [CrossRef]
- Carpi-Santos, R.; de Melo Reis, R.A.; Gomes, F.C.A.; Calaza, K.C. Contribution of Müller Cells in the Diabetic Retinopathy Development: Focus on Oxidative Stress and Inflammation. Antioxidants 2022, 11, 617. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, Q.; Zeng, Y.; Zhang, Y.; Zhang, M. Regulations of Retinal Inflammation: Focusing on Müller Glia. Front. Cell Dev. Biol. 2022, 10, 898652. [Google Scholar] [CrossRef]
- Chang, K.-C.; Liu, P.-F.; Chang, C.-H.; Lin, Y.-C.; Chen, Y.-J.; Shu, C.-W. The Interplay of Autophagy and Oxidative Stress in the Pathogenesis and Therapy of Retinal Degenerative Diseases. Cell Biosci. 2022, 12, 1. [Google Scholar] [CrossRef]
- Qin, M.; Xie, Z.; Cao, T.; Wang, Z.; Zhang, X.; Wang, F.; Wei, W.; Jin, M.; Ma, J.; Zeng, L.; et al. Autophagy in Rat Müller Glial Cells Is Modulated by the Sirtuin 4/AMPK/MTOR Pathway and Induces Apoptosis under Oxidative Stress. Cells 2022, 11, 2645. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-X.; Chen, C.-L.; Yang, J.-S.; Zhou, Z.-L.; Song, Z.-M.; Wang, Z.-Y. PI3K-Mediated Glioprotective Effect of Epidermal Growth Factor under Oxidative Stress Conditions. Int. J. Ophthalmol. 2014, 7, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of Metabolism and Mitochondrial Homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vecchiotti, D.; Di Vito Nolfi, M.; Veglianti, F.; Dall’Aglio, F.; Khan, H.N.; Flati, I.; Verzella, D.; Capece, D.; Alesse, E.; Angelucci, A.; et al. A 3D Bioprinting Approach to Studying Retinal Müller Cells. Genes 2024, 15, 1414. https://doi.org/10.3390/genes15111414
Vecchiotti D, Di Vito Nolfi M, Veglianti F, Dall’Aglio F, Khan HN, Flati I, Verzella D, Capece D, Alesse E, Angelucci A, et al. A 3D Bioprinting Approach to Studying Retinal Müller Cells. Genes. 2024; 15(11):1414. https://doi.org/10.3390/genes15111414
Chicago/Turabian StyleVecchiotti, Davide, Mauro Di Vito Nolfi, Francesca Veglianti, Francesca Dall’Aglio, Hafiz Nadeem Khan, Irene Flati, Daniela Verzella, Daria Capece, Edoardo Alesse, Adriano Angelucci, and et al. 2024. "A 3D Bioprinting Approach to Studying Retinal Müller Cells" Genes 15, no. 11: 1414. https://doi.org/10.3390/genes15111414
APA StyleVecchiotti, D., Di Vito Nolfi, M., Veglianti, F., Dall’Aglio, F., Khan, H. N., Flati, I., Verzella, D., Capece, D., Alesse, E., Angelucci, A., & Zazzeroni, F. (2024). A 3D Bioprinting Approach to Studying Retinal Müller Cells. Genes, 15(11), 1414. https://doi.org/10.3390/genes15111414






