Identification and Characterization of ABCG15—A Gene Required for Exocarp Color Differentiation in Pear
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. SNP Detection and Annotation
2.3. Genome-Wide Association Study
2.4. Transcriptome Analysis
2.5. Expression Analysis Using qRT–PCR
2.6. Metabolomics Analysis
3. Results
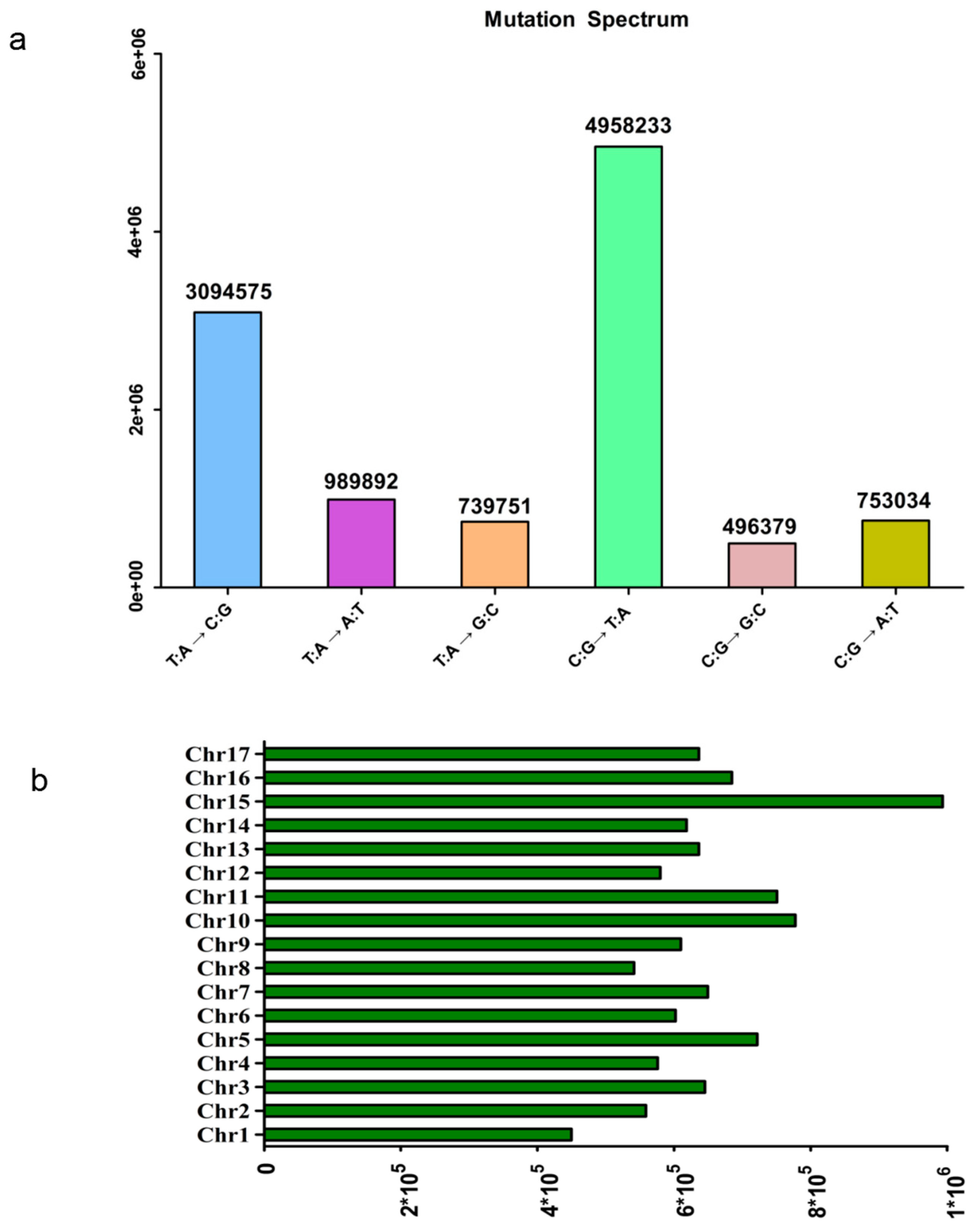
3.1. Genotyping of 447 Pear Accessions
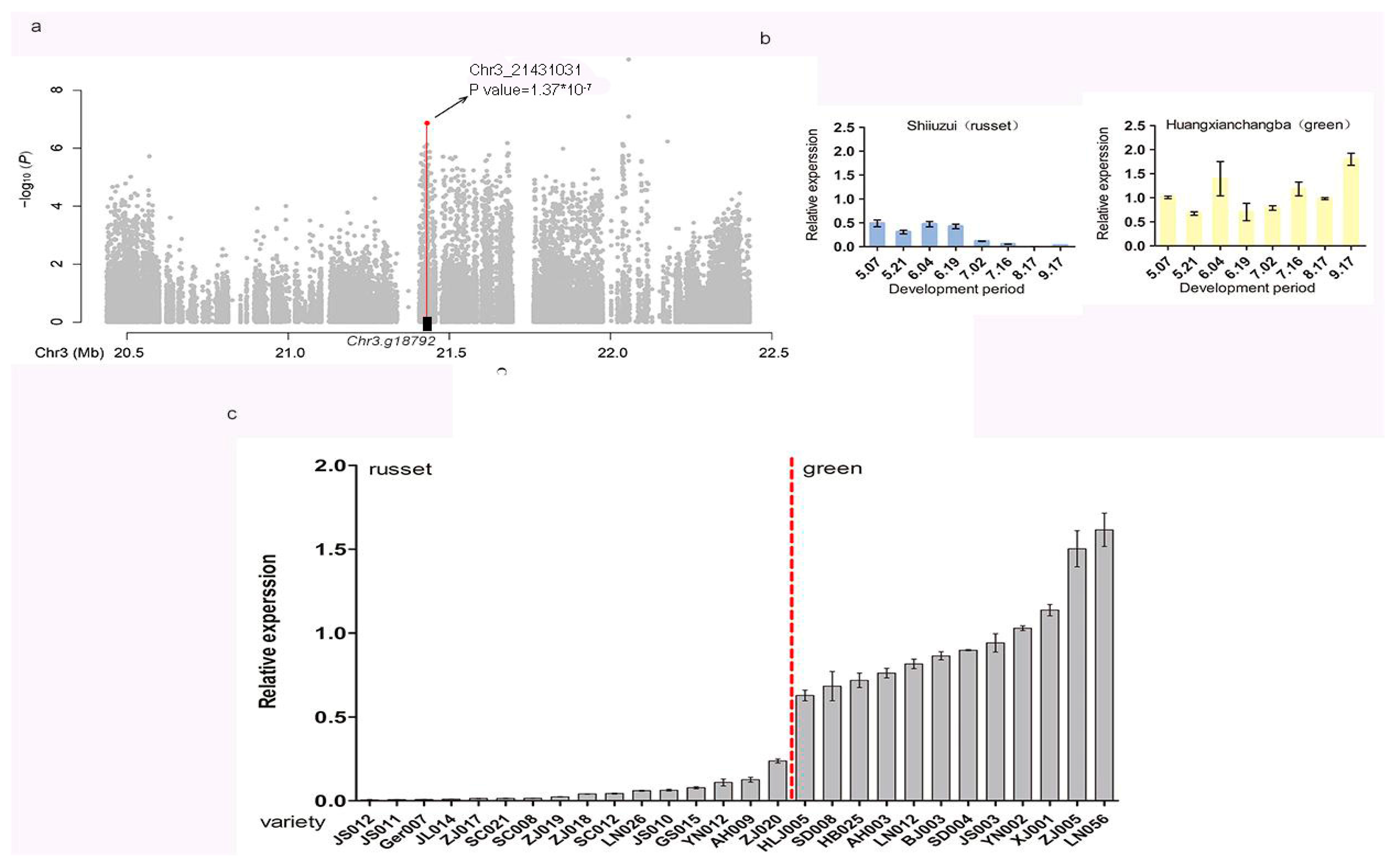
3.2. GWAS Analysis
3.3. Identification of Putative Candidate Genes for Exocarp Color
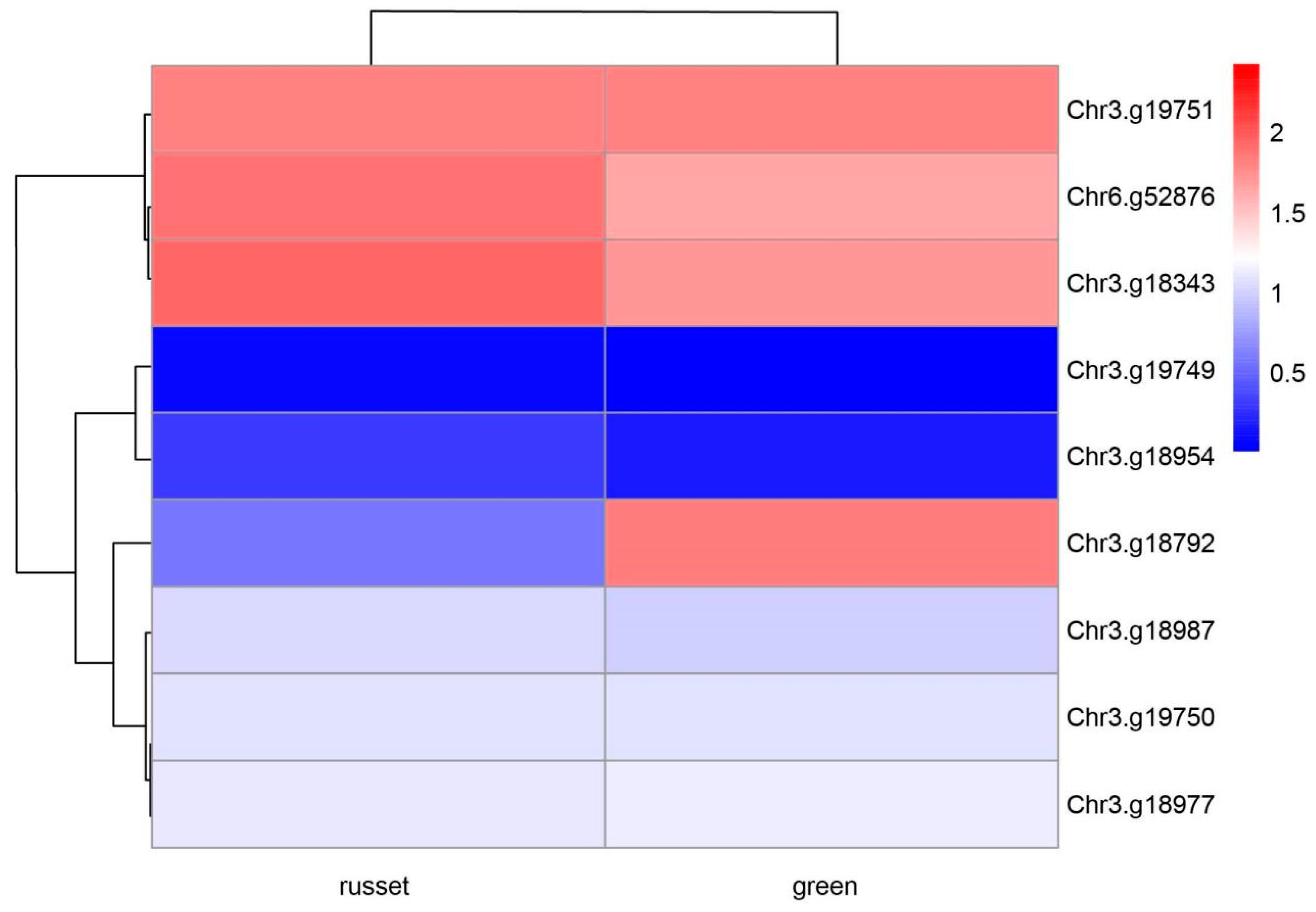
3.4. Transcriptome Profiles for the Exocarp Color of Pear Fruit
3.5. qRT-PCR Analysis
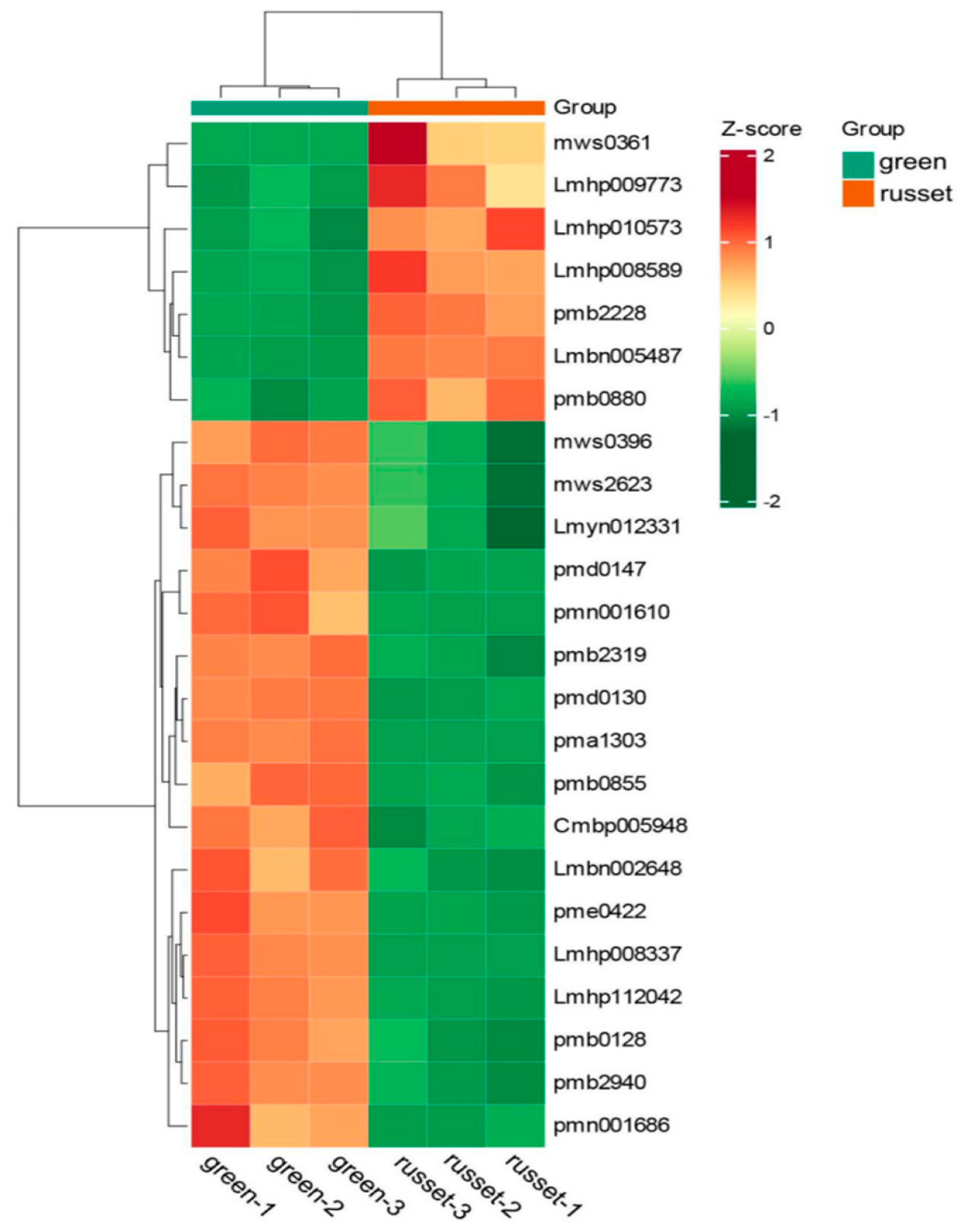
3.6. Comparison of Different Expression Metabolites between the Green and Russet Group
3.7. The Pathways Enriched by the 24 Differentially Expressed Metabolites Related to Exocarp Color
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, Y.; Shen, L.; Ma, K.; Wang, Y.; Ji, L.; Chen, J. Preliminary study of the ‘peel’ structure of cv. ‘Yali’ Pear. In Proceedings of the Thirteenth Annual Conference of the Fruit Tree Society, Baoding, China, 7 December 1995. [Google Scholar]
- Watanabe, S. Historical studies on the cause of apple russeting. Bull. Yamagata. Univ. 1969, 5, 205–267. [Google Scholar]
- Skene, D.S. The development of russet, rough russet and cracks on the fruit of the apple Cox’s Orange Pippin during the course of the season. J. Hortic. Sci. 1982, 57, 165–174. [Google Scholar] [CrossRef]
- Suh, M.C.; Samuels, A.L.; Jetter, R.; Kunst, L.; Pollard, M.; Ohlrogge, J.; Beisson, F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005, 139, 1649–1665. [Google Scholar] [CrossRef] [PubMed]
- Faust, M.; Shear, C.B. Russeting of apples, an interpretive review. Hortscience 1972, 7, 233–235. [Google Scholar] [CrossRef]
- Wertheim, S.J. Fruit russeting in apple as affected by various gibberellins. J. Hortic. Sci. 1982, 57, 283–288. [Google Scholar] [CrossRef]
- Curry, E.A.; Physiologist, P. Growth-Induced Microcracking and Repair Mechanisms of Fruit Cuticles. In Proceedings of the SEM Annual Conference, Albuquerque, NM, USA, 1–4 June 2009. [Google Scholar]
- Gamble, J.; Jaeger, S.R.; Harker, F.R. Preferences in pear appearance and response to novelty among Australian and New Zealand consumers. Postharvest Biol. Technol. 2006, 41, 38–47. [Google Scholar] [CrossRef]
- Legay, S.; Guerriero, G.; Deleruelle, A.; Lateur, M.; Evers, D.; André, C.M.; Hausman, J.-F. Apple russeting as seen through the RNA-seq lens: Strong alterations in the exocarp cell wall. Plant Mol. Biol. 2015, 88, 21–40. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Dai, M.S.; Zhang, S.J.; Shi, Z.B. Exploring candidate genes for pericarp russet pigmentation of sand pear (Pyrus pyrifolia) via RNA-seq data in two genotypes contrasting for pericarp color. PLoS ONE 2014, 9, e83675. [Google Scholar] [CrossRef]
- Heng, W.; Wang, Z.T.; Jiang, X.H.; Jia, B.; Liu, P.; Liu, L.; Ye, Z.F.; Zhu, L.W. The role of polyamines during exocarp formation in a russet mutant of ‘dangshansuli’ pear (pyrus bretschneideri rehd.). Plant Cell Rep. 2016, 35, 1841–1852. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, M.; Cai, D.; Shi, Z. Proteome and transcriptome profile analysis reveals regulatory and stress-responsive networks in the russet fruit skin of sand pear. Hortic. Res. 2020, 7, 15. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Zhang, S.; Dai, M.S.; Shi, Z.B. Pigmentation in sand pear (pyrus pyrifolia) fruit: Biochemical characterization, gene discovery and expression analysis with exocarp pigmentation mutant. Plant Mol. Biol. 2014, 85, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Bird, D.; Beisson, F.; Brigham, A.; Shin, J.; Greer, S.; Jetter, R.; Kunst, L.; Wu, X.; Yephremov, A.; Samuels, L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 2010, 52, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.C.; Liu, C.J. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Kim, K.Y.; Zhang, J.; Yamaoka, Y.; Gao, P.; Kim, H.; Hwang, J.U.; Suh, M.C.; Kang, B.; Lee, Y. Arabidopsisseedling establishment under waterlogging requires abcg5-mediated formation of a dense cuticle layer. New Phytol. 2021, 229, 156–172. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. Atabcg29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef]
- Kikuti, A. On the origin of Japanese pears and the inheritance of the skin colors of their fruits. Jpn. J. Genet. 1924, 3, 1–21. [Google Scholar] [CrossRef]
- Mori, H. Genetic study on major phenotypes in deciduous fruits trees. II. Genetics in Japanese pear. Bull. Natl. Inst. Agric. Sci. E2 1953, 2, 34–53. (In Japanese) [Google Scholar]
- Inoue, E.; Kasumi, M.; Sakuma, F.; Anzai, H.; Amano, K.; Hara, H. Identification of RAPD marker linked to fruit skin color in Japanese pear (pyrus pyrifolia nakai). Sci. Hortic. 2006, 107, 254–258. [Google Scholar] [CrossRef]
- Song, W.; Wang, C.H.; Tian, Y.K.; Tian, W.; Yin, H. SSR molecular markers linked to the fruit russet skin of pear. Acta Hortic. Sin. 2010, 37, 1325–1328. [Google Scholar]
- Kim, D.; Hwang, J.H.; Shin, Y.U.; Shin, I.S.; Lee, H.J.; Hong, S.S.; Kang, S. Development of molecular markers linked to several fruit traits in oriental pear. Acta Hortic. 2005, 671, 315–321. [Google Scholar] [CrossRef]
- Yamamoto, T.; Terakami, S.; Takada, N.; Nishio, S.; Saito, T. Identification of QTLs controlling harvest time and fruit skin color in Japanese pear (pyrus pyrifolia nakai). Breed. Sci. 2014, 64, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef]
- Wang, W.; Guo, W.; Le, L.; Yu, J.; Wu, Y.; Li, D.; Wang, Y.; Wang, H.; Lu, X.; Qiao, H.; et al. Integration of high-throughput phenotyping, GWAS, and predictive models reveals the genetic architecture of plant height in maize. Molecular Plant 2023, 16, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Sun, S.; Jin, C.; Su, J.; Wei, J.; Luo, X.; Wen, J.; Wei, T.; Sahu, S.K.; et al. GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet. Nat. Commun. 2022, 13, 1–17. [Google Scholar] [CrossRef]
- Hu, J.; Chen, B.; Zhao, J.; Zhang, F.; Xie, T.; Xu, K.; Gao, G.; Yan, G.; Li, H.; Li, L.; et al. Genomic selection and genetic architecture of agronomic traits during modern rapeseed breeding. Nat. Genet. 2022, 54, 694–704. [Google Scholar] [CrossRef]
- Wang, M.; Qi, Z.; Thyssen, G.N.; Naoumkina, M.; Jenkins, J.N.; McCarty, J.C.; Xiao, Y.; Li, J.; Zhang, X.; Fang, D.D. Genomic interrogation of a MAGIC population highlights genetic factors controlling fiber quality traits in cotton. Commun. Biol. 2022, 5, 60. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Xue, C.; Hu, H.; Li, J.; Xue, Y.; Wang, R. Genome-wide association studies provide insights into the genetic determination of fruit traits of pear. Nat. Commun. 2021, 12, 1144. [Google Scholar] [CrossRef]
- Cao, Y.F.; Liu, F.; Hu, H.; Zhang, B. Description and Data Standard for Pear (Pyrus spp.); China Agriculture Press: Beijing, China, 2006. [Google Scholar]
- Cao, Y.F.; Liu, F.; Hu, H.; Zhang, B. Descriptors for Pear Germplasm; Chinese Agriculture Press: Beijing, China, 2016. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Dong, X.; Wang, Z.; Tian, L.; Zhang, Y.; Qi, D.; Huo, H.; Xu, J.; Li, Z.; Liao, R.; Shi, M.; et al. De novo assembly of a wild pear (Pyrus betuleafolia) genome. Plant Biotechnol. J. 2020, 18, 581–595. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. Te variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Lippert, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Wei, S. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Park, J.M.; Park, C.J.; Lee, S.B.; Ham, B.K.; Shin, R.; Paek, K.H. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in Tobacco. Plant Cell 2001, 13, 1035–1046. [Google Scholar]
- Wang, M.; Yang, J.; Huang, H.; Zhu, L.; Heng, W. Cloning and expression analysis of two erf genes in pericarp of russet mutant of ‘dangshansuli’ pear. J. Trop. Subtrop. Bot. 2015, 23, 379–385. [Google Scholar]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of abc transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Chauhan, A.S.; Yusuf, M.A.; Sanyal, I.; Chauhan, P.S. Transcriptome sequencing of chickpea (Cicer arietinum L.) genotypes for identification of drought-responsive genes under drought stress condition. Plant Mol. Biol. Report. 2019, 37, 186–203. [Google Scholar] [CrossRef]
- Bianchetti, R.E.; Cruz, A.B.; Oliveira, B.S.; Demarco, D.; Purgatto, E.; Peres, L.E.P.; Rossi, M.; Freschi, L. Phytochromobilin deficiency impairs sugar metabolism through the regulation of cytokinin and auxin signaling in tomato fruits. Sci. Rep. 2017, 7, 7822. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS proteins, scarecrow-like21 and phytochrome a signal transduction1, function cooperatively in phytochrome a signal transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef]
- Cui, H.; Kong, D.; Liu, X.; Hao, Y. Scarecrow, scr-like 23 and short-root control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 2014, 78, 319–327. [Google Scholar] [CrossRef]
- Jung, Y.J.; Lee, M.C.; Kang, K.K. A transcription factor “osnac075” is essential for salt resistance in rice (Oryza sativa L.). J. Plant Biotechnol. 2011, 38, 94–104. [Google Scholar] [CrossRef]
- Qin, P.; Tu, B.; Wang, Y.; Deng, L.; Quilichini, T.D.; Li, T.; Wang, H.; Ma, B.; Li, S. ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 2013, 54, 138–154. [Google Scholar] [CrossRef]
- Xu, Y.; Hou, X.; Feng, J.; Khalil-Ur-Rehman, M.; Tao, J. Transcriptome sequencing analyses reveals mechanisms of eliminated russet by applying GA 3 and CPPU on ‘Shine Muscat’ grape. Sci. Hortic. 2019, 250, 94–103. [Google Scholar] [CrossRef]
- Xiao, Q.; Bai, X.; Zhang, C.; He, Y. Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review. J. Adv. Res. 2022, 35, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Sallam, A.; Baenziger, P.S.; Börner, A. GWAS: Fast-forwarding gene identification and characterization in temperate cereals: Lessons from barley–A review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A.; Cunningham, P.; Dean, M. The ABC transporter gene family of daphnia pulex. BMC Genom. 2009, 10, 1–18. [Google Scholar] [CrossRef]
- Dhara, A.R.A. Abcg transporter proteins with beneficial activity on plants. Phytochemistry 2021, 184, 112663. [Google Scholar] [CrossRef]
- Panikashvili, D.; Savaldi-Goldstein, S.; Mandel, T.; Yifhar, T.; Franke, R.B.; Hofer, R.; Schreiber, L.; Chory, J.; Aharoni, A. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 2007, 145, 1345–1360. [Google Scholar] [CrossRef]
- Pighin, J.A.; Zheng, H.; Balakshin, L.J.; Goodman, I.P.; Western, T.L.; Jetter, R.; Kunst, L.; Samuels, A.L. Plant cuticular lipid export requires an ABC transporter. Science 2004, 306, 702–704. [Google Scholar] [CrossRef]
- Banerjee, A.; Moreno, A.; Pata, J.; Falson, P.; Prasad, R. ABCG: A new fold of ABC exporters and a whole new bag of riddles. Adv. Protein Chem. Struct. Biol. 2021, 123, 163–191. [Google Scholar]
- Landgraf, R.; Smolka, U.; Altmann, S.; Eschen-Lippold, L.; Senning, M.; Sonnewald, S.; Weigel, B.; Frolova, N.; Strehmel, N.; Hause, G.; et al. The ABC transporter ABCG1 is required for suberin formation in potato tuber periderm. Plant Cell 2014, 26, 3403–3415. [Google Scholar] [CrossRef]
- Hou, Z.; Jia, B.; Li, F.; Liu, P.; Liu, L.; Ye, Z.; Zhu, L.; Wang, Q.; Heng, W. Characterization and expression of the ABC family (G group) in ‘Dangshansuli’ pear (Pyrus bretschneideri Rehd.) and its russet mutant. Genet. Mol. Biol. 2018, 41, 37–144. [Google Scholar] [CrossRef]


| Pbe-SD | Dangshansuli | Genebank (Dangshansuli) | |
|---|---|---|---|
| Mapping rate | 81.73–85% | 70.78–73.58% | 70.78–73.58% |
| Coverage | 81.6–88.71% | 70.29–74.34% | 70.29–74.34% |
| Gene ID | Description | Case Vsctrl. Log (Fold Change) | Case Vsctrl. P Value | Case Vsctrl. Q | Case Vsctrl. Significant |
|---|---|---|---|---|---|
| Chr6.g52876 | ethylene-responsive transcription factor 2-like | 0.43 | 0.32 | 0.73 | FALSE |
| Chr3.g19750 | abc transporter g family member 3-like | –0.08 | 0.72 | 0.94 | FALSE |
| Chr3.g19751 | cell division cycle protein 48 homolog | –0.07 | 0.73 | 0.94 | FALSE |
| Chr8.g54414 | cytochrome p450 81e8-like | – | – | – | – |
| Chr3.g18954 | hexokinase-3-like | 1.03 | 0.14 | 0.49 | FALSE |
| Chr3.g18987 | phytochromobilin:ferredoxinchloroplastic | 0.08 | 0.78 | 0.96 | FALSE |
| Chr3.g19749 | udp-glycosyltransferase 86a1-like | 2.27 | 0.01 | 0.10 | FALSE |
| Chr3.g18977 | scarecrow-like protein 3 | –0.18 | 0.47 | 0.85 | FALSE |
| Chr3.g18792 | abc transporter g family member 15-like | –4.73 | 1.24 × 10−45 | 1.90 × 10−42 | DOWN |
| Chr3.g18343 | nac domain-containing protein 72-like | 0.50 | 0.17 | 0.56 | FALSE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Xu, J.; Zhang, Y.; Cao, Y. Identification and Characterization of ABCG15—A Gene Required for Exocarp Color Differentiation in Pear. Genes 2023, 14, 1827. https://doi.org/10.3390/genes14091827
Zhang S, Xu J, Zhang Y, Cao Y. Identification and Characterization of ABCG15—A Gene Required for Exocarp Color Differentiation in Pear. Genes. 2023; 14(9):1827. https://doi.org/10.3390/genes14091827
Chicago/Turabian StyleZhang, Simeng, Jiayu Xu, Ying Zhang, and Yufen Cao. 2023. "Identification and Characterization of ABCG15—A Gene Required for Exocarp Color Differentiation in Pear" Genes 14, no. 9: 1827. https://doi.org/10.3390/genes14091827
APA StyleZhang, S., Xu, J., Zhang, Y., & Cao, Y. (2023). Identification and Characterization of ABCG15—A Gene Required for Exocarp Color Differentiation in Pear. Genes, 14(9), 1827. https://doi.org/10.3390/genes14091827





