Association Study of a Comprehensive Panel of Neuropeptide-Related Polymorphisms Suggest Potential Roles in Verbal Learning and Memory
Abstract
1. Introduction
2. Experimental Procedures
2.1. Participants
2.2. Phenotyping
2.2.1. Learning and Memory
2.2.2. IQ
2.3. Sample Collection and DNA Extraction
2.4. Genotyping and Gene Selection
2.5. Statistical Analysis
3. Results
3.1. Demographics and Phenotype Analysis
3.2. Genotype Association
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CCK | Cholecystokinin |
| eQTL | Expression quantitative trait loci |
| GALR1 | Galanin Receptor 1 |
| GEC | Genetic Type 1 error calculator |
| GLM | Generalized Linear Model |
| GTEx | Genotype-Tissue Expression |
| HVLT-R | Hopkins Verbal Learning Test—Revised |
| HWE | Hardy Weinberg Equilibrium |
| IGF1R | Insulin-Like Growth Factor 1 Receptor |
| IQ | Intelligence quotient |
| LD | Linkage Disequilibrium |
| MAF | Minor Allele Frequency |
| NPY | Neuropeptide Y |
| OXT | Oxytocin |
| OXTR | Oxytocin Receptor |
| PC1 | Principle Component 1 |
| PC2 | Principle Component 2 |
| PCA | Principle Component Analysis |
| SCG5 | Secretogranin V |
| SNP | Single-Nucleotide Polymorphism |
| VIPR1 | Vasoactive intestinal polypeptide receptor 1 |
| WASI | Wechsler Abbreviated Scale of Intelligence |
References
- Gurd, J.M.; Kischka, U.; Marshall, J.C. Handbook of Clinical Neuropsychology, 2nd ed.; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Kandel, E.R.; Dudai, Y.; Mayford, M.R. The molecular and systems biology of memory. Cell 2014, 157, 163–186. [Google Scholar] [CrossRef] [PubMed]
- Tulving, E. Episodic memory: From mind to brain. Annu. Rev. Psychol. 2002, 53, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Toader, A.C.; Regalado, J.M.; Li, Y.R.; Terceros, A.; Yadav, N.; Kumar, S.; Satow, S.; Hollunder, F.; Bonito-Oliva, A.; Rajasethupathy, P. Anteromedial thalamus gates the selection and stabilization of long-term memories. Cell 2023, 186, 1369–1381.e17. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, B.A.; Buckner, R.L. Functional-anatomic correlates of individual differences in memory. Neuron 2006, 51, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.S.; Quenzer, L.F. Psychopharmacology: Drugs, the Brain, and Behavior; Sinauer Associates, Inc.: Sunderland, MA, USA, 2013. [Google Scholar]
- Burbach, J.P. Neuropeptides from concept to online database www.neuropeptides.nl. Eur. J. Pharmacol. 2010, 626, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Shahjahan, M.; Kitahashi, T.; Parhar, I.S. Central pathways integrating metabolism and reproduction in teleosts. Front. Endocrinol. 2014, 5, 36. [Google Scholar] [CrossRef]
- Kormos, V.; Gaszner, B. Role of neuropeptides in anxiety, stress, and depression: From animals to humans. Neuropeptides 2013, 47, 401–419. [Google Scholar] [CrossRef]
- Gotzsche, C.R.; Woldbye, D.P. The role of NPY in learning and memory. Neuropeptides 2016, 55, 79–89. [Google Scholar] [CrossRef]
- Ogren, S.O.; Kuteeva, E.; Elvander-Tottie, E.; Hokfelt, T. Neuropeptides in learning and memory processes with focus on galanin. Eur. J. Pharmacol. 2010, 626, 9–17. [Google Scholar] [CrossRef]
- Borbely, E.; Scheich, B.; Helyes, Z. Neuropeptides in learning and memory. Neuropeptides 2013, 47, 439–450. [Google Scholar] [CrossRef]
- Chaudhury, D.; Loh, D.H.; Dragich, J.M.; Hagopian, A.; Colwell, C.S. Select cognitive deficits in vasoactive intestinal peptide deficient mice. BMC Neurosci. 2008, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Lei, G.; Jackson, M.F.; Macdonald, J.F. The involvement of PACAP/VIP system in the synaptic transmission in the hippocampus. J. Mol. Neurosci. 2010, 42, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Lindenberg, A.; Domes, G.; Kirsch, P.; Heinrichs, M. Oxytocin and vasopressin in the human brain: Social neuropeptides for translational medicine. Nat. Rev. Neurosci. 2011, 12, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Panizzon, M.S.; Lyons, M.J.; Jacobson, K.C.; Franz, C.E.; Grant, M.D.; Eisen, S.A.; Xian, H.; Kremen, W.S. Genetic architecture of learning and delayed recall: A twin study of episodic memory. Neuropsychology 2011, 25, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, N.; Kaprio, J.; Rinne, J.O.; Vuoksimaa, E. Immediate verbal recall and familial dementia risk: Population-based study of over 4000 twins. J. Neurol. Neurosurg. Psychiatry 2019, 90, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Bearden, C.E.; Glahn, D.C. Cognitive genomics: Searching for the genetic roots of neuropsychological functioning. Neuropsychology 2017, 31, 1003–1019. [Google Scholar] [CrossRef]
- Inui, A. Neuropeptide gene polymorphisms and human behavioural disorders. Nat. Rev. Drug Discov. 2003, 2, 986–998. [Google Scholar] [CrossRef]
- Montag, C.; Reuter, M. Disentangling the molecular genetic basis of personality: From monoamines to neuropeptides. Neurosci. Biobehav. Rev. 2014, 43, 228–239. [Google Scholar] [CrossRef]
- Tost, H.; Kolachana, B.; Hakimi, S.; Lemaitre, H.; Verchinski, B.A.; Mattay, V.S.; Weinberger, D.R.; Meyer-Lindenberg, A. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc. Natl. Acad. Sci. USA 2010, 107, 13936–13941. [Google Scholar] [CrossRef]
- Tops, M.; van Ijzendoorn, M.H.; Riem, M.M.; Boksem, M.A.; Bakermans-Kranenburg, M.J. Oxytocin receptor gene associated with the efficiency of social auditory processing. Front. Psychiatry 2011, 2, 60. [Google Scholar] [CrossRef][Green Version]
- Lucht, M.J.; Barnow, S.; Sonnenfeld, C.; Rosenberger, A.; Grabe, H.J.; Schroeder, W.; Volzke, H.; Freyberger, H.J.; Herrmann, F.H.; Kroemer, H.; et al. Associations between the oxytocin receptor gene (OXTR) and affect, loneliness and intelligence in normal subjects. Prog. Neuropsychopharmacol. Biol. Psychiatry 2009, 33, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Li, Z.; Su, Y. The association between oxytocin receptor gene polymorphism (OXTR) and trait empathy. J. Affect. Disord. 2012, 138, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, Y.; Li, R.; Broster, L.S.; Zhou, C.; Yang, S. Association of Oxytocin Receptor Gene (OXTR) rs53576 Polymorphism with Sociality: A Meta-Analysis. PLoS ONE 2015, 10, e0131820. [Google Scholar] [CrossRef] [PubMed]
- Lerer, E.; Levi, S.; Salomon, S.; Darvasi, A.; Yirmiya, N.; Ebstein, R.P. Association between the oxytocin receptor (OXTR) gene and autism: Relationship to Vineland Adaptive Behavior Scales and cognition. Mol. Psychiatry 2008, 13, 980–988. [Google Scholar] [CrossRef] [PubMed]
- de Wied, D.; Diamant, M.; Fodor, M. Central nervous system effects of the neurohypophyseal hormones and related peptides. Front. Neuroendocrinol. 1993, 14, 251–302. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, D.; Zhang, P.; Li, W.; Qin, W.; Liu, F.; Xu, J.; Xu, Q.; Wang, J.; Ye, Z.; et al. Neural mechanisms of AVPR1A RS3-RS1 haplotypes that impact verbal learning and memory. Neuroimage 2020, 222, 117283. [Google Scholar] [CrossRef]
- Redrobe, J.P.; Dumont, Y.; Herzog, H.; Quirion, R. Characterization of neuropeptide Y, Y2 receptor knockout mice in two animal models of learning and memory processing. J. Mol. Neurosci. 2004, 22, 159–166. [Google Scholar] [CrossRef]
- Itokawa, M.; Arai, M.; Kato, S.; Ogata, Y.; Furukawa, A.; Haga, S.; Ujike, H.; Sora, I.; Ikeda, K.; Yoshikawa, T. Association between a novel polymorphism in the promoter region of the neuropeptide Y gene and schizophrenia in humans. Neurosci. Lett. 2003, 347, 202–204. [Google Scholar] [CrossRef]
- Brandt, J. The hopkins verbal learning test: Development of a new memory test with six equivalent forms. Clin. Neuropsychol. 1991, 5, 125–142. [Google Scholar] [CrossRef]
- Kim, Y.; Bark, S.; Hook, V.; Bandeira, N. NeuroPedia: Neuropeptide database and spectral library. Bioinformatics 2011, 27, 2772–2773. [Google Scholar] [CrossRef]
- Kaufman, A.S.; Lichtenberger, E.O.; McLean, J.E. Two- and three-factor solutions of the WAIS-III. Assessment 2001, 8, 267–280. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lei, L.; Massart, R.; Suderman, M.J.; Machnes, Z.; Elgbeili, G.; Laplante, D.P.; Szyf, M.; King, S. DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm. PLoS ONE 2014, 9, e107653. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, H.; Wang, W.; Yang, L.; Xie, L.L.; Huang, W. Association of insulin growth factor-1 receptor gene polymorphisms with genetic susceptibility to idiopathic short stature. Genet. Mol. Res. 2013, 12, 4768–4779. [Google Scholar] [CrossRef] [PubMed]
- Counts, S.E.; Perez, S.E.; Ginsberg, S.D.; Mufson, E.J. Neuroprotective role for galanin in Alzheimer’s disease. Galanin 2010, 102, 143–162. [Google Scholar]
- Crawley, J.N. Galanin impairs cognitive abilities in rodents: Relevance to Alzheimer's disease. Cell Mol. Life Sci. 2008, 65, 1836–1841. [Google Scholar] [CrossRef]
- Sankar, R. Galanin and epilepsy: Promises with nuances. Epilepsy Curr. 2005, 5, 78–80. [Google Scholar] [CrossRef]
- Kinney, J.W.; Sanchez-Alavez, M.; Barr, A.M.; Criado, J.R.; Crawley, J.N.; Behrens, M.M.; Henriksen, S.J.; Bartfai, T. Impairment of memory consolidation by galanin correlates with in vivo inhibition of both LTP and CREB phosphorylation. Neurobiol. Learn. Mem. 2009, 92, 429–438. [Google Scholar] [CrossRef]
- Jackson, K.J.; Chen, X.; Miles, M.F.; Harenza, J.; Damaj, M.I. The neuropeptide galanin and variants in the GalR1 gene are associated with nicotine dependence. Neuropsychopharmacology 2011, 36, 2339–2348. [Google Scholar] [CrossRef] [PubMed]
- Grove, T.B.; Burghardt, K.J.; Kraal, A.Z.; Dougherty, R.J.; Taylor, S.F.; Ellingrod, V.L. Oxytocin Receptor (OXTR) Methylation and Cognition in Psychotic Disorders. Mol. Neuropsychiatry 2016, 2, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.; Hoffmann, T.J.; Wigg, K.; Jenkins, J.M. Association between the oxytocin receptor (OXTR) gene and children’s social cognition at 18 months. Genes. Brain Behav. 2014, 13, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Willmott, M.; Vetuz, G.; Toye, C.; Kirley, A.; Hawi, Z.; Brookes, K.J.; Gill, M.; Kent, L. Evidence that genetic variation in the oxytocin receptor (OXTR) gene influences social cognition in ADHD. Prog. Neuropsychopharmacol. Biol. Psychiatry 2010, 34, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Israel, S.; Lerer, E.; Shalev, I.; Uzefovsky, F.; Riebold, M.; Laiba, E.; Bachner-Melman, R.; Maril, A.; Bornstein, G.; Knafo, A.; et al. The oxytocin receptor (OXTR) contributes to prosocial fund allocations in the dictator game and the social value orientations task. PLoS ONE 2009, 4, e5535. [Google Scholar] [CrossRef] [PubMed]
- Dauge, V.; Lena, I. CCK in anxiety and cognitive processes. Neurosci. Biobehav. Rev. 1998, 22, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Namjou, B.; Keddache, M.; Marsolo, K.; Wagner, M.; Lingren, T.; Cobb, B.; Perry, C.; Kennebeck, S.; Holm, I.A.; Li, R.; et al. EMR-linked GWAS study: Investigation of variation landscape of loci for body mass index in children. Front. Genet. 2013, 4, 268. [Google Scholar] [CrossRef]
- Su, J.; Huang, F.; Tian, Y.; Tian, R.; Qianqian, G.; Bello, S.T.; Zeng, D.; Jendrichovsky, P.; Lau, C.G.; Xiong, W.; et al. Entorhinohippocampal cholecystokinin modulates spatial learning by facilitating neuroplasticity of hippocampal CA3-CA1 synapses. Cell Rep. 2023, 42, 113467. [Google Scholar] [CrossRef]
- Zaben, M.; Sheward, W.J.; Shtaya, A.; Abbosh, C.; Harmar, A.J.; Pringle, A.K.; Gray, W.P. The neurotransmitter VIP expands the pool of symmetrically dividing postnatal dentate gyrus precursors via VPAC2 receptors or directs them toward a neuronal fate via VPAC1 receptors. Stem Cells 2009, 27, 2539–2551. [Google Scholar] [CrossRef]
- Nunan, R.; Sivasathiaseelan, H.; Khan, D.; Zaben, M.; Gray, W. Microglial VPAC1R mediates a novel mechanism of neuroimmune-modulation of hippocampal precursor cells via IL-4 release. Glia 2014, 62, 1313–1327. [Google Scholar] [CrossRef]
- Cunha-Reis, D.; Aidil-Carvalho Mde, F.; Ribeiro, J.A. Endogenous inhibition of hippocampal LTD and depotentiation by vasoactive intestinal peptide VPAC1 receptors. Hippocampus 2014, 24, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Debette, S.; Ibrahim Verbaas, C.A.; Bressler, J.; Schuur, M.; Smith, A.; Bis, J.C.; Davies, G.; Wolf, C.; Gudnason, V.; Chibnik, L.B.; et al. Genome-wide studies of verbal declarative memory in nondemented older people: The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Biol. Psychiatry 2015, 77, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Arpawong, T.E.; Pendleton, N.; Mekli, K.; McArdle, J.J.; Gatz, M.; Armoskus, C.; Knowles, J.A.; Prescott, C.A. Genetic variants specific to aging-related verbal memory: Insights from GWASs in a population-based cohort. PLoS ONE 2017, 12, e0182448. [Google Scholar] [CrossRef] [PubMed]
- Lahti, J.; Tuominen, S.; Yang, Q.; Pergola, G.; Ahmad, S.; Amin, N.; Armstrong, N.J.; Beiser, A.; Bey, K.; Bis, J.C.; et al. Genome-wide meta-analyses reveal novel loci for verbal short-term memory and learning. Mol. Psychiatry 2022, 27, 4419–4431. [Google Scholar] [CrossRef]
- Geng, C.H.; Wang, C.; Yang, J.; Wang, H.; Ma, R.Q.; Liu, X.; Wang, C.H. Arginine vasopressin improves the memory deficits in Han Chinese patients with first-episode schizophrenia. Peptides 2017, 97, 8–15. [Google Scholar] [CrossRef]
| Gene Family | Gene | Gene Name |
|---|---|---|
| Acyl-CoA Binding Protein | DBI | diazepam binding inhibitor |
| Adipose neuropeptides | ADIPOQ | adiponectin |
| LEP | leptin | |
| LEPR | leptin receptor | |
| NUCB2 | nucleoblindin 2 | |
| RETN | resistin | |
| RETNLB | resistin-like beta | |
| UBL5 | ubiquitin-like 5 | |
| AVIT (prokineticin) | PROK1 | prokineticin 1 |
| PROK2 | prokineticin 2 | |
| Bombesin-like peptides | GRP | gastrin-releasing peptide |
| NMB | neuromedin B | |
| NMBR | neuromedin B receptor | |
| Calcitonin | CALCA | calcitonin-related polypeptide alpha |
| CALCB | calcitonin-related polypeptide alpha | |
| IAPP | islet amyloid peptide | |
| Cholecystokinin/gastrin | CCK | cholecystokinin |
| CCKAR | cholecystokinin A receptor | |
| CCKBR | cholecystokinin B receptor | |
| Cerebellins | CBLN1 | cerebellin 1 precursor |
| CBLN2 | cerebellin 2 precursor | |
| CBLN3 | cerebellin 3 precursor | |
| CBLN4 | cerebellin 4 precursor | |
| Corticotropin-releasing hormone | CRHBP | corticotropin-releasing hormone binding protein |
| CRHR1 | corticotropin-releasing hormone receptor 1 | |
| CRHR2 | corticotropin-releasing hormone receptor 2 | |
| Corticotropin-releasing factor | UCN2 | urocortin 2 |
| UCN3 | urocortin 3 | |
| UTS2 | urotensin 2 | |
| Endothelin | EDN1 | endothelin 1 |
| EDN2 | endothelin 2 | |
| EDN3 | endothelin 3 | |
| F- and Y- amides | NPFF | neuropeptide FF-amide peptide precursor |
| NPFFR1 | neuropeptide FF receptor 1 | |
| NPFFR2 | neuropeptide FF receptor 1 | |
| NPY | neuropeptide Y | |
| NPY1R | neuropeptide Y receptor Y1 | |
| NPY2R | neuropeptide Y receptor Y2 | |
| NPY5R | neuropeptide Y receptor Y5 | |
| PPY | pancreatic polypeptide | |
| PYY | peptide YY | |
| QRFP | pyroglutamylated RFamide peptide | |
| QRFPR | pyroglutamylated RFamide peptide receptor | |
| Galanin | GAL | galanin and GMAP prepropeptide |
| GALP | galanin-like peptide | |
| GALR1 | galanin receptor 1 | |
| Glucagon/secretin | ADCYAP1 | adenylate cyclase activating polypeptide 1 |
| ADCYAP1R1 | ADCYAP receptor type 1 | |
| GCG | glucagon | |
| GHRH | growth hormone-releasing hormone | |
| GHRHR | growth hormone-releasing hormone receptor | |
| GIP | gastric inhibitory polypeptide | |
| GIPR | gastric inhibitory polypeptide receptor | |
| SCTR | secretin receptor | |
| VIP | vasoactive intestinal peptide | |
| VIPR1 | vasoactive intestinal peptide receptor 1 | |
| VIPR2 | vasoactive intestinal peptide receptor 2 | |
| Gonadotropin-releasing hormone | GNRH1 | gonadotropin-releasing hormone 1 |
| GNRH2 | gonadotropin-releasing hormone 2 | |
| APLNR | apelin receptor | |
| HCRTR1 | hypocretin receptor 1 | |
| HCRTR2 | hypocretin receptor 2 | |
| Granins | CHGA | chromogranin A |
| CHGB | chromogranin B | |
| SCG2 | secretogranin II | |
| SCG3 | secretogranin III | |
| SCG5 | secretogranin V | |
| Insulin | IGF1 | insulin-like growth factor 1 |
| IGF1R | insulin-like growth factor 1 receptor | |
| IGF2R | insulin-like growth factor 2 receptor | |
| INS-IGF2 | INS-IGF2 read-through | |
| INSR | insulin receptor | |
| RLN1 | relaxin 1 | |
| RLN2 | relaxin 2 | |
| RLN3 | relaxin 2 | |
| Kinin and tensin | TAC1 | tachykinin precursor 1 |
| TAC3 | tachykinin precursor 3 | |
| Kisspeptin | KISS1 | KISS-1 metastasis-suppressor |
| KISS1R | KISS-1 receptor | |
| Motilin | GHRL | ghrelin and obestatin prepropeptide |
| MLN | motilin | |
| MLNR | motilin receptor | |
| Natriuretic factors | NPPA | natriuretic peptide A |
| NPPB | natriuretic peptide B | |
| NPPC | natriuretic peptide C | |
| Neurexophilins | NXPH1 | neurexophilin 1 |
| NXPH2 | neurexophilin 2 | |
| NXPH3 | neurexophilin 3 | |
| NXPH4 | neurexophilin 1 | |
| Neuromedins | NMS | neuromedin S |
| NMU | neuromedin U | |
| NMUR1 | neuromedin U receptor 1 | |
| NMUR2 | neuromedin U receptor 2 | |
| Neuropeptide B/W | NPW | neuropeptide W |
| NPBWR2 | neuropeptides Band W receptor 2 | |
| NPS | neuropeptide S | |
| NPSR1 | neuropeptide S receptor 1 | |
| No-family neuropeptides | CARTPT | CART prepropeptide |
| Opioid | PDYN | prodynorphin |
| PENK | prokephalin | |
| PNOC | prepronociceptin | |
| POMC | propiomelanocortin | |
| Parathyroid hormone | PTHLH | parathyroid hormone-like hormone |
| Somatostatin | SST | somatostatin |
| SSTR1 | Somatostatin receptor 1 | |
| SSTR2 | Somatostatin receptor 2 | |
| SSTR3 | Somatostatin receptor 3 | |
| SSTR4 | Somatostatin receptor 4 | |
| SSTR5 | Somatostatin receptor 5 | |
| Somatotrophin/prolactin | PRL | prolactin |
| PRLHR | prolactin-releasing hormone receptor | |
| PRLR | prolactin receptor | |
| Tensins and kinins | AGT | angiotensin |
| AGTR1 | angiotensin II receptor type 1 | |
| KNG1 | kininogen 1 | |
| NTS | neurotensin | |
| NTSR1 | neurotensin receptor 1 | |
| Thyrotropin-releasing hormone | TRH | thyrotropin-releasing hormone |
| TRHR | thyrotropin-releasing hormone receptor | |
| Vasopressin/oxytocin | OXTR | oxytocin receptor |
| AVP | arginine vasopressin | |
| AVPI1 | arginine vasopressin-induced 1 | |
| AVPR1A | arginine vasopressin receptor 1A | |
| AVPR1B | arginine vasopressin receptor 1B |
| Variable | Participants (n = 597) N (%) |
|---|---|
| Age group (years) | |
| 16–25 | 465 (77.9) |
| 26–35 | 79 (13.2) |
| 36–45 | 36 (6.0) |
| 46–55 | 10 (1.7) |
| 56–65 | 7 (1.2) |
| Gender | |
| Male | 171 (28.6) |
| Female | 426 (71.4) |
| Ethnicity | |
| Caucasian | 446 (74.7) |
| Other | 151 (25.3) |
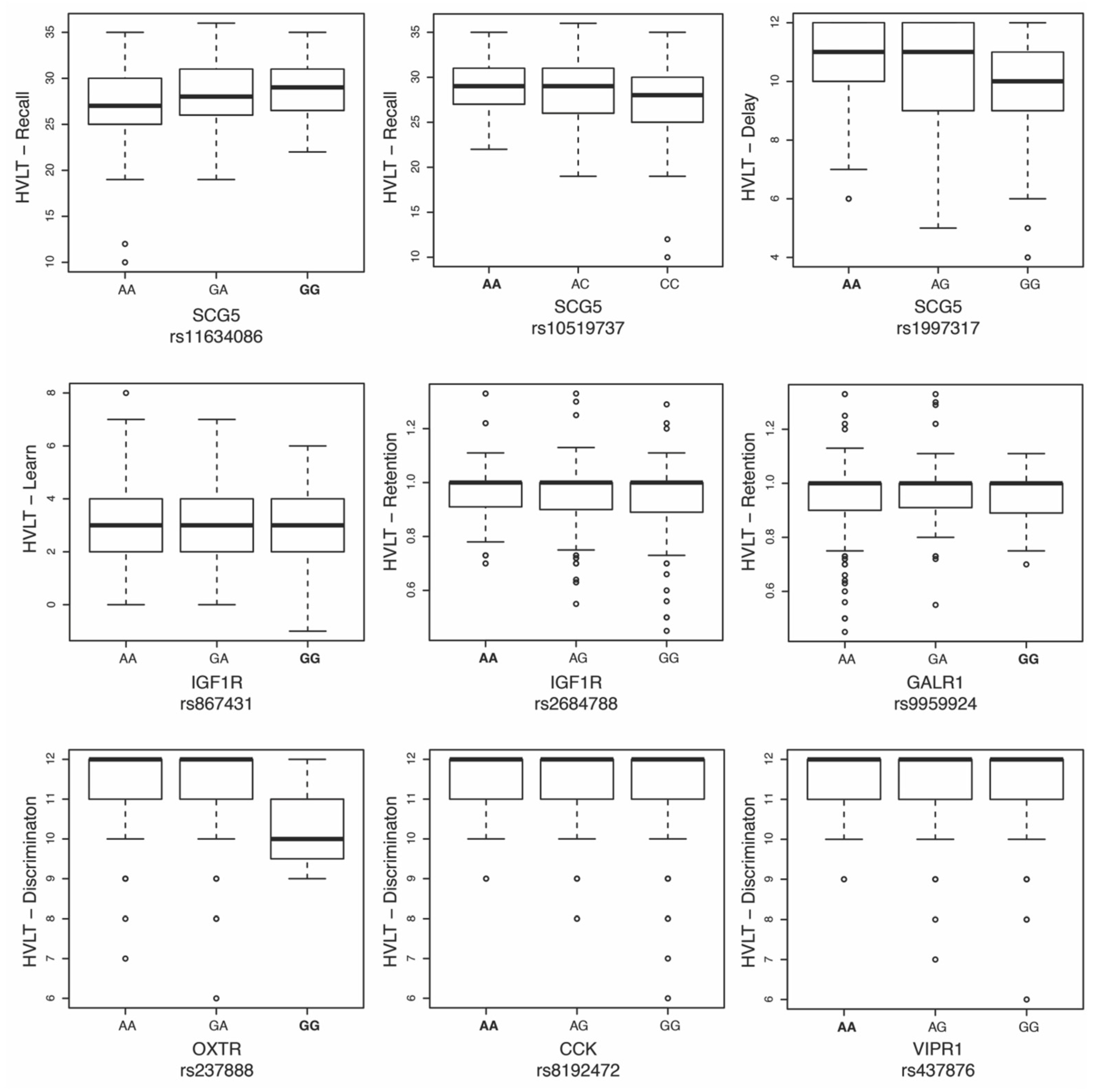
| Gene | Chr | Location | SNP | Region | β | t | p * | Phenotype |
|---|---|---|---|---|---|---|---|---|
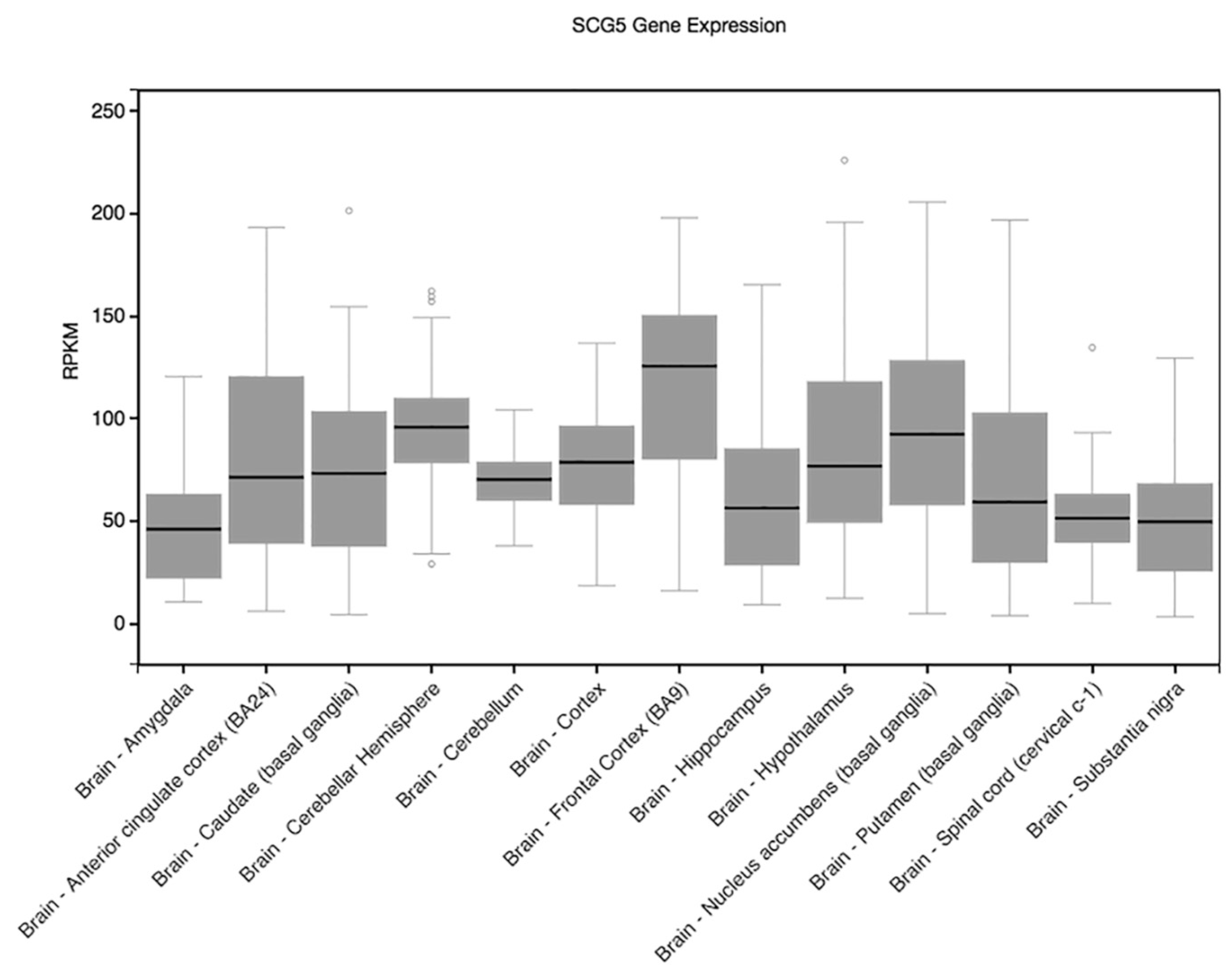
| SCG5 | 15 | 32965630 | rs11634086 | Intron | 0.6918 | 3.47 | 0.0005587 | Recall |
| SCG5 | 15 | 32969457 | rs10519737 | Intron | 0.7417 | 3.288 | 0.001069 | Recall |
| SCG5 | 15 | 32985954 | rs1997317 | Intron | 0.3512 | 4.021 | 6.55 × 10−5 | Delay |
| IFG1R | 15 | 99449683 | rs867431 | Intron | −0.297 | −3.438 | 0.0006276 | Learning |
| IFG1R | 15 | 99504437 | rs2684788 | 3’UTR | 0.0254 | 3.813 | 0.0001518 | Retention |
| GALR1 | 18 | 74967894 | rs9959924 | Intron | 0.0363 | 3.345 | 0.0008761 | Retention |
| OXTR | 3 | 8797095 | rs237888 | Intron | −0.347 | −3.36 | 0.0008307 | Discrimination |
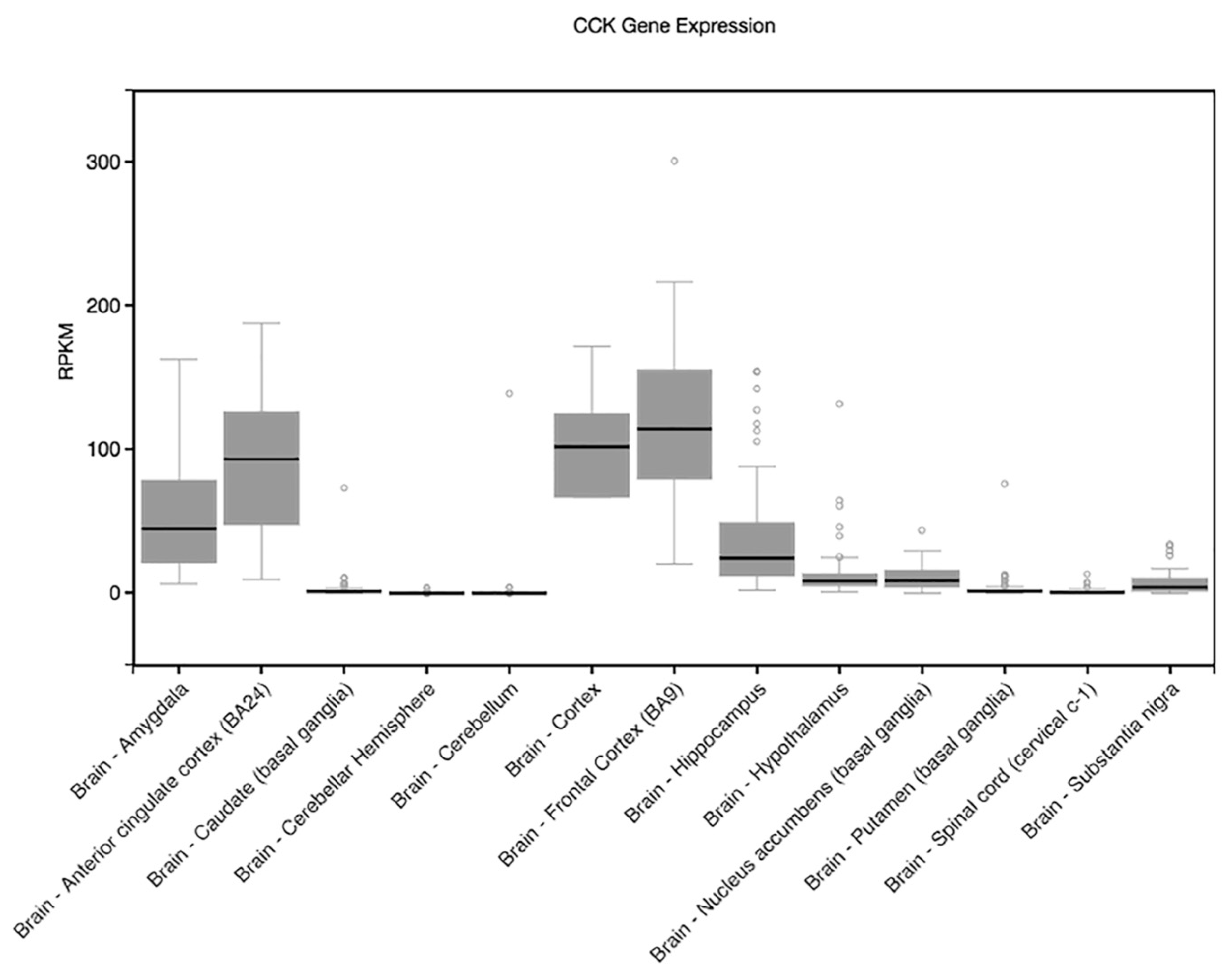
| CCK | 3 | 42299870 | rs8192472 | Intron | 0.1716 | 3.312 | 0.0009836 | Discrimination |
| VIPR1 | 3 | 42568440 | rs437876 | Intron | 0.1755 | 3.332 | 0.0009171 | Discrimination |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avgan, N.; Sutherland, H.G.; Lea, R.A.; Haupt, L.M.; Shum, D.H.K.; Griffiths, L.R. Association Study of a Comprehensive Panel of Neuropeptide-Related Polymorphisms Suggest Potential Roles in Verbal Learning and Memory. Genes 2024, 15, 30. https://doi.org/10.3390/genes15010030
Avgan N, Sutherland HG, Lea RA, Haupt LM, Shum DHK, Griffiths LR. Association Study of a Comprehensive Panel of Neuropeptide-Related Polymorphisms Suggest Potential Roles in Verbal Learning and Memory. Genes. 2024; 15(1):30. https://doi.org/10.3390/genes15010030
Chicago/Turabian StyleAvgan, Nesli, Heidi G. Sutherland, Rod A. Lea, Larisa M. Haupt, David H. K. Shum, and Lyn R. Griffiths. 2024. "Association Study of a Comprehensive Panel of Neuropeptide-Related Polymorphisms Suggest Potential Roles in Verbal Learning and Memory" Genes 15, no. 1: 30. https://doi.org/10.3390/genes15010030
APA StyleAvgan, N., Sutherland, H. G., Lea, R. A., Haupt, L. M., Shum, D. H. K., & Griffiths, L. R. (2024). Association Study of a Comprehensive Panel of Neuropeptide-Related Polymorphisms Suggest Potential Roles in Verbal Learning and Memory. Genes, 15(1), 30. https://doi.org/10.3390/genes15010030







