Abstract
Light and temperature are key factors influencing the accumulation of anthocyanin in fruit crops. To assess the effects of fruit bagging during development and high post-ripening temperature on ‘Hongyang’ kiwifruit, we compared the pigmentation phenotypes and expression levels of anthocyanin-related genes between bagged and unbagged treatments, and between 25 °C and 37 °C postharvest storage temperatures. Both the bagging and 25 °C treatments showed better pigmentation phenotypes with higher anthocyanin concentrations. The results of the qRT-PCR analysis revealed that the gene expression levels of LDOX (leucoanthocyanidin dioxygenase), F3GT (UDP-flavonoid 3-O-glycosyltransferase ), AcMYB10, and AcbHLH42 were strongly correlated and upregulated by both the bagging treatment and 25 °C storage. The results of bimolecular fluorescence complementation and luciferase complementation imaging assays indicated an interaction between AcMYB10 and AcbHLH42 in plant cells, whereas the results of a yeast one-hybrid assay further demonstrated that AcMYB10 activated the promoters of AcLODX and AcF3GT. These results strongly suggest that enhanced anthocyanin synthesis is caused by the promoted expression of AcLODX and AcF3GT, regulated by the complex formed by AcMYB10–AcbHLH42.
1. Introduction
Kiwifruit (Actinidia chinensis), a woody vine-borne edible berry belonging to the Actinidiaceae family, is one of the most valuable fruit crops because of its diverse nutritional and health benefits for humans [1]. The Actinidia genus encompasses approximately 75 species, exhibiting a diverse array of colors in both the skin and flesh, which is primarily determined by the accumulation of chlorophyll, carotenoids, anthocyanins, and their various combinations [2]. The current commercial kiwifruit (A. chinensis) cultivars available on the market are commonly categorized into green-, yellow-, and red-flesh varieties [3]. Among them, ‘Hongyang’, the most prominent and renowned, red-fleshed kiwifruit cultivar in China, exhibits a star-shaped accumulation of anthocyanins in the inner pericarp. This unique characteristic increases its appeal to customers and breeders compared with green-fleshed varieties due to the widely accepted biofunctionality of anthocyanins [4,5]. In cultivation practice, various environmental factors influence the accumulation of anthocyanins in fruit, such as temperature, levels of plant nutrients, shading, and concentrations of endogenous hormones [6], which create challenges in the cultivation of red-fleshed kiwifruit.
Fruit bagging is an important pomological technique employed during fruit development to optimize the visual appearance and internal quality of fruits by effectively manipulating the microenvironment surrounding the fruit, including controlling light intensity, temperature, and humidity levels. Fruit bagging also serves as a protective measure against abiotic and biotic stressors [7,8]. Given the extensive research on the photoregulation of anthocyanin production in the plant kingdom [9], fruit bagging has emerged as an effective technique for enhancing the peel pigmentation in various fruits, including apples [10,11], peaches [12], grapes [13], and pears [14]. Moreover, fruit bagging management is common in the production of kiwifruit, aiming to increase fruit quality. It has been applied with green-fleshed ‘Jinkui’ (A. chinensis) [15], yellow-fleshed ‘Jinyan’ (A. chinensis) [16], as well as A. eriantha lines “G19”, “G21”, and “G28” [17]. In ‘Hongbaoshixing’ (a whole-fruit pigmented A. arguta cultivar), bagging treatment throughout fruit development substantially restricted the proper coloration of peel [18]. Although fruit bagging can enhance the fruit storability, flesh color, as well as flavonoid and anthocyanin contents of ‘Hongyang’, the underlying mechanism of these effects remains poorly understood [19,20,21]. Low temperatures stimulate anthocyanin accumulation in the plant kingdom [22,23], whereas high temperatures suppress anthocyanin accumulation [24,25]. In terms of the cultivation practices used for ‘Hongyang’, the fruits harvested in warmer temperature regions commonly exhibit poor flesh pigmentation compared with those harvested in cooler areas [26]. Although our previous study revealed that 25 °C might serve as the threshold for anthocyanin accumulation during the fruit-ripening process of ‘Hongyang’ on the tree, and we hypothesized that the kiwifruit-ripening process could tolerate temperatures as high as 35–40 °C, the impact of relatively high temperatures on postharvest anthocyanin accumulation remains unclear, as most commercial kiwifruit cultivars require early harvesting, with subsequent postharvest ripening.
In terms of anthocyanin biosynthesis, the key structural genes have been extensively documented, most of which have been profiled, mined, isolated, cloned, and function-verified by various multi-omic approaches and molecular methods. These genes include CHS (encoding chalcone synthase), F3H (favanone 3-hydroxylase), DFR (dihydrofavonol 4-reductase), LDOX/ANS (leucoanthocyanidin dioxygenase), and UFGT (UDP-flavonoid 3-O-glycosyltransferase) [27,28,29,30,31]. Since the early 2000s, accumulating evidence has indicated that changes in the expression of these structural genes, which are regulated by transcription factors (TFs), are responsible for light- and temperature-induced anthocyanin accumulation in fruit crops [24,32]. Among the key TFs involved in regulating anthocyanin biosynthesis, R2R3-MYB genes are the most crucial family regulating fruit coloration through functionating with the MYB–bHLH–WD40 complex [33,34]. In kiwifruit, more than 150 MYBs have been documented [35,36]. Among them, AcMYB110 [37], AcMYBF110 [2], AcMYB75 [23], AcMYB10 [23,36], and AcMYB123 [38] are involved in the regulation of anthocyanin synthesis. Moreover, in our previous study, we demonstrated that AcMRP and AcMYB1 mediate the downregulation of anthocyanin biosynthesis in ‘Hongyang’ under high-temperature conditions [26]. Additionally, as a related paralog of AcMYB1, we verified that AcMYB10 is a key regulator participating in light- and temperature-induced anthocyanin biosynthesis in vitro [36]. Therefore, whether AcMYB10 is involved in the regulation of anthocyanin synthesis induced by bagging and high temperatures during the post-ripening period is worth investigating. This investigation contributes towards achieving a comprehensive understanding of anthocyanin accumulation in ‘Hongyang’ kiwifruit, thereby providing a theoretical foundation for molecular breeding, standardized cultivation practices, and postharvest technologies aimed at enhancing flesh coloration.
To evaluate the impact of bagging treatment and high postharvest temperatures on anthocyanin synthesis in ‘Hongyang’, we conducted bagging treatments during fruit development and subjected the fruits to 37/25 °C treatments during the post-ripening period. Subsequently, we compared the pigmentation of the fruits and expression levels of the relevant genes. Additionally, we performed bimolecular fluorescence complementation (BiFC) and bimolecular luminescence complementation (BiLC) assays to investigate the interaction between AcMYB10 and AcbHLH42. Furthermore, a yeast one-hybrid (Y1H) assay was employed to confirm the promoter achievability of AcMYB10 and AcbHLH42 on AcLODX and AcF3GT. This study provides valuable insights into elucidating the mechanism underlying light- and temperature-regulated anthocyanin biosynthesis, as well as developing pre- and postharvest treatment strategies for enhancing the fruit coloration of red-fleshed kiwifruit.
2. Results
2.1. Effects of Fruit Bagging on Anthocyanin Accumulation in ‘Hongyang’ Kiwifruit
After bagging treatment, the inner pericarp of ‘Hongyang’ kiwifruit exhibited significantly deeper pigmentation phenotypes than unbagged fruits (Figure 1A). Additionally, bagged fruits displayed significantly higher total anthocyanin (0.29 ± 0.08 mg/100 g FW) and lower chlorophyll (0.85 ± 0.03 mg/100 g FW) levels than unbagged fruits (0.17 ± 0.03 mg/100 g FW and 2.06 ± 0.18 mg/100 g FW). Regarding gene expression, no significant difference was observed in the expression level of the light response gene HY5 between bagged and unbagged fruits. However, with respect to anthocyanin synthesis, bagged kiwifruits exhibited significantly higher expression levels of two key transcription factor genes (bHLH42 and MYB10) as well as seven important structural genes (CHS, CHI, F3H, F3′H, DFR, LDOX, and F3GT), than unbagged ones (Figure 1C). Furthermore, two chlorophyll-biosynthesis-related genes, CBR and GLUTR, showed decreased expression levels in bagged fruit, whereas two chlorophyll-degradation-related genes, RBCS and PPH, demonstrated increased expression levels in bagged fruits relative to unbagged fruits; these changes may contribute to the enhanced green pigmentation observed in unbagged kiwifruits. Nevertheless, no significant differences were detected in the expression levels of PAO, SGR, or CAO between bagging-treated and untreated kiwifruits. Collectively, these results indicate that the application of bagging can effectively accelerate anthocyanin synthesis within the pericarp tissue of ‘Hongyang’ kiwifruit and suggest the involvement of regulation-related genes bHLH42 and MYB10 in transcriptional regulation processes associated with anthocyanin synthesis induced using this treatment.
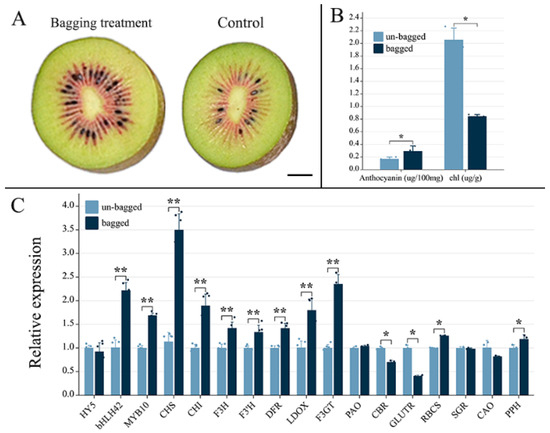
Figure 1.
Phenotypes, anthocyanin and chlorophyll contents, and related gene expression of bagged and unbagged ‘Hongyang’ kiwifruit: (A) bisected fruits of bagged and unbagged kiwifruit harvested at 130 days after flower (DAF); (B) the contents of anthocyanin and total chlorophyll (chl) in the flesh of bagged and unbagged kiwifruit harvested at 130 DAF; and (C) expression level of light response gene (HY5), anthocyanin biosynthesis and transcriptional regulation factor genes (CHS, CHI, F3H, F3′H, DFR, LDOX, F3GT, bHLH42, and MYB10), and chlorophyll biosynthesis (PAO, CBR, GLUTR) and degradation-related genes (RBCS, SGR, CAO and PPH) of bagged and unbagged kiwifruit harvested at 130 DAF. Abbreviation: HY5, (ELONGATED HYPOCOTYL5); CHI, chalcone isomerase; CHS, chalcone synthase; F3H, flavonoid 3-hydroxylase; F3′H, flavonoid3′-hydroxylase; DFR, dihydroflavonol 4-reductase; LDOX, leucoanthocyanidin dioxygenase; F3GT, UDP-flavonoid 3-O-glycosyltransferase. CAO, chlorophyll-a oxygenase; CBR, chlorophyll-b reductase; GLUTR, glutamyl tRNA reductase; PAO, pheophorbide a oxygenase; PPH, pheophytin pheophorbide hydrolase; RBCS, small subunit of ribulose-1,5-bisphosphate carboxylase; SGR, stay-green. *, p ≤ 0.05; **, p ≤ 0.01.
2.2. Effect of High Postharvest Temperature on Anthocyanin Accumulation in ‘Hongyang’ Kiwifruit
To investigate the impact of high postharvest temperatures on anthocyanin accumulation in ‘Hongyang’ kiwifruit during storage, two temperature treatments (25 °C and 37 °C) were implemented based on our previous study [26]. As depicted in Figure 2A, the red pigment in the inner pericarp of ‘Hongyang’ kiwifruit gradually darkened with increasing storage time under 25 °C treatments, whereas at 37 °C, the red pigmentation gradually decreased. The anthocyanin content in ‘Hongyang’ kiwifruit significantly increased during storage at 25 °C starting at 4 days, but slightly decreased at 37 °C (Figure 2B). Significant differences in the anthocyanin content between the treatments at 25 and 37 °C were observed on days 4, 6, and 8 of storage.
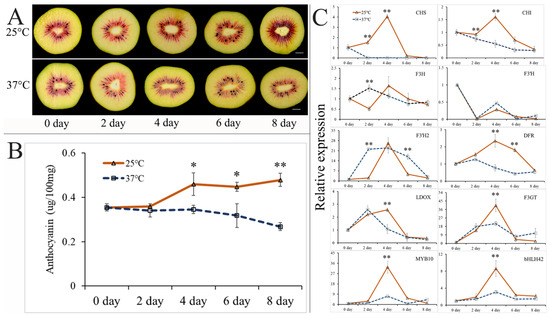
Figure 2.
Phenotypes, anthocyanin contents, and gene expression levels of ‘Hongyang’ kiwifruit stored at 25 °C and 37 °C after harvest: (A) cross-cutting phenotypes of ‘Hongyang’ kiwifruit after 0, 2, 4, 6, and 8 days of storage at 25 °C and 37 °C. Scale represents 1.0 cm; (B) anthocyanin contents in ‘Hongyang’ kiwifruit after 0, 2, 4, 6, and 8 days of storage at 25 °C and 37 °C; and (C) expression levels of genes involved in anthocyanin metabolism in ‘Hongyang’ kiwifruit after 0, 2, 4, 6, and 8 days of storage at 25 °C and 37 °C. Results are presented as the mean ± SD of three replicates. *, p ≤ 0.05; **, p ≤ 0.01.
To identify the key genes involved in the downregulation of anthocyanin synthesis induced by high postharvest temperature, qRT-PCR was performed to detect the expression levels of eight key structural genes in the anthocyanin biosynthesis pathway (AcCHS, AcCHI, AcF3H, AcF3′H, AcF3′H2, AcDFR, AcLDOX, and AcF3GT) and two important transcription factor genes (AcMYB10 and AcbHLH42) (Figure 2C). During the 25 °C storage period, the expression levels of all the genes initially increased, followed by a decline, reaching their peak expression after 4 days of storage, except for AcCHI, AcF3H, and AcF3′H; during 37 °C storage, no consistent gene expression pattern was observed, suggesting disordered anthocyanin synthesis when the postharvest temperature is high. Notably, significantly higher expression levels of AcCHS, AcCHI, AcDFR, AcLDOX, AcF3GT, AcMYB10, and AcbHLH42 were observed after 4 days of storage at 25 °C than at 37 °C (Figure 2C). As the first significant difference in anthocyanin content between the two temperature treatments was detected on day 4 of storage (Figure 2B), these findings suggest that a critical timing for the response of kiwifruit to high postharvest temperature occurs within the initial 4 days of storage. Furthermore, only one significant difference being found in the expression levels of AcLDOX, AcF3GT, AcMYB10, and AcbHLH42 on day 4 of storage between the two temperature treatments implies their role in responding to high-temperature stress and influencing postharvest anthocyanin synthesis. To further detect the interaction among these anthocyanin-related genes, Pearson’s correlation coefficient (r) test was performed using the qRT-PCR data (Figure S2). In general, the expression levels of AcCHS, AcCHI, AcF3H, AcDFR, AcLDOX, AcF3GT, AcMYB10 and AcbHLH42 exhibited statistically significant correlations (p < 0.05). Notably, the expression level of AcMYB10 exhibited a strong positive correlation with that of AcbHLH42 (r = 0.96), while their expressions also demonstrated strong positive correlations with AcCHS (r = 0.87 and 0.82, respectively) and AcF3GT (r = 0.89 and 0.84, respectively). Collectively, these results indicate that elevated postharvest temperatures have a detrimental impact on the synthesis of anthocyanins in ‘Hongyang’ kiwifruit; AcLDOX, AcF3GT, AcMYB10, and AcbHLH42 may play crucial roles in regulating the temperature-induced accumulation of anthocyanins.
2.3. Interaction between AcMYB10 and AcbHLH42
Considering the significantly correlated expression of AcbHLH42 and AcMYB10 observed, we investigated the interactions between AcMYB10 and AcbHLH42 using bimolecular fluorescence complementation (BiFC) assays. Two fusion protein vectors, pSPYNE-AcMYB10 (or pSPYNE-AcbHLH42) and pSPYCE-AcbHLH42 (or pSPYCE- AcMYB10), were constructed and co-transformed into onion epidermal cells (Figure 3). As a result, a strong yellow fluorescent signal was observed in the nucleus transformed with both pSPYNE-AcMYB10 + pSPYCE-AcbHLH42 and pSPYNE-AcbHLH42 + pSPYCE-AcMYB10, as well as nuclear localization. No fluorescent signal was detected in the cells co-transformed with negative controls (pSPYNE-AcMYB10 + pSPYCE, pSPYNE-AcbHLH42 + pSPYCE, pSPYCE-AcbHLH42 + pSPYNE and pSPYCE-AcMYB10 + pSPYNE) or the two empty vectors (pSPYCE + pSPYNE) (Figure 4). These analyses indicate that both AcMYB10 and AcbHLH42 are nuclear proteins and are able to physically interact in plant cells.
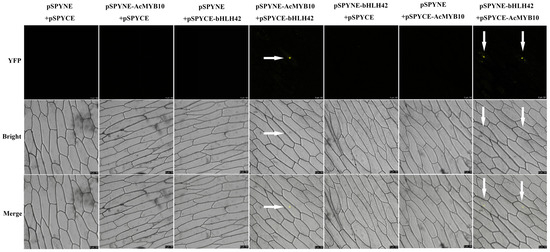
Figure 3.
Bimolecular fluorescence complementation (BiFC) illustration of the interaction between AceMYB10 and AcbHLH42 in onion (Allium cepa) epidermal cells. Note: pSPYNE and pSPYCE vectors were used as the negative controls; YFP fluorescence was detected 2 days after transfection; photos were taken on a fluorescence microscope at a magnification of 200×; arrows point to the locations of the yellow fluorescence.
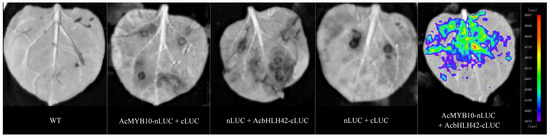
Figure 4.
Bimolecular luminescence complementation (BiLC) illustration of the interactions between AcMYB10 and AcbHLH42 in tobacco (N. benthamiana) leaves. Note: The four constructs were used as negative controls: WT, AcMYB10-nLUC+cLUC, nLUC+AcbHLH42-cLUC, and nLUC+cLUC. AcMYB10-nLUC and AcbHLH42-cLUC were combined at 1:1 (v/v). The images were captured with a charge-coupled device camera at 3 days post inoculation (dpi); scale at the bottom right of the picture represents 1 cm.
To confirm this interaction, AcMYB10 was fused to the N-terminal of LUC (AcMYB10-nLUC), and AcbHLH42 was fused to the C-terminal of LUC (AcbHLH42-cLUC). Then, the constructs were transiently expressed in Nicotiana benthamiana leaves. The results showed that only leaves co-transformed with AcMYB10-nLUC and AcbHLH42-cLUC produced a strong LUC signal (Figure 4). No fluorescence signal was detected in the WT (blank control) or in AcMYB10-nLUC+cLUC, nLUC+AcbHLH42-cLUC or nLUC+cLUC) (Figure 4). The results further confirmed the interaction between AcMYB10 and AcbHLH42.
2.4. AcMYB10 and AcbHLH42 Regulate Promoter Activity of AcLDOX and AcF3GT
Given the synergistic expression patterns of AcMYB10, AcbHLH42, AcLDOX and AcF3GT, Y1H assays were performed to investigate the transcriptional activation of AcMYB10 and AcbHLH42 on the promoters of AcLDOX and AcF3GT (Figure 5). The results showed that all of the yeast cells grew well on SD/-Trp/-Ura media, whereas only the positive control and bait vectors AcLDOXpro::Lacz and AcF3GTpro::Lacz co-transformed with the prey vector pJG-AcMYB10 had blue cells on media supplemented with X-gal, showing the AcMYB10-promoted expression of LacZ driven by the AcLDOX and AcF3GT promoters (Figure 5A). AcbHLH42 could not directly bind to the promoters of AcLDOX or AcF3GT (Figure 5B). From the above results, we concluded that AcMYB10 and AcbHLH42 bind to each other and AcMYB10 could directly binds to the promoter of AcLDOX and AcF3GT, thereby regulating anthocyanin biosynthesis.
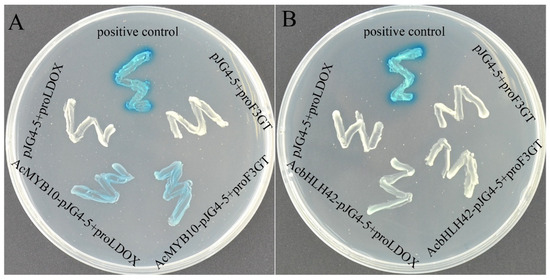
Figure 5.
Y1H assay of AcMYB10 and AcbHLH42 on the promoters of AcLDOX and AcF3GT: (A) Y1H assay showing AcMYB10 binds to the promoters of AcLDOX (AcLDOX pro::Lacz) and AcF3GT (AcF3GTpro::Lacz); and (B) Y1H assay showing bHLH42 does not bind the promoters of AcLDOX (AcLDOX pro::Lacz) and AcF3GT (AcF3GTpro::Lacz). Note: positive control: “p53::LacZ + pJG-p53”; negative control: “bait + pJG4-5” and co-transformants (bait + prey) on SD/-Trp/-Ura medium supplemented with X-gal for 3 days.
3. Discussion
In this study, our findings demonstrated that bagging treatment significantly increased anthocyanin accumulation, reduced chlorophyll content, and promoted the expression level of anthocyanin synthesis genes in the inner pericarp of ‘Hongyang’ kiwifruit, which is in accordance with the previous studies on kiwifruit [39], apple [40] and peach [41]. However, despite being reported as a light-responsive and versatile transcription factor that directly binds to, and induces, the expression of certain MYB transcription factors, or that activates the expression of biosynthetic enzyme genes in conjunction with PIF3 activity along with other MYB factors [24,42,43,44,45], HY5 did not demonstrate any significant synergistic effect on fruit-bagging-induced anthocyanin synthesis in ‘Hongyang’. This suggests that the observed increase in anthocyanin synthesis in bagged fruit may be independent of light. Therefore, we propose that bagging treatments might enhance anthocyanin content through comprehensively modifying the microenvironment surrounding ‘Hongyang’, including reducing ambient temperature and humidity. In addition to anthocyanin accumulation, the effect of bagging treatment on the other qualities of ‘Hongyang’ kiwifruit needs more in-depth and comprehensive research, as previously reported [16].
While the positive regulatory role of low temperature in promoting anthocyanin synthesis in horticultural crops has been widely acknowledged [46,47,48,49,50,51,52], relatively limited attention has been devoted to investigating the impact of high temperature on anthocyanin accumulation [53]. The findings from our previous study [26] demonstrated that maintaining a temperature ≤25 °C during fruit ripening on the tree ensures the functional biosynthesis of anthocyanins in ‘Hongyang’. However, the microenvironment surrounding kiwifruit during development is inherently unstable, and kiwifruit is commonly harvested prematurely and subjected to postharvest ripening treatments in most commercial contexts in China. Based on this, we investigated the impact of high postharvest temperatures on anthocyanin synthesis. The results demonstrated that, similar to preharvest conditions, high postharvest temperatures negatively affect anthocyanin synthesis. Although the results of gene expression analysis in our study suggested that high temperatures negatively regulate the genes involved in anthocyanin synthesis, further research is required to determine whether these elevated temperatures induce anthocyanin degradation. Additionally, considering the predominant green-fleshed nature of most kiwifruit cultivars, further investigations are warranted to determine whether post-harvest high-temperature conditions delay (as observed in Thai lime) [54] or enhance (as seen in banana) [55] chlorophyll degradation.
Currently, accumulating evidence strongly supports the dominant role of the MYB–bHLH–WD40 complex in regulating anthocyanin biosynthesis across the plant kingdom [34]. In kiwifruit, the interactions between AcMYB123 and AcbHLH42 [38], AcMYBF110 and AcbHLH1/4/5 [56], as well as AcMYB10 and AcbHLH42 in our present study [36] positively regulate the expressions of AcF3GT and AcANS (AcLDOX), thereby facilitating anthocyanin biosynthesis in the inner pericarp of ‘Hongyang’ kiwifruit. Therefore, based on the findings of this study, we hypothesize that the regulation of enhanced anthocyanin biosynthesis induced via fruit bagging or via 25 °C postharvest storage treatment (compared with 37 °C) may also involve a similar mechanism (Figure S3). However, the regulation of individual or specific anthocyanin accumulation remains poorly understood, necessitating the utilization of advanced measurement techniques (such as HPLC, LCMS, or metabolome profiling) in future investigations. Furthermore, given the abundance of MYB orthologs and paralogs in plant genomes, further investigation is warranted to elucidate their synergistic, distinct, redundant, or competitive roles in anthocyanin synthesis. Additionally, it is crucial to explore their regulatory mechanisms including the identification of interacting bHLH partners.
4. Materials and Methods
4.1. Plant Materials and Sample Treatment
The bagging experiment was conducted on 8-year-old female vines of A. chinensis cv. ‘Hongyang’ at the Cangxi Kiwifruit Research Institute in Sichuan Province, China (105.96° E, 31.76° N). Nine vines with similar growth vigor and yield potential were divided into three replicates, each containing three vines. At 40 days after flowering (DAF), thirty fruits from each plant were carefully enclosed in custom-made kiwifruit paper bags (Jinguonong Packaging Materials Co., Ltd., Yantai, China) with a single layer of yellow outer surface and a black inner surface (150 mm × 180 mm). The fruits were collected at 130DAF while the unbagged fruits were collected at the same time and served as controls. The flesh of each fruit (excluding the fruit axis) was diced and immediately frozen using liquid nitrogen before being securely stored at −80 °C for further analysis. For the postharvest treatment, a total of 180 fruits, harvested at 130DAF and exhibiting identical ripeness and size, without any physical damage or bacterial infection, were sampled. The fruits were randomly divided into two treatment groups: one was stored at 37 °C and the other at 25 °C, with three replicates for each temperature condition. Fruit samples were collected at 0, 2, 4, 6, and 8 days of storage for each treatment group, respectively, with six fruits per time point. The flesh (excluding the fruit axis) of each fruit was chopped and treated with liquid nitrogen before being stored at −80 °C for subsequent measurement.
4.2. Determination of Anthocyanin and Chlorophyll Content
The total anthocyanin content was extracted and determined following the method described by Shin et al. [45] and Lim et al. [57]. In brief, the sample was ground in liquid nitrogen, weighed accurately (0.10 g), and mixed with 600 µL of 1% hydrochloric methanol buffer. The mixture was then shaken overnight (8 h) at 4 °C in darkness using a shaker (TS-2102C, JTLIANGYOU, Changzhou, China). To each sample, 200 µL of double-distilled water and 200 µL of chloroform were added before centrifugation at 12,000 rpm for 10 min. The absorbance values were measured at wavelengths of 530 nm and 657 nm using a multifunctional full-wavelength enzyme labeler (Infinite M200 PRO, TECAN, Männedorf, Switzerland) by an absorbing supernatant with a volume of 200 µL. Finally, the anthocyanin content was calculated as A530 − 0.33 × A657.
Chlorophyll a and b were extracted and determined using the methods described by Hiscox and Israelstam [58]. In summary, the sample was triturated with liquid nitrogen in mortars. A precise weight of 0.10 g (±0.04 g) of powder was diluted to 5.0 mL dimethyl sulfoxide in a 15 mL centrifuge tube and transferred to a vibration incubator at 28 °C under dark conditions and at a speed of 200 rpm/min for 72 h. After centrifugation, the absorbance values of the supernatant extract were measured at wavelengths of 663 nm and 645 nm using a spectrophotometer. The content of Chl a, Chl b, and total Chl were calculated as follows: Chl a = (12.7 × OD663 − 2.69 × OD645) × VT/W × 1000 × VS; Chl b = (22.9 × OD645 − 4.68 × OD663) × VT/W × 1000 × VS; Chl = Chla + Chlb.
4.3. Determination of Gene Expression Levels
The total RNA was extracted using a plant RNA extraction kit (RC401, Vazyme, Nanjing, China) according to the manufacturer’s instructions. Briefly, 1 μg of RNA was treated with DNase I and reverse transcribed at 37 °C using a reverse transcription kit (AE341-02, TransGen Biotech, Beijing, China). qRT-PCR was performed on a Quant Studio 6 Flex system (ThermoFisher Scientific, Carlsbad, CA, USA), following the manufacturer’s instructions for AceQ qPCR SYBR Green Master Mix (Q131-02, Vazyme, Nanjing, China). Based on the previous reports [2,24,36,38], we selected seventeen genes involved in light response, anthocyanin synthesis, and chlorophyll synthesis and degradation for determination of their expression levels (Table S1). Among them, HY5 is a key gene related to light response [24]; bHLH42 and MYB10 are transcription factors that regulate anthocyanin synthesis; CHS, CHI, F3H, F3′H, DFR, LDOX and F3GT are structure genes on the anthocyanin biosynthesis pathway [59]; and PAO, CBR, GLUTR, RBCS, SGR, CAO and PPH are structural genes related to chlorophyll synthesis and degradation. Achn107181 (Actin) was conducted as the endogenous control [60]. All the primers used in this study are listed in Table S1. The qRT-PCR procedure and expression level determination method (2−∆∆ Ct) were performed following the workflow reported by Yu et al. [36].
4.4. Bimolecular Fluorescence Complementation (BiFC) Assay
The BiFC assay was performed based on the procedures previously described [61]. Briefly, the coding sequences (CDS) of AcMYB10 and AcbHLH42 were cloned into a pSPYNE vector, resulting in pSPYNE-AcMYB10 and pSPYNE-AcbHLH42 encoding 155 amino acids at the N-terminal of yellow fluorescent protein (YFP). The sequences were then inserted into a pSPYCE carrier containing YFP C-terminal 83 amino acids, resulting in the generation of pSPYCE-AcbHLH42 and pSPYCE-AcMYB10. The recombinant or control vectors were transformed into A. tumefaciens strain EHA105 and then co-transformed into onion epidermal cells using an infiltration buffer. After 48 h invasion, the yellow fluorescence signal of YFP in the onion nucleus was observed using a confocal three-dimensional scanning microscope with excitation wavelengths set at YFP:510 nm and DAPI:488 nm (Leica-Microsystems TCS-SP8, Wetzlar, Germany).
4.5. Bimolecular Luminescence Complementation (BiLC) Assay
The BiLC assay was performed as previously described [62]. Briefly, the full-length ORF of AcbHLH42, excluding the stop codon, was cloned into the pCAMBIA-nLUC vector to create the fusion vector pCAMBIA-AcbHLH42-nLUC. Similarly, the complete ORF of AcMYB10 was cloned into the pCAMBIA-cLUC vector to generate the fusion vector pCAMBIA-AcMYB10-cLUC. Agrobacterium cultures containing these constructs were co-transformed into tobacco leaves at a 1:1 ratio. After incubating in darkness for 12 h, plants were transferred to light conditions at 25 °C for 48 h. Transformed tobacco leaves were then soaked in a solution of 0.15 mg mL−1 D-Luciferin potassium (115144-35-9, Coolaber, Beijing, China) for 2–3 min before observing LUC activity using a Chemiluminescence Imaging System (NightSHADE LB985, Berthold, Germany).
4.6. Yeast One-Hybrid (Y1H) Assay
The Y1H assay was performed as previously described [61]. Briefly, the ~1.5 kb promoter sequence of AcLODX and the ~1.1 kb promoter sequence of AcF3GT (Figure S1) were amplified and inserted into pLacZi plasmids (Clontech; TaKaRa Bio Inc., Shiga, Japan) using EcoRI and kpnI restriction sites to generate AcLODX pro::LacZ and AcF3GT pro::LacZ constructs. The full-length CDS of AcMYB10 and AcbHLH42 were cloned into pJG4-5 vectors (Clontech; TaKaRa Bio Inc.) using EcoRI and XhoI restriction sites, creating pJG-AcMYB10 and pJG-AcbHLH42, respectively. The NcoI-digested AcLODX pro::LacZ and AcF3GT pro::LacZ vectors were co-transformed with the pJG-AcMYB10 and pJG-AcbHLH42 vectors into yeast strain EGY48 using a yeast transformation kit (SK2400-200T, Coolaber, Beijing, China). The transformants were cultivated on SD/-Trp-Ura dropout medium at 30 °C for a duration of 2 days. Subsequently, single colonies were streaked onto an SD chromogenic medium (X-gal) and incubated at 28 °C for a period of 2 days.
5. Conclusions
The application of a fruit bagging treatment significantly enhanced anthocyanin accumulation and reduced chlorophyll accumulation in the pericarp of ‘Hongyang’ kiwifruit during fruit development. The results of gene expression analysis suggested that AcMYB10, bHLH42, AcCHS, AcCHI, AcF3H, AcF3′H, AcDFR, AcLDOX, and AcF3GT are responsible for upregulating anthocyanin levels, whereas AcCBR, AcGLUTR, AcRBCS, and AcPPH may contribute to downregulating chlorophyll levels. The high-temperature postharvest treatment (37 °C) significantly suppressed anthocyanin synthesis compared with the control at 25 °C, as evidenced by noticeably decreased anthocyanin accumulation starting from day 4 of storage. This inhibitory effect could be attributed to the downregulation of key genes and TFs, including AcCHS, AcCHI, AcF3H, AcDFR, AcLDOX, AcF3GT, AcMYB10, and AcbHLH42. The nuclear expression of AcMYB10 and AcbHLH42, as well as the interaction between their translated proteins, was confirmed through BiFC and BiLC analyses. The results of a yeast one-hybrid assay revealed that AcMYB10 can activate the promoters of AcLDOX and AcF3GT, potentially elucidating their high expression synergy in fruit bagging and postharvest experiments. This study provides a further step in elucidating the mechanisms regulating anthocyanin synthesis induced by fruit bagging and temperature treatments in ‘Hongyang’ kiwifruit.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15010097/s1, Table S1. Primer sequences used in this study; Figure S1. Promoter sequences of AcF3GT and AcLDOX; Figure S2. Correlation analysis of gene expression between anthocyanin synthesis genes and regulatory genes; Figure S3. Regulation model of anthocyanin accumulation pathway in bagging and storage of kiwifruit.
Author Contributions
Conceptualization, Y.W. and X.L.; methodology, M.Y.; software, M.Y.; validation, J.X., K.D. and C.Z.; formal analysis, M.Y.; investigation, X.Q.; resources, K.D.; data curation, H.G.; writing—original draft preparation, M.Y.; writing—review and editing, C.Z.; visualization, D.Q.; supervision, J.H.; project administration, C.Z.; funding acquisition, M.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by ‘National Key R&D Program of China (grant number 2022YFD1600500)’, ‘Heilongjiang Postdoctoral Science Foundation (grant number LBH-Z22087)’, and ‘Young Talents Foundation of NEAU (grant number 21QC27)’.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are included in the article and supplementary materials. Additional related data can be obtained by contacting the corresponding authors upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Guo, J.; Yuan, Y.H.; Dou, P.; Yue, T.L. Multivariate statistical analysis of the polyphenolic constituents in kiwifruit juices to trace fruit varieties and geographical origins. Food Chem. 2017, 232, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, B.; Qi, Y.; Chen, X.; Liu, C.; Liu, Z.; Ren, X. Expression differences of pigment structural genes and transcription factors explain flesh coloration in three contrasting kiwifruit cultivars. Front. Plant Sci. 2017, 8, 1507. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Huang, W.; Wang, Z.; Li, L.; Li, D.; Zhang, Q.; Zhao, T.; Zhang, P. The breeding progress and development status of the kiwifruit industry in China. In Proceedings of the X International Symposium on Kiwifruit 1332, Yalova, Turkey, 27–30 September 2021; pp. 445–454. [Google Scholar]
- Han, X.; Zhang, Y.; Zhang, Q.; Ma, N.; Liu, X.; Tao, W.; Lou, Z.; Zhong, C.; Deng, X.W.; Li, D. Two haplotype-resolved, gap-free genome assemblies for Actinidia latifolia and Actinidia chinensis shed light on the regulatory mechanisms of vitamin C and sucrose metabolism in kiwifruit. Mol. Plant 2023, 16, 452–470. [Google Scholar] [CrossRef]
- Liu, Y.; Qi, Y.; Chen, X.; He, H.; Liu, Z.; Zhang, Z.; Ren, Y.; Ren, X. Phenolic compounds and antioxidant activity in red-and in green-fleshed kiwifruits. Food Res. Int. 2019, 116, 291–301. [Google Scholar] [CrossRef]
- Jaakola, L. Phenylpropanoid metabolism and biosynthesis of anthocyanins. Mol. Biol. Biochem. Fruit Ripening 2013, 5, 117–134. [Google Scholar] [CrossRef]
- Guo, S.-H.; Xu, T.-F.; Shi, T.-C.; Jin, X.-Q.; Feng, M.-X.; Zhao, X.-H.; Zhang, Z.-W.; Meng, J.-F. Cluster bagging promotes melatonin biosynthesis in the berry skins of Vitis vinifera cv. Cabernet Sauvignon and Carignan during development and ripening. Food Chem. 2020, 305, 125502. [Google Scholar] [CrossRef]
- Zhang, B.B.; Guo, J.Y.; Ma, R.J.; Cai, Z.X.; Yan, J.; Zhang, C.H. Relationship between the bagging microenvironment and fruit quality in ‘Guibao’ peach [Prunus persica (L.) Batsch]. J. Hortic. Sci. Biotechnol. 2015, 90, 303–310. [Google Scholar] [CrossRef]
- Mancinelli, A.L. Interaction between Light Quality and Light Quantity in the Photoregulation of Anthocyanin Production. Plant Physiol. 1990, 92, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, H.; Li, J.; Qin, S.; Niu, Z.; Qiao, X.; Yang, B. Influence of genetic background, growth latitude and bagging treatment on phenolic compounds in fruits of commercial cultivars and wild types of apples (Malus spp.). Eur. Food Res. Technol. 2021, 247, 1149–1165. [Google Scholar] [CrossRef]
- Jing, C.; Feng, D.; Zhao, Z.; Wu, X.; Chen, X. Effect of environmental factors on skin pigmentation and taste in three apple cultivars. Acta Physiol. Plant. 2020, 42, 69. [Google Scholar] [CrossRef]
- Zhu, M.; Fang, W.; Chen, C.; Wang, L.; Cao, K. Effects of shading by bagging on carotenoid accumulation in peach fruit flesh. J. Plant Growth Regul. 2021, 40, 1912–1921. [Google Scholar] [CrossRef]
- Sun, R.-Z.; Cheng, G.; Li, Q.; Zhu, Y.-R.; Zhang, X.; Wang, Y.; He, Y.-N.; Li, S.-Y.; He, L.; Chen, W. Comparative physiological, metabolomic, and transcriptomic analyses reveal developmental stage-dependent effects of cluster bagging on phenolic metabolism in Cabernet Sauvignon grape berries. BMC Plant Biol. 2019, 19, 583. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Cao, P.; Wu, X.; Yuan, Y.; Yu, P.; Tao, S.; Zhang, S. Effects of light quality on fruit quality and absorption of mineral elements in’Dangshan Suli’pear fruit development. Photosynthetica 2018, 45, 1173–1184. [Google Scholar] [CrossRef]
- Han, F.; Liu, X.; Zhong, C. Effects of different types of fruit bags on the quality of kiwifruit ‘Jinkui’. China Fruits 2017, 3, 45–49. (In Chinese) [Google Scholar] [CrossRef]
- Xu, Y.; Liu, Y.; Li, W.; Yang, C.; Lin, Y.; Wang, Y.; Chen, C.; Wan, C.; Chen, J.; Gan, Z. The Effects of Bagging on Color Change and Chemical Composition in ‘Jinyan’ Kiwifruit (Actinidia chinensis). Horticulturae 2022, 8, 478. [Google Scholar] [CrossRef]
- Liao, G.; He, Y.; Li, X.; Zhong, M.; Huang, C.; Yi, S.; Liu, Q.; Xu, X. Effects of bagging on fruit flavor quality and related gene expression of AsA synthesis in Actinidia eriantha. Sci. Hortic. 2019, 256, 108511. [Google Scholar] [CrossRef]
- Li, Y.; Qi, X.; Cui, W.; Lin, M.; Qiao, C.; Zhong, Y.; Fang, J.; Hu, C. Restraint of bagging on fruit skin coloration in on-tree kiwifruit (Actinidia arguta). J. Plant Growth Regul. 2021, 40, 603–616. [Google Scholar] [CrossRef]
- Ma, C.; Cao, S.; Li, W.; Du, J.; Han, Z.; Li, L.; Wang, R. Effects of different bagging on main quality and storability indices of ‘Hongyang’ kiwifruits. Food Ferment. Ind. 2019, 45, 202–208. (In Chinese) [Google Scholar] [CrossRef]
- Jue, D.-W.; Sang, X.-L.; Li, Z.-X.; Zhang, W.-L.; Liao, Q.-H.; Tang, J. Determination of the effects of pre-harvest bagging treatment on kiwifruit appearance and quality via transcriptome and metabolome analyses. Food Res. Int. 2023, 173, 113276. [Google Scholar] [CrossRef]
- Li, Y.; Qi, X.; Lin, M.; Li, Z.; Fang, J. Effect of bagging on fruit pigmentation in two types of red-fleshed kiwifruit. J. Fruit Sci. 2016, 33, 1492–1501. [Google Scholar] [CrossRef]
- Carmona, L.; Alquézar, B.; Marques, V.V.; Peña, L. Anthocyanin biosynthesis and accumulation in blood oranges during postharvest storage at different low temperatures. Food Chem. 2017, 237, 7–14. [Google Scholar] [CrossRef]
- Li, W.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 16861. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.-Y.; Paik, I.; Nguyen, T.T.; Kim, J.; Oh, E. High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 2017, 8, 1787. [Google Scholar] [CrossRef]
- Niu, J.; Zhang, G.; Zhang, W.; Goltsev, V.; Sun, S.; Wang, J.; Li, P.; Ma, F. Anthocyanin concentration depends on the counterbalance between its synthesis and degradation in plum fruit at high temperature. Sci. Rep. 2017, 7, 7684. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.P.; Wang, Y.C.; Li, Z.Z.; Jiang, Z.W.; Yang, H.L.; Gong, J.J.; He, S.S.; Wu, S.Q.; Yang, Z.Q.; Zheng, J. High-temperature inhibition of biosynthesis and transportation of anthocyanins results in the poor red coloration in red-fleshed Actinidia chinensis. Physiol. Plant. 2015, 153, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qi, Y.; Zhang, A.; Wu, H.; Liu, Z.; Ren, X. Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis). Plant Mol. Biol. 2019, 100, 451–465. [Google Scholar] [CrossRef]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L. A key structural gene, AaLDOX, is involved in anthocyanin biosynthesis in all red-fleshed kiwifruit (Actinidia arguta) based on transcriptome analysis. Gene 2018, 648, 31–41. [Google Scholar] [CrossRef]
- Montefiori, M.; Espley, R.V.; Stevenson, D.; Cooney, J.; Datson, P.M.; Saiz, A.; Atkinson, R.G.; Hellens, R.P.; Allan, A.C. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 2011, 65, 106–118. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Wei, L.; Cheng, J.; Xiang, J.; Zheng, T.; Wu, J. Transcriptome and proteome analysis of the fig (Ficus carica L.) cultivar Orphan and its mutant Hongyan based on the fruit peel colour in South China. Czech J. Genet. Plant Breed. 2022, 59, 33–42. [Google Scholar] [CrossRef]
- Yang, J.; Li, B.; Shi, W.; Gong, Z.; Chen, L.; Hou, Z. Transcriptional activation of anthocyanin biosynthesis in developing fruit of blueberries (Vaccinium corymbosum L.) by preharvest and postharvest UV irradiation. J. Agric. Food Chem. 2018, 66, 10931–10942. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Uddin, N.; Khan, M.K.U.; Ali, N.; Ali, K.; Jones, D.A. Diverse role of basic Helix-Loop-Helix (bHLH) transcription factor superfamily genes in the fleshy fruit-bearing plant species. Czech J. Genet. Plant Breed. 2022, 59, 1–13. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Yu, M.; Man, Y.; Lei, R.; Lu, X.; Wang, Y. Metabolomics Study of Flavonoids and Anthocyanin-Related Gene Analysis in Kiwifruit (Actinidia chinensis) and Kiwiberry (Actinidia arguta). Plant Mol. Biol. Report. 2020, 38, 353–369. [Google Scholar] [CrossRef]
- Yu, M.; Man, Y.; Wang, Y. Light-and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. Int. J. Mol. Sci. 2019, 20, 5228. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.G.; Seal, A.G.; Montefiori, M.; McGhie, T.K.; Tsang, G.K.; Datson, P.M.; Hilario, E.; Marsh, H.E.; Dunn, J.K.; Hellens, R.P.; et al. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genom. 2013, 14, 28. [Google Scholar] [CrossRef]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Y.; Liao, G.; Xu, X.; Jia, D.; Zhong, M.; Wang, H.; Ye, B. Transcriptome and Metabolome reveal AsA regulatory network between metabolites and genes after fruit shading by bagging in kiwifruit (Actinidia eriantha). Sci. Hortic. 2022, 302, 111184. [Google Scholar] [CrossRef]
- Feng, F.; Li, M.; Ma, F.; Cheng, L. The effects of bagging and debagging on external fruit quality, metabolites, and the expression of anthocyanin biosynthetic genes in ‘Jonagold’ apple (Malus domestica Borkh.). Sci. Hortic. 2014, 165, 123–131. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, M.; Wu, H.; Yuan, C.; Li, H.; Zhang, Y. Effects of fruit bagging on anthocyanin accumulation and related gene expression in peach. J. Am. Soc. Hortic. Sci. 2021, 146, 217–223. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Feng, F.; Li, M.; Ma, F.; Cheng, L. Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Biochem. 2013, 69, 54–61. [Google Scholar] [CrossRef]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 50, 933. [Google Scholar] [CrossRef]
- Fang, Z.; Lin-Wang, K.; Jiang, C.; Zhou, D.; Lin, Y.; Pan, S.; Espley, R.V.; Ye, X. Postharvest temperature and light treatments induce anthocyanin accumulation in peel of ‘Akihime’ plum (Prunus salicina Lindl.) via transcription factor PsMYB10. 1. Postharvest Biol. Technol. 2021, 179, 111592. [Google Scholar] [CrossRef]
- Yamane, T.; Jeong, S.T.; Goto-Yamamoto, N.; Koshita, Y.; Kobayashi, S. Effects of temperature on anthocyanin biosynthesis in grape berry skins. Am. J. Enol. Vitic. 2006, 57, 54–59. [Google Scholar] [CrossRef]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Lo Piero, A.R.; Puglisi, I.; Rapisarda, P.; Petrone, G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J. Agric. Food Chem. 2005, 53, 9083–9088. [Google Scholar] [CrossRef]
- de Rosas, I.; Ponce, M.T.; Malovini, E.; Deis, L.; Cavagnaro, B.; Cavagnaro, P. Loss of anthocyanins and modification of the anthocyanin profiles in grape berries of Malbec and Bonarda grown under high temperature conditions. Plant Sci. 2017, 258, 137–145. [Google Scholar] [CrossRef]
- Movahed, N.; Pastore, C.; Cellini, A.; Allegro, G.; Valentini, G.; Zenoni, S.; Cavallini, E.; D’Incà, E.; Tornielli, G.B.; Filippetti, I. The grapevine VviPrx31 peroxidase as a candidate gene involved in anthocyanin degradation in ripening berries under high temperature. J. Plant Res. 2016, 129, 513–526. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagne, D.; Rowan, D.D.; Troggio, M. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, M.J.; Alquezar, B.; Alos, E.; Lado, J.; Zacarias, L. Biochemical bases and molecular regulation of pigmentation in the peel of Citrus fruit. Sci. Hortic. 2013, 163, 46–62. [Google Scholar] [CrossRef]
- Kaewsuksaeng, S.; Tatmala, N.; Srilaong, V.; Pongprasert, N. Postharvest heat treatment delays chlorophyll degradation and maintains quality in Thai lime (Citrus aurantifolia Swingle cv. Paan) fruit. Postharvest Biol. Technol. 2015, 100, 1–7. [Google Scholar] [CrossRef]
- Yang, X.-t.; Zhang, Z.-q.; Joyce, D.; Huang, X.-m.; Xu, L.-y.; Pang, X.-q. Characterization of chlorophyll degradation in banana and plantain during ripening at high temperature. Food Chem. 2009, 114, 383–390. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, K.; Qi, Y.; Lv, G.; Ren, X.; Liu, Z.; Ma, F. Transcriptional Regulation of Anthocyanin Synthesis by MYB-bHLH-WDR Complexes in Kiwifruit (Actinidia chinensis). J. Agric. Food Chem. 2021, 69, 3677–3691. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-H.; Kim, D.-H.; Kim, J.K.; Lee, J.-Y.; Ha, S.-H. A Radish Basic Helix-Loop-Helix Transcription Factor, RsTT8 Acts a Positive Regulator for Anthocyanin Biosynthesis. Front. Plant Sci. 2017, 8, 1917. [Google Scholar] [CrossRef]
- Hiscox, J.; Israelstam, G. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979, 57, 1332–1334. [Google Scholar] [CrossRef]
- Peng, Y.; Lin-Wang, K.; Cooney, J.M.; Wang, T.; Espley, R.V.; Allan, A.C. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic. Res. 2019, 6, 3. [Google Scholar] [CrossRef]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef]
- Liu, X.; Wu, R.; Bulley, S.M.; Zhong, C.; Li, D. Kiwifruit MYBS1-like and GBF3 transcription factors influence l-ascorbic acid biosynthesis by activating transcription of GDP-L-galactose phosphorylase 3. New Phytol. 2022, 234, 1782–1800. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).