Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sequencing
2.2. Assembly and Annotation of Organelle Genomes
2.3. Repeat Sequence Detection and Codon Analysis
2.4. RNA Editing Site Analysis
2.5. Transfer Fragments and Collinearity Analysis
2.6. Organellar Phylogenetical Inference
3. Results
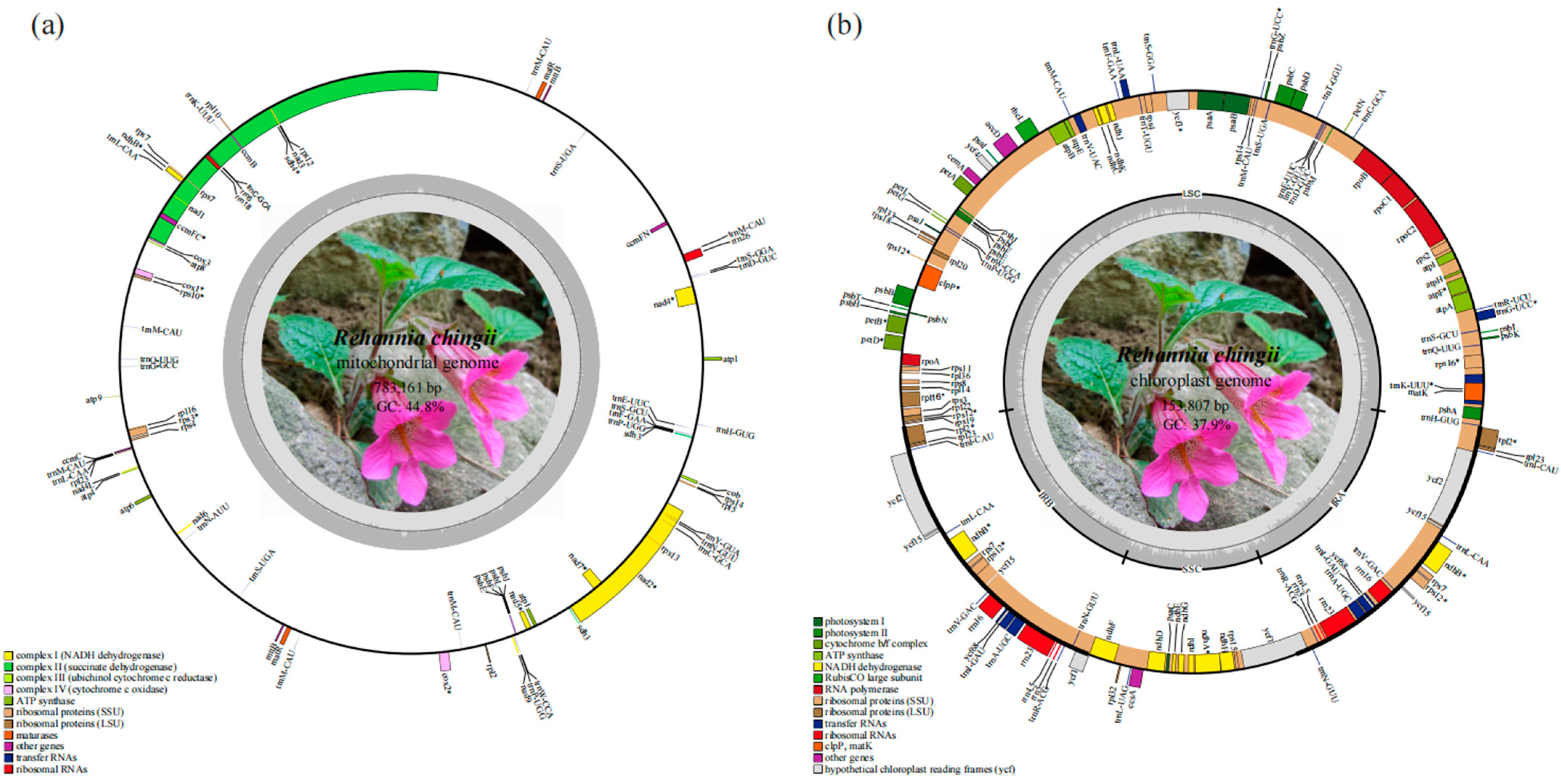
3.1. Genome Assembly and Characterization
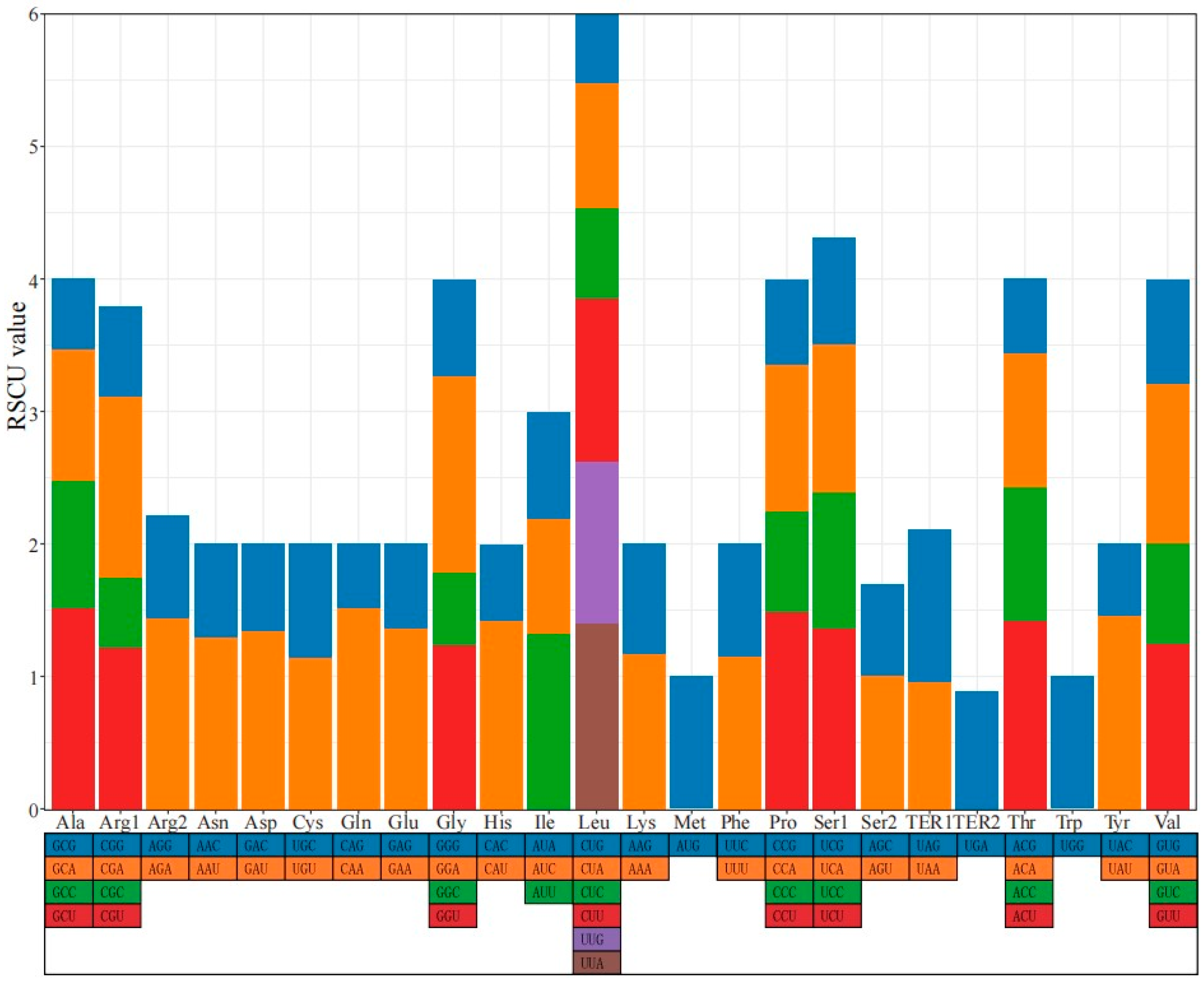
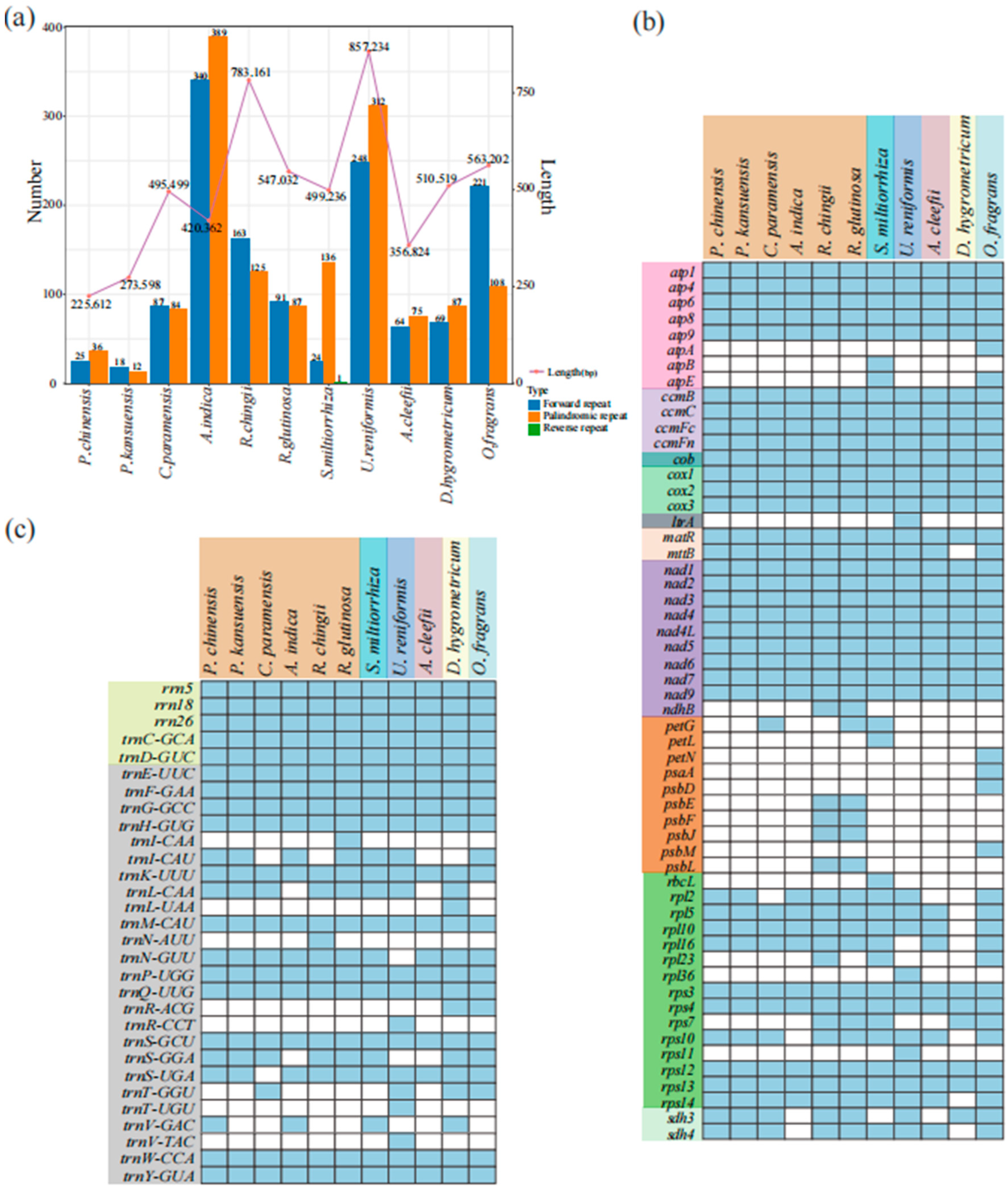
3.2. Codon and Repeat Sequence Analysis
3.3. RNA Editing Site Analysis
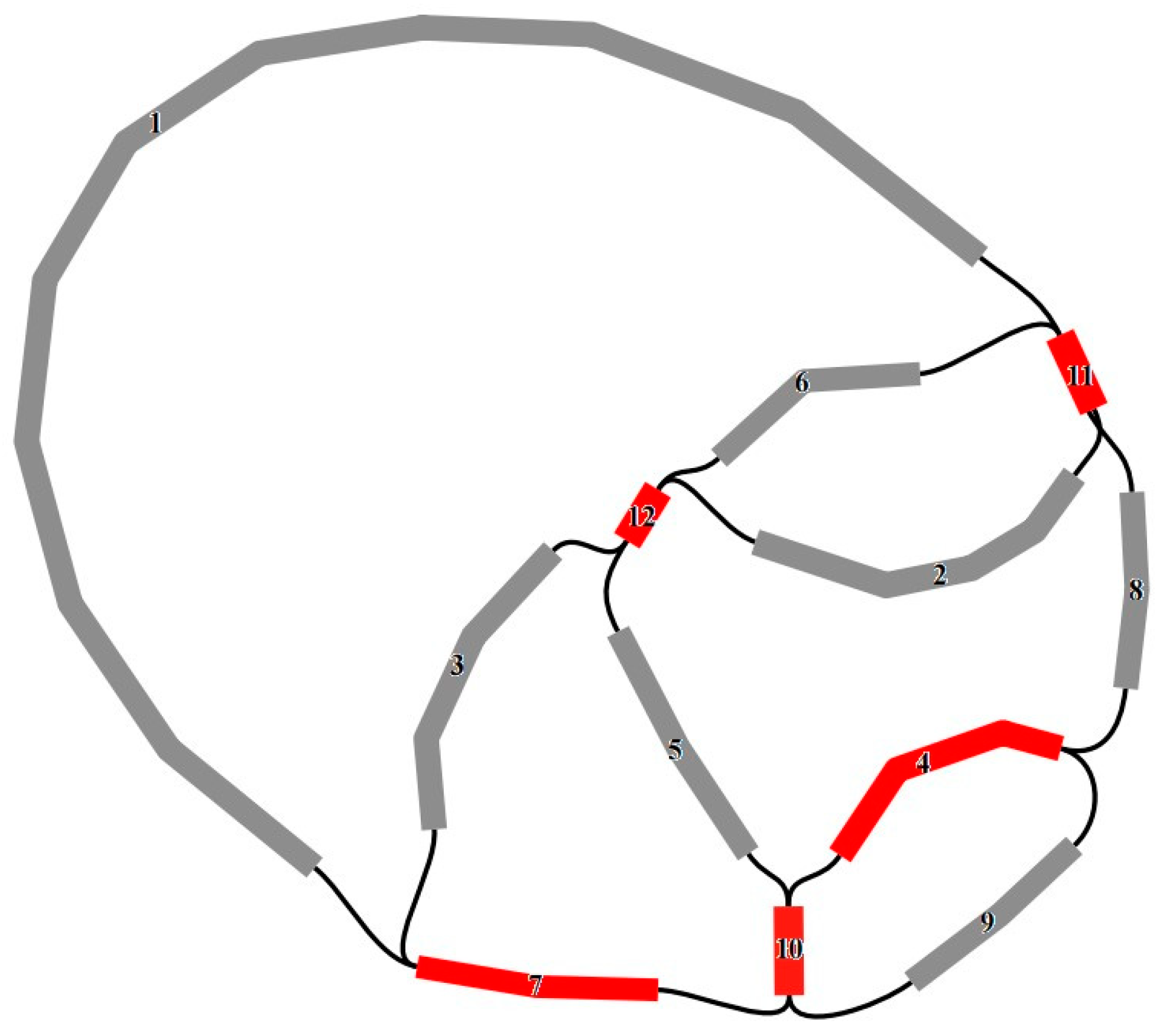
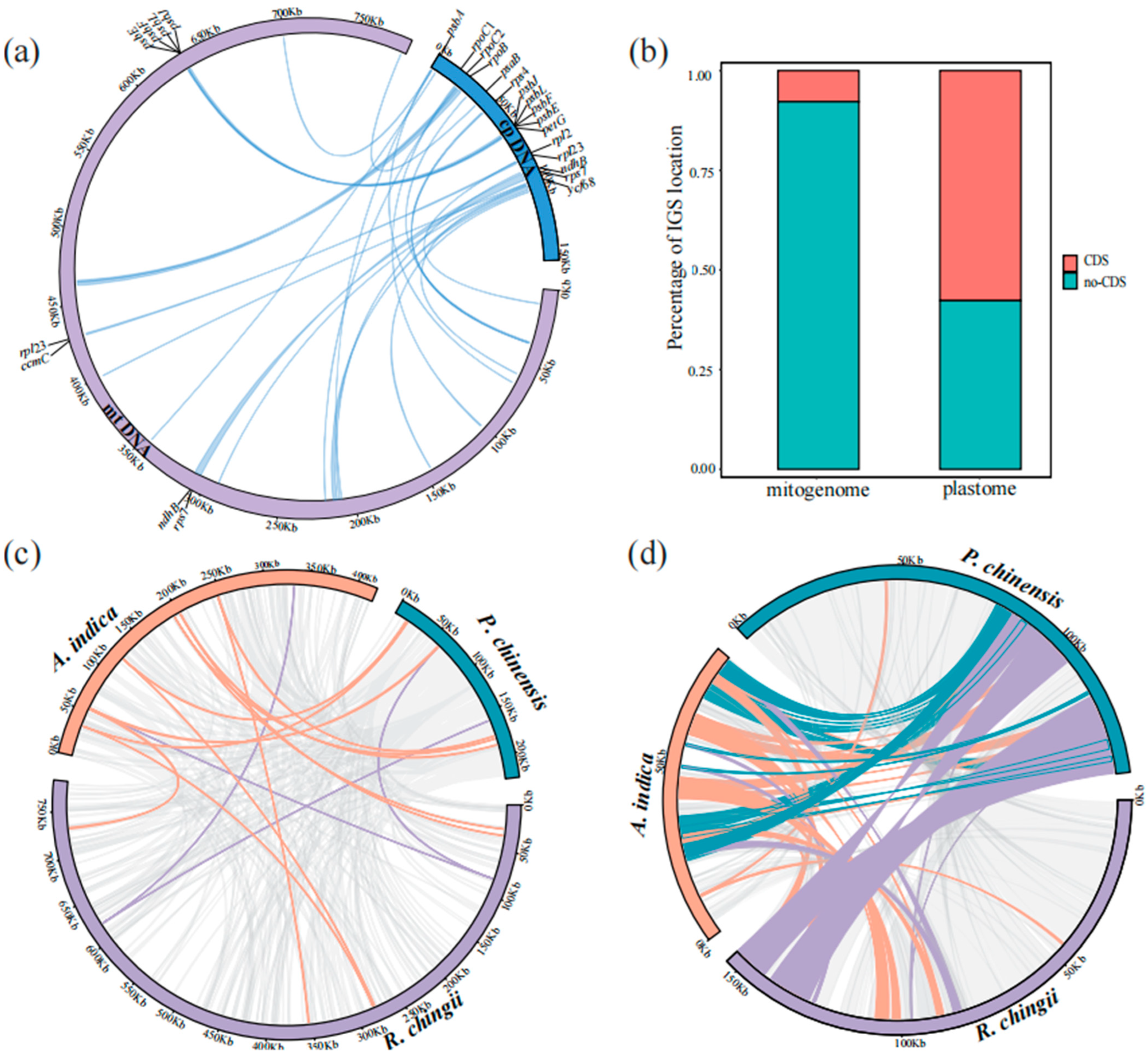
3.4. Transfer Fragments Detection and Collinearity Analysis
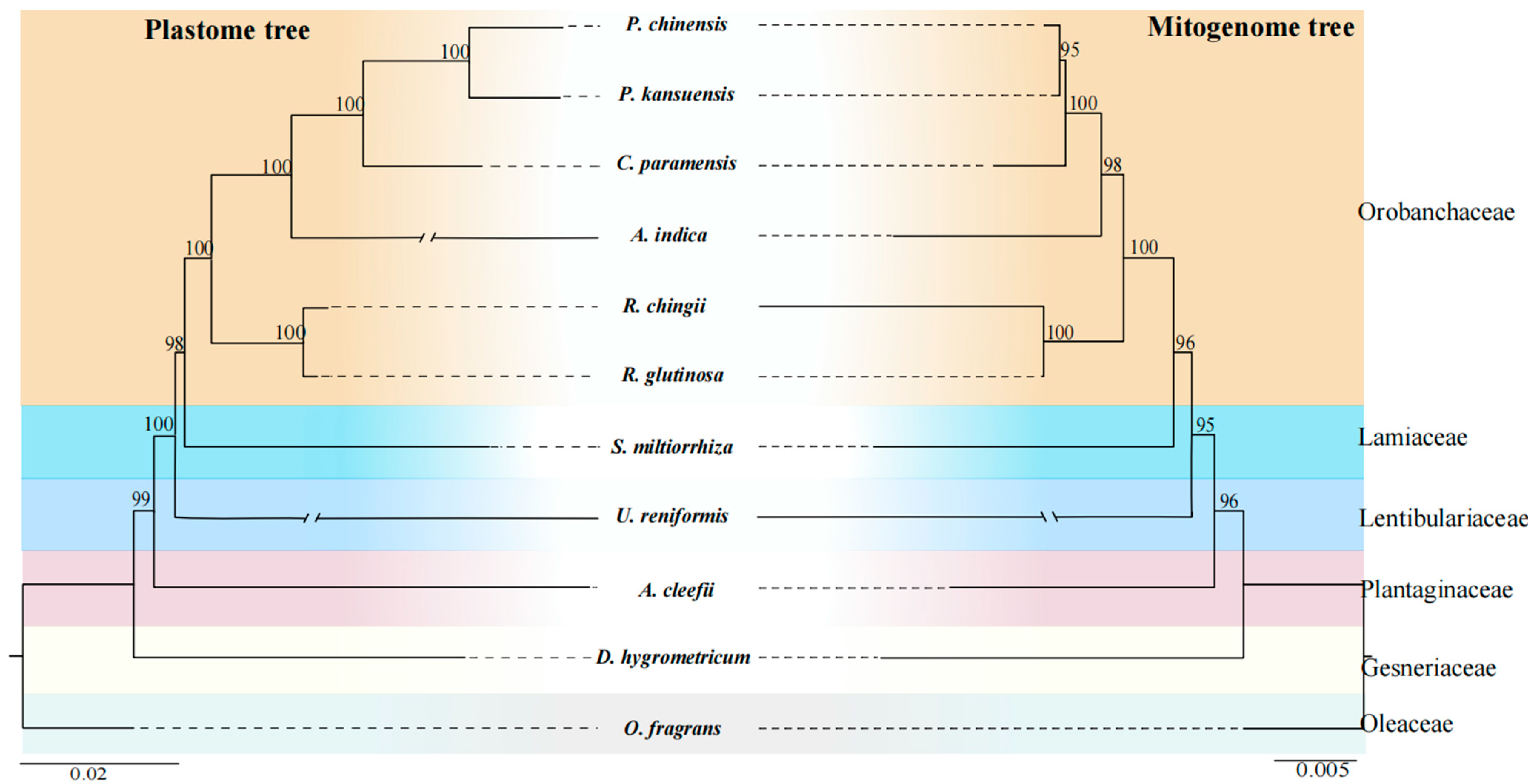
3.5. Phylogenetic Analysis of R. chingii
3.6. Comparison of Genomic Features with Ten Other Lamiales mitogenomes
4. Discussion
4.1. Characterization of the R. chingii Mitogenome
4.2. MTPTs in the R. chingii Mitogenome
4.3. RNA Editing in the R. chingii Mitogenome
4.4. Phylogenetic Relationships and Comparison of Genomic Features in Orobanchaceae Mitogenomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.-Q.; Liao, X.-Z.; Zhang, X.-N.; Tembrock, L.R.; Broz, A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2020, 60, 160–168. [Google Scholar] [CrossRef]
- Yao, G.; Jin, J.-J.; Li, H.-T.; Yang, J.-B.; Mandala, V.S.; Croley, M.; Mostow, R.; Douglas, N.A.; Chase, M.W.; Christenhusz, M.J.M.; et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenetics Evol. 2019, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.-H.; Cai, J.; Tian, X.-Y.; Han, L.-H.; Song, Y. Characteristics of the Complete Plastid Genome Sequences of the Monotypic Genus Dodecadenia (Family: Lauraceae) and Its Phylogenomic Implications. Forests 2022, 13, 1240. [Google Scholar] [CrossRef]
- Hajdari, A.; Pulaj, B.; Schmiderer, C.; Mala, X.; Wilson, B.; Lluga-Rizani, K.; Mustafa, B. A phylogenetic analysis of the wild Tulipa species (Liliaceae) of Kosovo based on plastid and nuclear DNA sequence. Adv. Genet. 2021, 2, e202100016. [Google Scholar] [CrossRef] [PubMed]
- Picault, N.; Hodges, M.; Palmieri, L.; Palmieri, F. The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 2004, 9, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Shao, X.-F.; Wei, Y.-Y.; Jiang, S.; Xu, F.; Wang, H.-F. Quantitative proteomics reveals that tea tree oil effects Botrytis cinerea mitochondria function. Pestic. Biochem. Physiol. 2020, 164, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, N.; Li, S.-S.; Grover, C.E.; Nie, H.-S.; Wendel, J.F.; Hua, J.-P. Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit. Rev. Plant Sci. 2017, 36, 55–69. [Google Scholar] [CrossRef]
- Yurina, N.P.; Odintsova, M.S. Mitochondrial genome structure of photosynthetic eukaryotes. Biochemistry 2016, 81, 101–113. [Google Scholar] [CrossRef]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef]
- Lai, C.-J.; Wang, J.; Kan, S.-L.; Zhang, S.; Li, P.; Reeve, W.G.; Wu, Z.-Q.; Zhang, Y.-H. Comparative analysis of mitochondrial genomes of Broussonetia spp. (Moraceae) reveals heterogeneity in structure, synteny, intercellular gene transfer, and RNA editing. Front. Plant Sci. 2022, 13, 1052151. [Google Scholar] [CrossRef]
- Putintseva, Y.A.; Bondar, E.I.; Simonov, E.P.; Sharov, V.V.; Oreshkova, N.V.; Kuzmin, D.A.; Konstantinov, Y.M.; Shmakov, V.N.; Belkov, V.I.; Sadovsky, M.G.; et al. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genom. 2020, 21, 1–12. [Google Scholar] [CrossRef]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (β vulgaris L.) reveals a novel gene for tRNACys(GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Clifton, S.W.; Minx, P.; Fauron, C.M.-R.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Brad, B.W.; Suman, K.; Catherine, T.; et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004, 136, 3486–3503. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- Li, J.; Tang, H.; Luo, H.; Tang, J.; Zhong, N.; Xiao, L. Complete mitochondrial genome assembly and comparison of Camellia sinensis var. Assamica cv. Duntsa. Front. Plant Sci. 2023, 14, 1117002. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Stein, D.B. Conservation of chloroplast genome structure among vascular plants. Curr. Genet. 1986, 10, 823–833. [Google Scholar] [CrossRef]
- Goremykin, V.V.; Hirsch-Ernst, K.I.; Wölfl, S.; Hellwig, F.H. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol. Biol. Evol. 2003, 20, 1499–1505. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–125. [Google Scholar] [CrossRef]
- Park, S.; Grewe, F.; Zhu, A.; Ruhlman, T.A.; Sabir, J.; Mower, J.P.; Jansen, R.K. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. N. Phytol. 2015, 208, 570–583. [Google Scholar] [CrossRef]
- He, W.-C.; Xiang, K.-L.; Chen, C.-J.; Wang, J.; Wu, Z.-Q. Master graph: An essential integrated assembly model for the plant mitogenome based on a graph-based framework. Brief. Bioinform. 2022, 24, bbac522. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-T.; Hou, Z.-Y.; Li, C.; Yang, J.-P.; Niu, Z.-T.; Xue, Q.-Y.; Liu, W.; Ding, X.-Y. Rapid structural evolution of Dendrobium mitogenomes and mito-nuclear phylogeny discordances in Dendrobium (Orchidaceae). J. Syst. Evol. 2022, 61, 790–805. [Google Scholar] [CrossRef]
- Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Nickrent, D.L. Parasitic angiosperms: How often and how many? Taxon 2020, 69, 5–27. [Google Scholar] [CrossRef]
- Westwood, J.H.; dePamphilis, C.W.; Das, M.; Fernández-Aparicio, M.; Honaas, L.A.; Timko, M.P.; Wafula, E.K.; Wickett, N.J.; Yoderl, J.I. The parasitic plant genome project: New tools for understanding the biology of Orobanche and Striga. Weed Sci. 2012, 60, 295–306. [Google Scholar] [CrossRef]
- Zhang, R.-T.; Xu, B.; Li, J.-F.; Zhao, Z.; Han, J.; Lei, Y.J.; Yang, Q.; Peng, F.-F.; Liu, Z.-L. Transit from autotrophism to heterotrophism: Sequence variation and evolution of chloroplast genomes in Orobanchaceae species. Front. Genet. 2020, 11, 542017. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Q.; Fu, W.-R.; Tang, Y.-W.; Zhang, W.-J.; Song, Z.-P.; Li, L.-F.; Yang, J.; Ma, H.; Yang, J.-H.; Zhou, C.; et al. Diverse trajectories of plastome degradation in holoparasitic Cistanche and genomic location of the lost plastid genes. J. Exp. Bot. 2020, 71, 877–892. [Google Scholar] [CrossRef]
- Xu, Y.-X.; Zhang, J.-X.; Ma, C.-R.; Lei, Y.-T.; Shen, G.-J.; Jin, J.-J.; Eaton, D.A.R.; Wu, J.-Q. Comparative genomics of Orobanchaceous species with different parasitic lifestyles reveals the origin and stepwise evolution of plant parasitism. Mol. Plant 2022, 15, 1384–1399. [Google Scholar] [CrossRef]
- Sun, J.; Sun, M.-Q.; Wang, D.-C.; Xu, K.-L.; Hu, R.-Y.; Zhang, Y.-H. Plastomes of two Rehmannia species:comparative genomic and phylogenetic analyses. Mitochondrial DNA Part B 2021, 6, 753–754. [Google Scholar] [CrossRef]
- Albach, D.C.; Li, H.-Q.; Zhao, N.; Jensen, S.R. Molecular systematics and phytochemistry of Rehmannia (Scrophulariaceae). Biochem. Syst. Ecol. 2006, 35, 293–300. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, C.; Li, Z.-H.; Li, J.; Liu, Z.-L. Development of twelve nuclear microsatellites in the endemic herb Rehmannia chingii using next generation sequencing. Conserv. Genet. Resour. 2015, 7, 223–224. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Shi, G.-R.; Wang, X.; Zhang, C.-L.; Wang, Y.; Chen, R.-Y.; Yu, D.-Q. Nine new compounds from the whole plants of Rehmannia chingii. J. Asian Nat. Prod. Res. 2016, 18, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-P.; Chen, J.; Shu, A.-M.; Liu, L.-P.; Wu, Q.; Wu, J.-S.; Song, S.-Y.; Fan, W.-P.; Zhu, Y.-H.; Xu, H.-Q.; et al. Combination of the herbs Radix rehmanniae and Cornus officinalis mitigated testicular damage from diabetes mellitus by enhancing glycolysis via the AGEs/RAGE/HIF-1α Axis. Front. Pharmacol. 2021, 12, 678300. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.-Y.; Zhao, J.-H.; Han, K.; Liu, Z.-L. Complete chloroplast genome sequences of Rehmannia chingii, an endemic and endangered herb. Conserv. Genet. Resour. 2016, 8, 407–409. [Google Scholar] [CrossRef]
- Zuo, X.; Miao, C.-Y.; Li, M.-M.; Gu, L.; Yang, X.; Song, C.; Li, M.-J.; Du, J.-F.; Xie, C.-X.; Liu, X.-Y.; et al. Purple Rehmannnia: Investigation of the activation of R2R3-MYB transcription factors involved in anthocyanin biosynthesis. Physiol. Plant. 2023, 175, e13920. [Google Scholar] [CrossRef]
- Pahlich, E.; Gerlitz, C. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry 1980, 19, 11–13. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F.-Q. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2017, 11, e0163962. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, M.V.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Heng, L. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar]
- Kolmogorov, M.; Bickhart, D.M.; Behsaz, B.; Gurevich, A.; Rayko, M.; Shin, S.B.; Kuhn, K.; Yuan, J.; Polevikov, E.; Smith, T.P.L.; et al. metaFlye: Scalable long-read metagenome assembly using repeat graphs. Nat. Methods 2020, 17, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2017, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; DePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Shi, L.-C.; Chen, H.-M.; Jiang, M.; Wang, L.-Q.; Wu, X.; Huang, L.-F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Gene Predict. Methods Protoc. 2019, 1962, 1–14. [Google Scholar]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2000; pp. 73–74. [Google Scholar]
- Chen, S.-F.; Zhou, Y.-Q.; Chen, Y.-R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, N.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 20078–22079. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. RNA editing in plant mitochondria: 20 years later. IUBMB Life 2009, 61, 1101–1104. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.-H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yu, R.-X.; Chen, J.-F.; Liu, Y.; Zhou, R. Highly active repeat-mediated recombination in the mitogenome of the holoparasitic plant Aeginetia indica. Front. Plant Sci. 2022, 13, 988368. [Google Scholar] [CrossRef]
- Chen, J.-F.; Yu, R.-X.; Dai, J.-H.; Liu, Y.; Zhou, R.-C. The loss of photosynthesis pathway and genomic locations of the lost plastid genes in a holoparasitic plant Aeginetia indica. BMC Plant Biol. 2020, 20, 199. [Google Scholar] [CrossRef]
- Zeng, S.-Y.; Zhou, T.; Han, K.; Yang, Y.-C.; Zhao, J.-H.; Liu, Z.-L.; Bell, C. The complete chloroplast genome sequences of six Rehmannia species. Genes 2017, 8, 103. [Google Scholar] [CrossRef]
- Qian, J.; Song, J.-Y.; Gao, H.-H.; Zhu, Y.-J.; Xu, J.; Pang, X.-H.; Yao, H.; Sun, C.; Li, X.-E.; Li, C.-Y.; et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef]
- Zhang, T.-W.; Zhang, X.-W.; Hu, S.-N.; Yu, J. An efficient procedure for plant organellar genome assembly, based on whole genome data from the 454 GS FLX sequencing platform. Plant Methods 2011, 7, 38. [Google Scholar] [CrossRef]
- Mower, J.P.; Hanley, L.; Wolff, K.; Pabón-Mora, N.; González, F. Complete mitogenomes of two Aragoa species and phylogeny of plantagineae (Plantaginaceae, Lamiales) using mitochondrial genes and the nuclear ribosomal RNA repeat. Plants 2021, 10, 2673. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Guo, W.; Partha, R.; Fan, W.; Levsen, N.; Wolff, K.; Nugent, J.M.; Pabón-Mora, N.; González, F. Plastomes from tribe Plantagineae (Plantaginaceae) reveal infrageneric structural synapormorphies and localized hypermutation for Plantago and functional loss of ndh genes from Littorella. Mol. Phylogenetics Evol. 2021, 162, 107217. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.-F.; Li, Y.-F.; Zhang, C.; Wang, X.-R.; Li, M.-Z. The complete chloroplast genome of sweet olive (Osmanthus fragrans Lour.). Mitochondrial DNA Part B 2019, 4, 1063–1064. [Google Scholar] [CrossRef]
- Swofford, D.L.; Sullivan, J. Phylogeny inference based on parsimony and other methods using PAUP*. Phylogenetic Handb. A Pract. Approach DNA Protein Phylogeny Cáp 2003, 7, 160–206. [Google Scholar]
- Xia, X.-H.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenetics Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Xiong, Y.-L.; Yu, Q.-Q.; Xiong, Y.; Zhao, J.-M.; Lei, X.; Liu, L.; Liu, W.; Peng, Y.; Zhang, J.-B.; Li, D.-X.; et al. The complete mitogenome of Elymus sibiricus and insights into its evolutionary pattern based on simple repeat sequences of seed plant mitogenomes. Front. Plant Sci. 2021, 12, 802321. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Sun, M.-Y.; Zhang, M.-Y.; Chen, X.-N.; Liu, Y.-Y.; Liu, B.-B.; Li, J.-M.; Wang, R.-Z.; Zhao, K.-J.; Wu, J. Rearrangement and domestication as drivers of Rosaceae mitogenome plasticity. BMC Biol. 2022, 20, 181. [Google Scholar] [CrossRef]
- Palmer, J.D.; Herbon, L.A. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988, 28, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Palmer, J.D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA 2009, 106, 16728–16733. [Google Scholar] [CrossRef]
- Tsunewaki, K. Interorganellar DNA transfer in wheat: Dynamics and phylogenetic origin. Proc. Jpn. Acad. Ser. B 2011, 87, 529–549. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, L.; Liu, T.-J.; Hao, G.; Ge, X.-J.; Yan, H.-F. Comparative analyses of three complete Primula mitogenomes with insights into mitogenome size variation in Ericales. BMC Genom. 2022, 23, 770. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.-Y.; Li, J.-L.; Zhang, X.; Yu, J. The complete mitochondrial genome of Amorphophallus albus and development of molecular markers for five Amorphophallus species based on mitochondrial DNA. Front. Plant Sci. 2023, 14, 1180417. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Park, S. Complete plastid and mitochondrial genomes of Aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic male sterility (CMS). Int. J. Mol. Sci. 2021, 22, 6143. [Google Scholar]
- Li, J.-H.; Li, J.-L.; Ma, Y.-B.; Kou, L.; Wei, J.-J.; Wang, W.-X. The complete mitochondrial genome of okra (Abelmoschus esculentus): Using nanopore long reads to investigate gene transfer from chloroplast genomes and rearrangements of mitochondrial DNA molecules. BMC Genom. 2022, 23, 481. [Google Scholar] [CrossRef]
- Bi, C.-W.; Paterson, A.H.; Wang, X.-L.; Xu, Y.-Q.; Wu, D.-Y.; Qu, Y.-S.; Jiang, A.-N.; Ye, Q.-L.; Ye, N. Corrigendum to “Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches”. BioMed Res. Int. 2019, 2019, 9691253. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Ceriotti, L.F.; Gatica-Soria, L.M.; Roulet, M.E.; Garcia, L.E.; Sato, H.A. Beyond parasitic convergence: Unraveling the evolution of the organellar genomes in holoparasites. Ann. Bot. 2023, mcad108. [Google Scholar] [CrossRef]
- Zhang, M.-Y.; Li, Z.; Wang, Z.-J.; Xiao, Y.; Bao, L.; Wang, M.; An, C.-J.; Gao, Y.-F. Exploring the RNA Editing Events and Their Potential Regulatory Roles in Tea Plant (Camellia sinensis L.). Int. J. Mol. Sci. 2022, 23, 13640. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Howad, W.; Tang, H.V.; Pring, D.R.; Kempken, F. Nuclear genes from Tx CMS maintainer lines are unable to maintain atp6 RNA editing in any anther cell-type in the sorghum bicolor A3 cytoplasm. Curr. Genet. 1999, 36, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.J.; Zhang, Q.-N.; Qin, X.-J.; Xu, Y.-H.; Ni, C.-Z.; Huang, J.-H.; Zhu, L.-L.; Zhong, F.-Y.; Liu, W.; Yao, G.-X.; et al. Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol. 2018, 220, 878–892. [Google Scholar] [CrossRef]
- Zhang, Q.-N.; Xu, Y.-H.; Huang, J.-S.; Zhang, K.; Xiao, H.-J.; Qin, X.-J.; Zhu, L.-L.; Zhu, Y.-G.; Hu, J. The rice pentatricopeptide repeat protein PPR756 is involved in pollen development by affecting multiple RNA editing in mitochondria. Front. Plant Sci. 2020, 11, 749. [Google Scholar] [CrossRef] [PubMed]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Wang, J.; Kan, S.-L.; Liao, X.-Z.; Zhou, J.-W.; Tembrock, L.R.; Daniell, H.; Jin, S.-X.; Wu, Z.-Q. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024. [Google Scholar] [CrossRef]

| Group of Genes | Name of Genes |
|---|---|
| Complex I | nad1 *, nad2 *, nad3, nad4 *, nad4L, nad5 *, nad6, nad7 *, nad9 |
| Complex II | sdh3 (×2), sdh4 |
| Complex III | cob |
| Complex IV | cox1 *, cox2 *, cox3 |
| Complex V | atp1 (×2), atp4, atp6, atp8, atp9 |
| Cytochrome c biogenesis | ccmB, ccmC, ccmFc *, ccmFn |
| Maturases | matR (×2) |
| Transport membrane protein | mttB (×2) |
| Ribosome | rpl2, rpl5, rpl10, rpl16, rpl23, rps3 *, rps4, rps7, rps10 *, rps12, rps13, rps14 |
| rRNA | rrn5, rrn18, rrn26 |
| tRNA | trnC-GCA (×2), trnD-GUG, trnE-UUC, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU, trnL-CAA (×2), trnM-CAU (×6), trnN-AUU, trnN-GUU, trnP-UGG (×2), trnQ-UUG, trnS-GCU, trnS-GGA, trnS-UGA (×2), trnW-CCA, trnY-GUA |
| plastid gene | ndhB *, psbE, psbF, psbJ, psbL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Feng, Y.-L.; Wang, J.; Zhu, S.-S.; Jin, X.-J.; Wu, Z.-Q.; Zhang, Y.-H. Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family. Genes 2024, 15, 98. https://doi.org/10.3390/genes15010098
Han Y, Feng Y-L, Wang J, Zhu S-S, Jin X-J, Wu Z-Q, Zhang Y-H. Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family. Genes. 2024; 15(1):98. https://doi.org/10.3390/genes15010098
Chicago/Turabian StyleHan, Ying, Yan-Lei Feng, Jie Wang, Shan-Shan Zhu, Xin-Jie Jin, Zhi-Qiang Wu, and Yong-Hua Zhang. 2024. "Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family" Genes 15, no. 1: 98. https://doi.org/10.3390/genes15010098
APA StyleHan, Y., Feng, Y.-L., Wang, J., Zhu, S.-S., Jin, X.-J., Wu, Z.-Q., & Zhang, Y.-H. (2024). Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family. Genes, 15(1), 98. https://doi.org/10.3390/genes15010098







