Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of the PHD-Finger Family Genes in Pepper
2.2. Analyses of Chromosomal Location, Phylogenetic Relationship, Gene Structure, Functional Domains, and Conserved Motifs of the CaPHD Gene Family
2.3. Identification of Cis-Acting Promoter Elements and Gene Expression Patterns
2.4. Plant Materials and Responses to Abiotic Stresses and Phytohormone Treatments
2.5. qRT-PCR Analysis
3. Results
3.1. Genome-Wide Identification and Chromosomal Location of CaPHD Gene Family in Pepper
3.2. Phylogenetic Analyses of the CaPHD Family
3.3. Gene Structure and Conserved Motif Analysis of the CaPHD Family
3.4. Analysis of Cis-Acting Regulatory Elements
3.5. Tissue-Specific Expression Patterns of CaPHD Genes
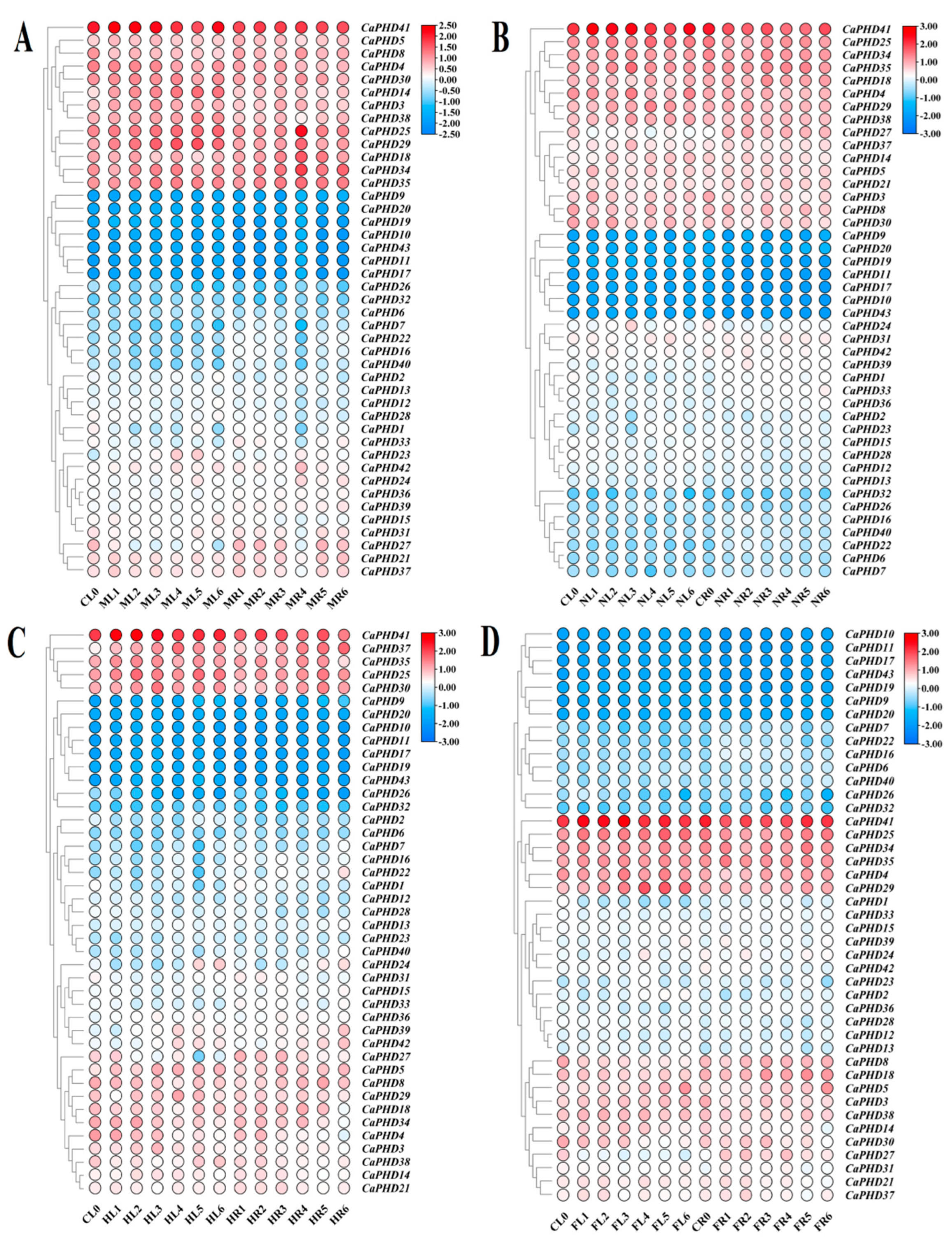
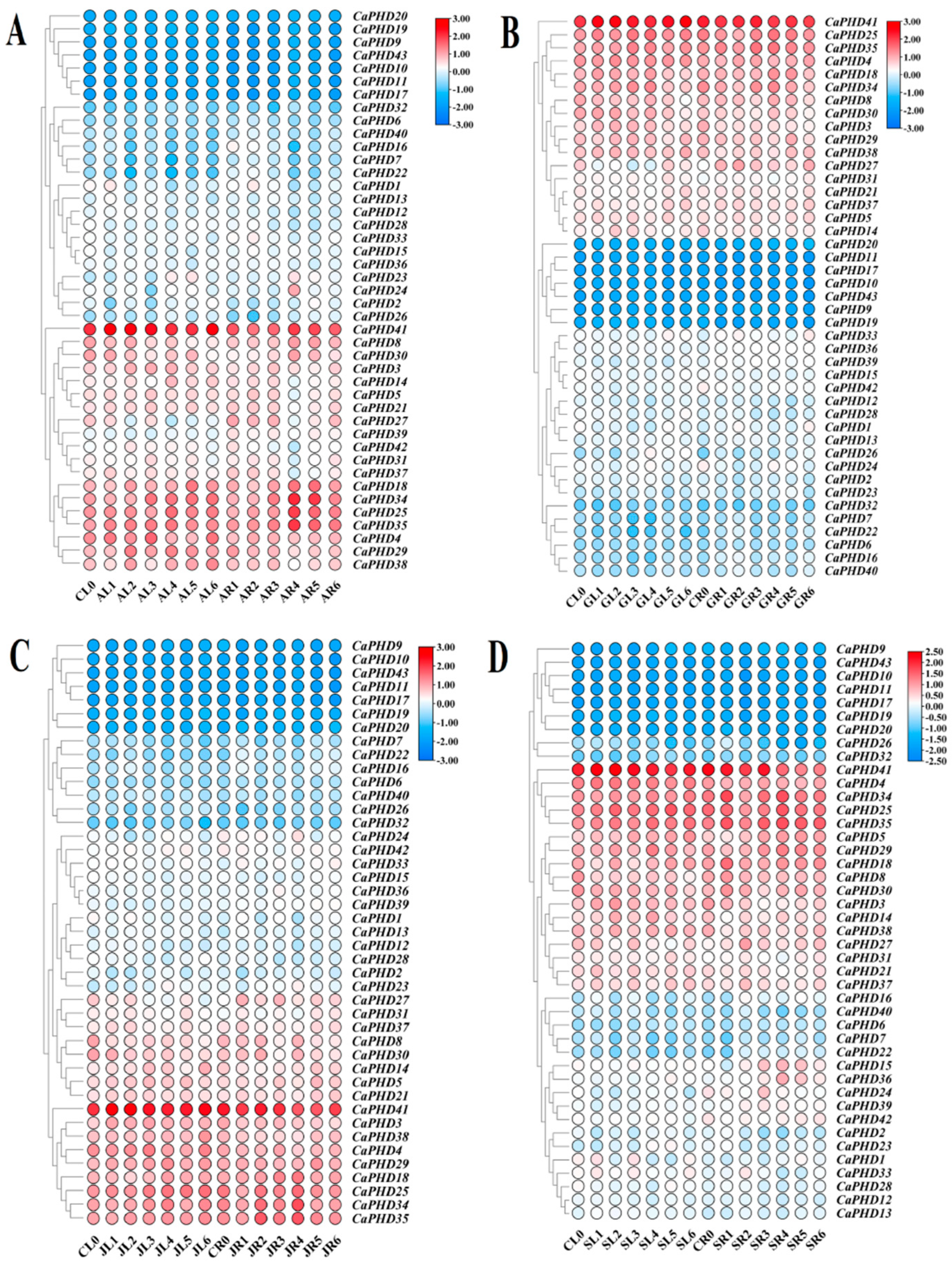
3.6. Expression Profiles of Stress-Related CaPHD Genes in Pepper
3.7. Expression Response of CaPHD Genes to Phytohormone Treatment
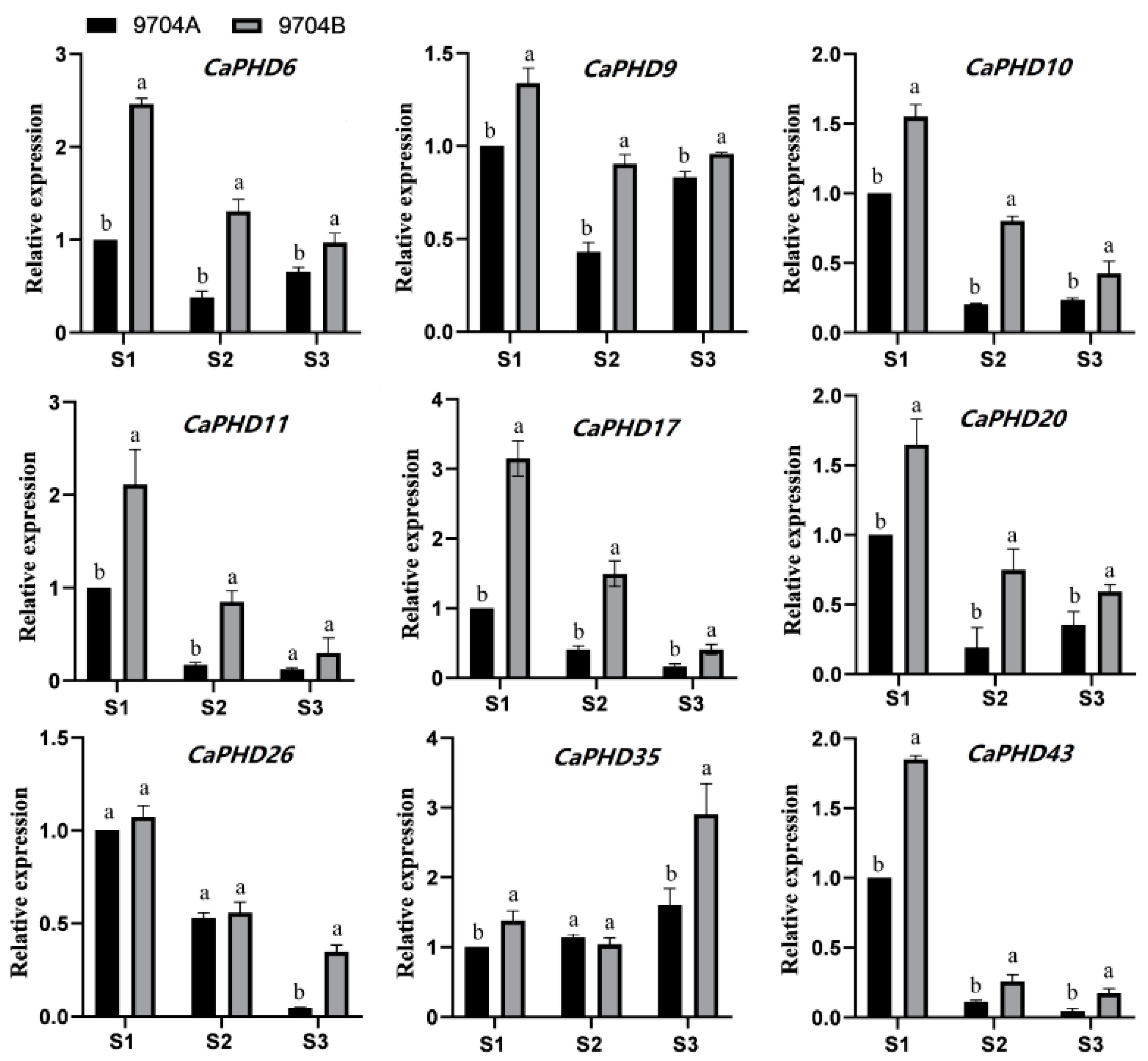
3.8. qRT-PCR Validation of Gene Expression Patterns
4. Discussion
4.1. Identification and Phylogenetic Relationships of the CaPHD Gene Family in Pepper
4.2. CaPHDs Play Roles in Response to Developmental and Abiotic Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar]
- Arnaud, D.; Déjardin, A.; Leplé, J.-C.; Lesage-Descauses, M.-C.; Pilate, G. Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 2007, 14, 103–116. [Google Scholar] [PubMed]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. CMLS 1998, 54, 582–596. [Google Scholar]
- Matthews, J.M.; Bhati, M.; Lehtomaki, E.; Mansfield, R.E.; Cubeddu, L.; Mackay, J.P. It takes two to tango: The structure and function of LIM, RING, PHD and MYND domains. Curr. Pharm. Des. 2009, 15, 3681–3696. [Google Scholar] [PubMed]
- Borden, K.L.; Freemont, P.S. The RING finger domain: A recent example of a sequence—Structure family. Curr. Opin. Struct. Biol. 1996, 6, 395–401. [Google Scholar]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995, 20, 56–59. [Google Scholar]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [PubMed]
- Wu, S.; Wu, M.; Dong, Q.; Jiang, H.; Cai, R.; Xiang, Y. Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa. Gene 2016, 575, 75–89. [Google Scholar]
- Sun, M.; Jia, B.; Yang, J.; Cui, N.; Zhu, Y.; Sun, X. Genome-Wide Identification of the PHD-Finger Family Genes and Their Responses to Environmental Stresses in Oryza sativa L. Int. J. Mol. Sci. 2017, 18, 2005. [Google Scholar]
- Chen, X.; Hayat, S.; Cheng, Z. Genome-Wide Identification and Analysis of Genes Encoding Phd-Finger Protein in Tomato. Pak. J. Bot. 2016, 48, 155–165. [Google Scholar]
- Wu, X.-J.; Li, M.-Y.; Que, F.; Wang, F.; Xu, Z.-S.; Xiong, A.-S. Genome-wide analysis of PHD family transcription factors in carrot (Daucus carota L.) reveals evolution and response to abiotic stress. Acta Physiol. Plant. 2016, 38, 67. [Google Scholar] [CrossRef]
- Bienz, M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006, 31, 35–40. [Google Scholar] [CrossRef]
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W.H. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014, 10, e1004091. [Google Scholar] [CrossRef]
- Ye, Y.; Gong, Z.; Lu, X.; Miao, D.; Shi, J.; Lu, J.; Zhao, Y. Germostatin resistance locus 1 encodes a PHD finger protein involved in auxin-mediated seed dormancy and germination. Plant J. 2016, 85, 3–15. [Google Scholar] [CrossRef]
- López-González, L.; Mouriz, A.; Narro-Diego, L.; Bustos, R.; Martínez-Zapater, J.M.; Jarillo, J.A.; Piñeiro, M. Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 2014, 26, 3922–3938. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Choi, D.; Lee, J. Fine mapping of the genic male-sterile ms1gene in Capsicum annuum L. Theor. Appl. Genet. 2018, 131, 183–191. [Google Scholar] [CrossRef]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D.; et al. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef]
- Thu, S.W.; Rai, K.M.; Sandhu, D.; Rajangam, A.; Balasubramanian, V.K.; Palmer, R.G.; Mendu, V. Mutation in a PHD-finger protein MS4 causes male sterility in soybean. BMC Plant Biol. 2019, 19, 378. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, L.; Zhang, P.; Zhang, Y.; Yu, P.; Liu, L.; Abbas, A.; Xiang, X.; Wu, W.; Zhan, X.; et al. TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice. Plant J. 2019, 99, 844–861. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.; Liang, W.; Zong, J.; Wilson, Z.A.; Zhang, D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef]
- Wei, W.; Huang, J.; Hao, Y.J.; Zou, H.F.; Wang, H.W.; Zhao, J.Y.; Liu, X.Y.; Zhang, W.K.; Ma, B.; Zhang, J.S.; et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE 2009, 4, e7209. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Pi, E.X.; Tsai, S.N.; Lam, H.M.; Sun, S.M.; Kwan, Y.W.; Ngai, S.M. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 2011, 11, 178. [Google Scholar] [CrossRef]
- Barlow, J.H.; Faryabi, R.B.; Callén, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.W.; McKinnon, P.; Wright, G.; et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef]
- Lv, J.; Liu, Z.; Liu, Y.; Ou, L.; Deng, M.; Wang, J.; Song, J.; Ma, Y.; Chen, W.; Zhang, Z.; et al. Comparative Transcriptome Analysis between Cytoplasmic Male-sterile Line and Its Maintainer During the Floral Bud Development of Pepper. Hortic. Plant J. 2020, 6, 89–98. [Google Scholar] [CrossRef]
- Chen, G.; Ye, X.; Zhang, S.; Zhu, S.; Yuan, L.; Hou, J.; Wang, C. Comparative Transcriptome Analysis between Fertile and CMS Flower Buds in Wucai (Brassica campestris L.). BMC Genom. 2018, 19, 908. [Google Scholar] [CrossRef]
- Chen, L.; Ning, M.; Ping-Yong, W.; Nan, F.; Huo-Lin, S. Transcriptome sequencing and De Novo analysis of a cytoplasmic male sterile line and its near-isogenic restorer line in chili pepper (Capsicum annuum L.). PLoS ONE 2013, 8, e65209. [Google Scholar]
- Yang, Z.; Liu, L.; Sun, L.; Yu, P.; Zhang, P.; Abbas, A.; Xiang, X.; Wu, W.; Zhang, Y.; Cao, L. OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 2019, 99, 175–191. [Google Scholar] [CrossRef]
- Fernández-Gómez, J.; Talle, B.; Wilson, Z.A. Increased expression of the MALE STERILITY1 transcription factor gene results in temperature-sensitive male sterility in barley. J. Exp. Bot. 2020, 71, 6328–6339. [Google Scholar] [CrossRef]
- Wu, H.; Zheng, L.; Qanmber, G.; Guo, M.; Wang, Z.; Yang, Z. Response of phytohormone mediated plant homeodomain (PHD) family to abiotic stress in upland cotton (Gossypium hirsutum spp.). BMC Plant Biol. 2021, 21, 13. [Google Scholar] [CrossRef] [PubMed]
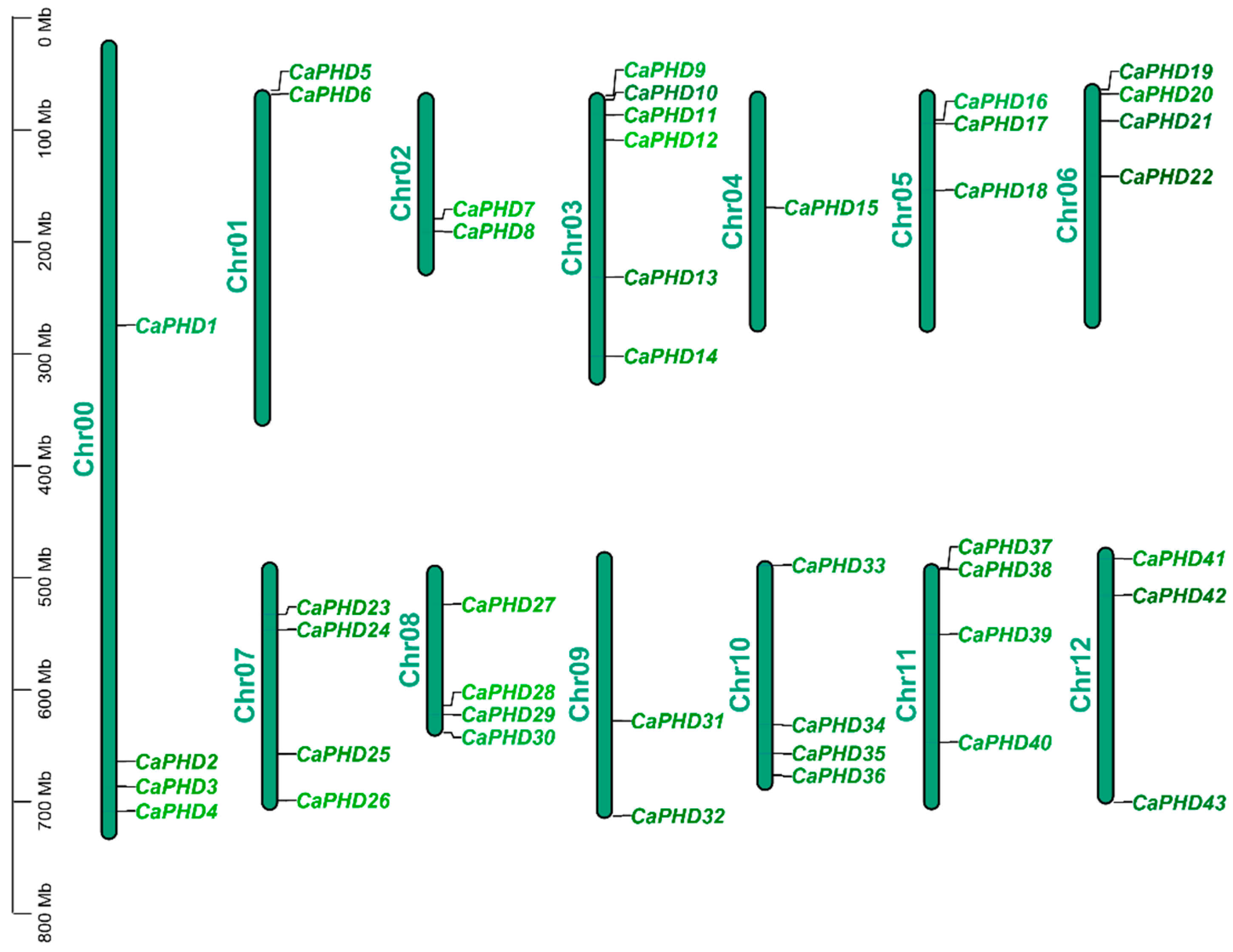
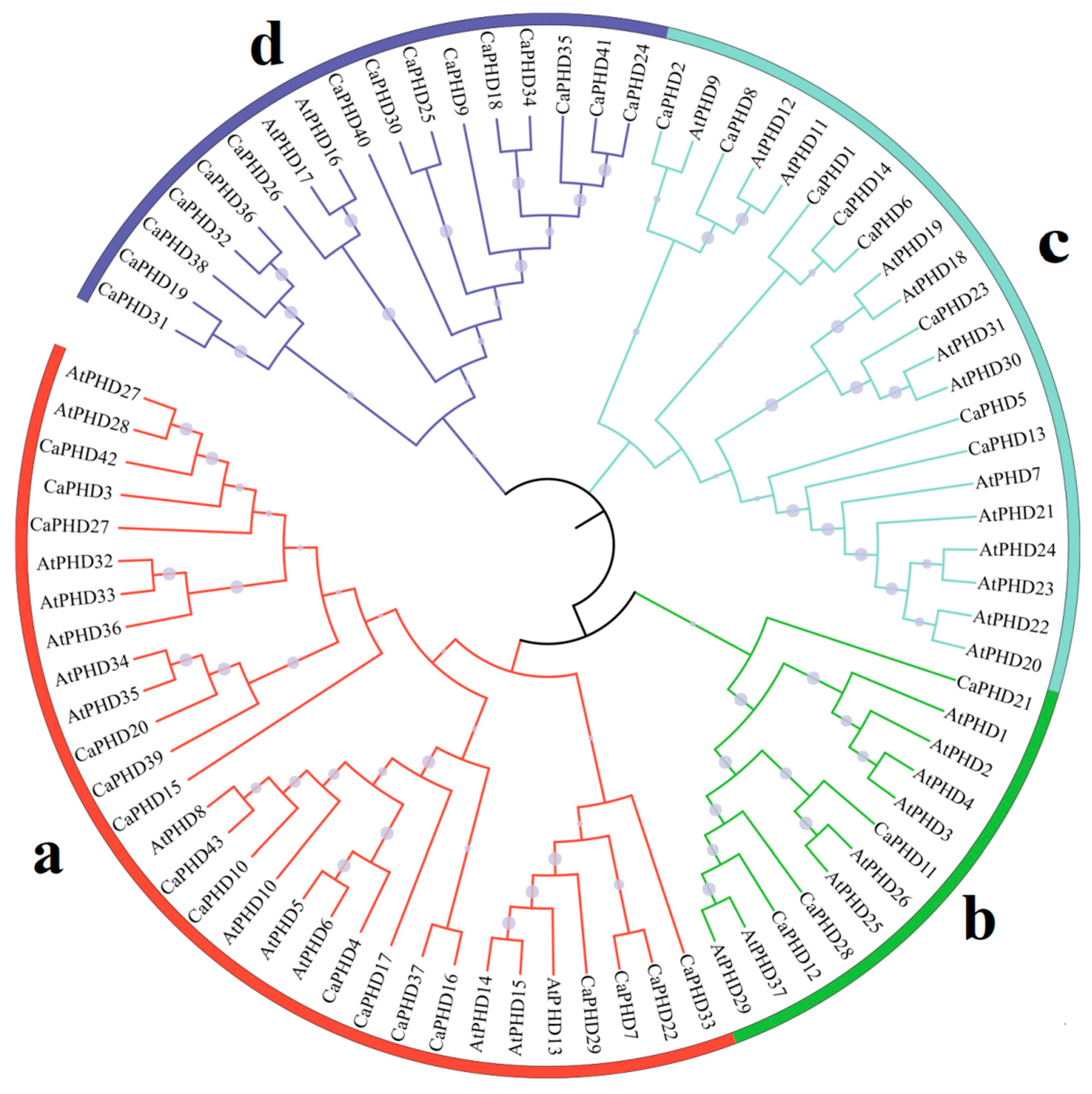
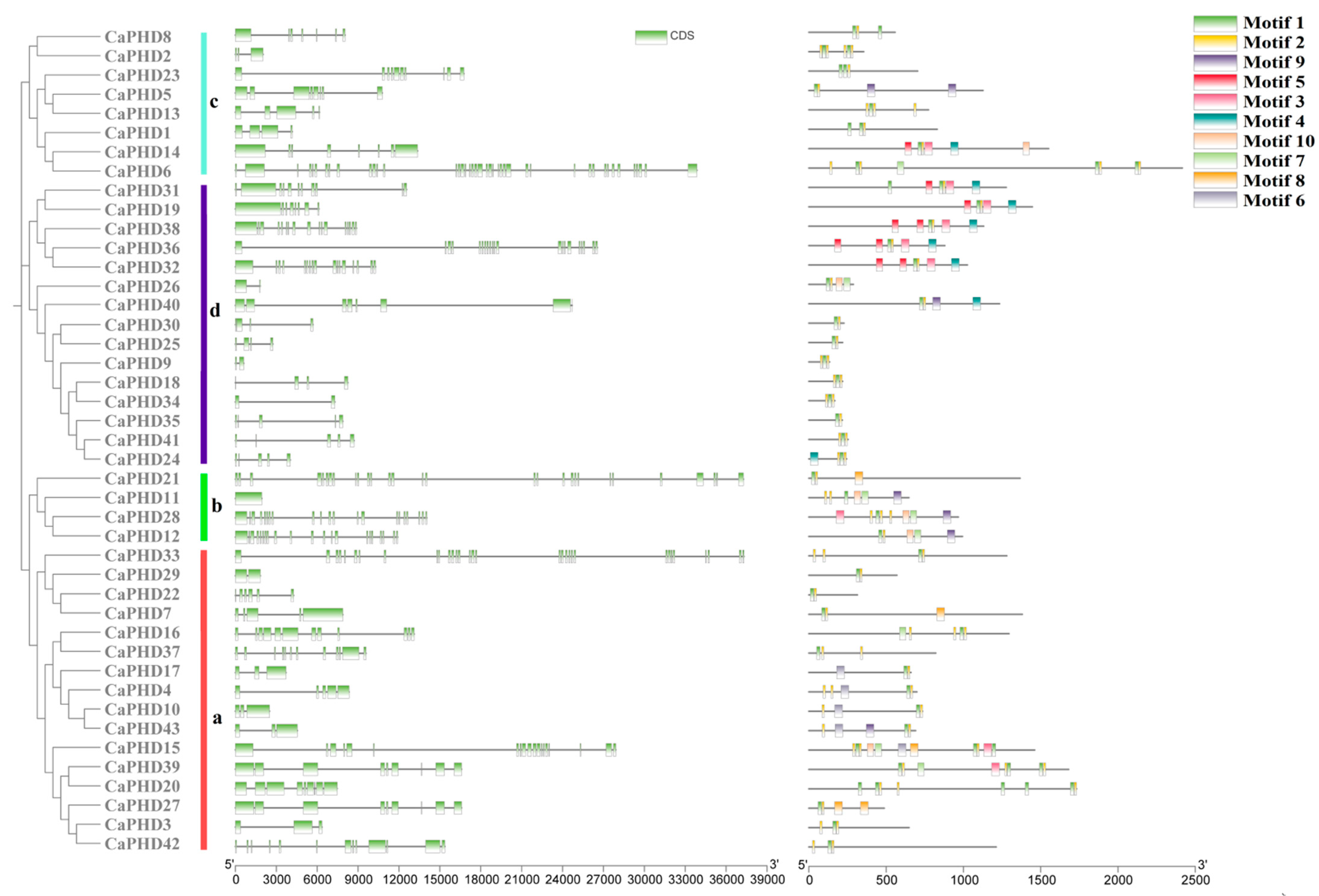

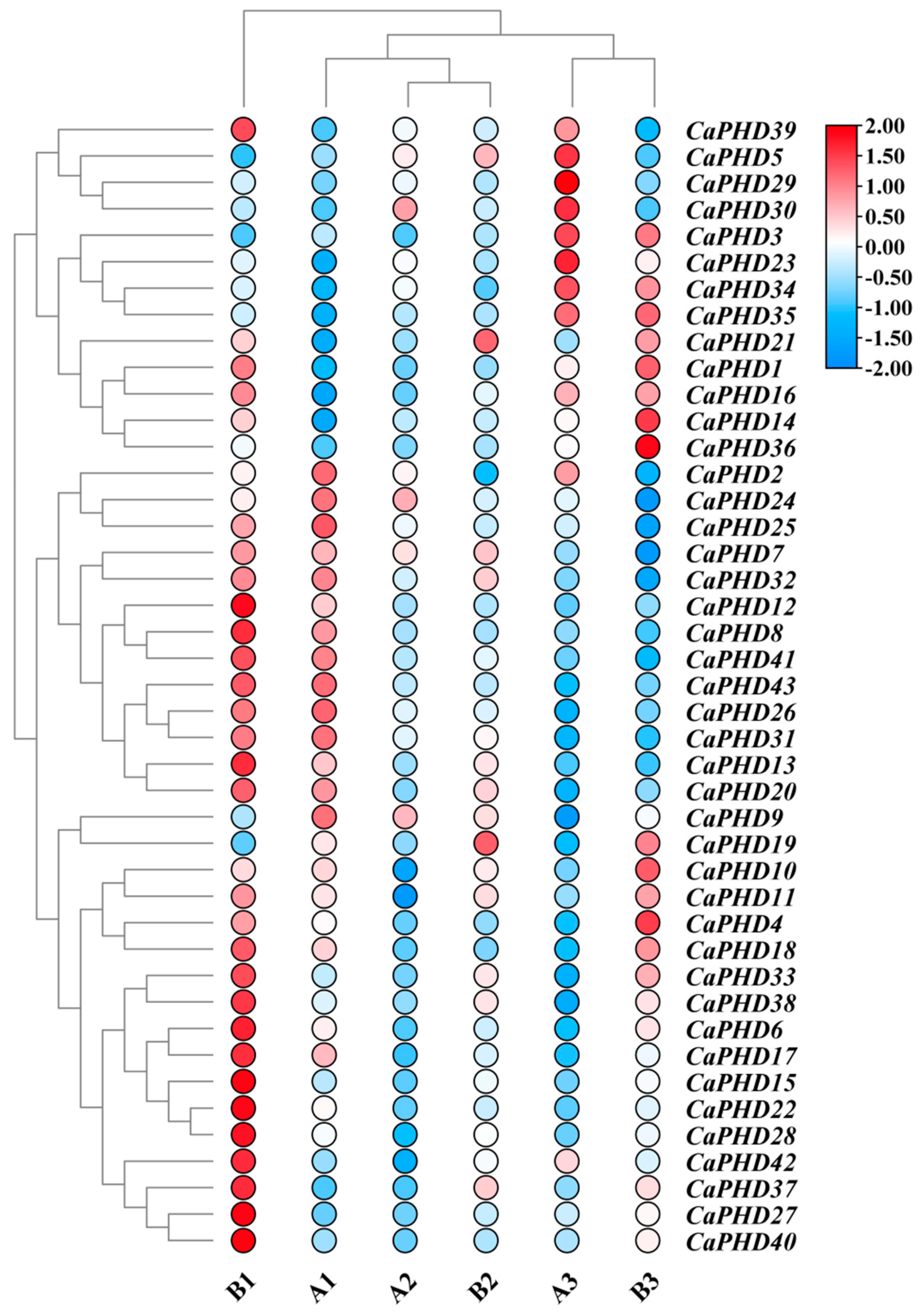
| Name | Gene ID | 5′ End | 3′ End | Protein Size (aa) | ORF (bp) | pI |
|---|---|---|---|---|---|---|
| CaPHD1 | Capana00g000483 | 254,466,020 | 254,470,192 | 830 | 2493 | 5.47 |
| CaPHD2 | Capana00g004445 | 644,022,103 | 644,024,135 | 355 | 1068 | 5.59 |
| CaPHD3 | Capana00g004699 | 666,200,574 | 666,206,931 | 648 | 1947 | 8.35 |
| CaPHD4 | Capana00g005010 | 688,470,993 | 688,479,334 | 699 | 2100 | 8.22 |
| CaPHD5 | Capana01g000006 | 177,703 | 188,477 | 1127 | 3384 | 8.7 |
| CaPHD6 | Capana01g000238 | 3,671,024 | 3,704,883 | 2416 | 7251 | 5.78 |
| CaPHD7 | Capana02g001155 | 112,144,118 | 112,151,993 | 1381 | 4146 | 8.34 |
| CaPHD8 | Capana02g001395 | 123,396,735 | 123,404,780 | 558 | 1677 | 5.23 |
| CaPHD9 | Capana03g000140 | 1,991,155 | 1,991,777 | 134 | 405 | 4.53 |
| CaPHD10 | Capana03g000419 | 5,842,921 | 5,845,422 | 735 | 2208 | 6.14 |
| CaPHD11 | Capana03g001155 | 19,520,907 | 19,522,850 | 647 | 1944 | 8.64 |
| CaPHD12 | Capana03g002025 | 41,777,984 | 41789894 | 994 | 2985 | 8.93 |
| CaPHD13 | Capana03g003071 | 164,384,287 | 164,390,463 | 774 | 2325 | 8.48 |
| CaPHD14 | Capana03g003707 | 235,051,566 | 235,064,929 | 1551 | 4656 | 5.5 |
| CaPHD15 | Capana04g001765 | 103,459,026 | 103,486,934 | 1461 | 4386 | 7.81 |
| CaPHD16 | Capana05g000721 | 26,527,365 | 26,540,480 | 1295 | 3888 | 8.29 |
| CaPHD17 | Capana05g000766 | 29,933,832 | 29,937,546 | 661 | 1986 | 8.11 |
| CaPHD18 | Capana05g001230 | 89,689,887 | 89,698,139 | 219 | 660 | 5.3 |
| CaPHD19 | Capana06g000344 | 4,800,170 | 4,806,304 | 1446 | 4341 | 5.51 |
| CaPHD20 | Capana06g000580 | 8,805,536 | 8,813,012 | 1732 | 5199 | 7.04 |
| CaPHD21 | Capana06g001447 | 32,753,142 | 32,790,414 | 1367 | 4104 | 5.53 |
| CaPHD22 | Capana06g002050 | 82,405,508 | 82,409,778 | 314 | 945 | 8.19 |
| CaPHD23 | Capana07g000572 | 46,259,688 | 46,276,459 | 704 | 2115 | 8.64 |
| CaPHD24 | Capana07g000653 | 59,977,020 | 59,981,053 | 245 | 738 | 5.29 |
| CaPHD25 | Capana07g001300 | 170,904,485 | 170,907,239 | 219 | 660 | 7.09 |
| CaPHD26 | Capana07g002056 | 212,461,334 | 212,463,160 | 288 | 867 | 5.13 |
| CaPHD27 | Capana08g000308 | 34,753,957 | 34,763,289 | 488 | 1467 | 8.96 |
| CaPHD28 | Capana08g001093 | 124,758,585 | 124,772,634 | 967 | 2904 | 8.41 |
| CaPHD29 | Capana08g001577 | 132,947,991 | 132,949,834 | 570 | 1713 | 6.6 |
| CaPHD30 | Capana08g002581 | 149,165,449 | 149,171,136 | 227 | 684 | 8.48 |
| CaPHD31 | Capana09g001374 | 150,627,608 | 150,640,165 | 1277 | 3834 | 6.69 |
| CaPHD32 | Capana09g002280 | 235,766,586 | 235,776,872 | 1026 | 3081 | 6.44 |
| CaPHD33 | Capana10g000226 | 3,810,195 | 3,847,501 | 1281 | 3846 | 6.22 |
| CaPHD34 | Capana10g001358 | 146,143,245 | 146,150,538 | 168 | 507 | 6.05 |
| CaPHD35 | Capana10g001653 | 171,574,802 | 171,582,681 | 216 | 651 | 4.98 |
| CaPHD36 | Capana10g001914 | 191,133,928 | 191,160,485 | 880 | 2643 | 9.35 |
| CaPHD37 | Capana11g000110 | 3,151,600 | 3,161,184 | 821 | 2466 | 4.78 |
| CaPHD38 | Capana11g000184 | 4,429,857 | 4,438,746 | 1131 | 3396 | 4.91 |
| CaPHD39 | Capana11g000907 | 62,802,424 | 62,819,020 | 1681 | 5046 | 7.92 |
| CaPHD40 | Capana11g001326 | 159,279,171 | 159,303,879 | 1234 | 3705 | 5.97 |
| CaPHD41 | Capana12g000480 | 9,701,874 | 9,710,579 | 254 | 765 | 5.07 |
| CaPHD42 | Capana12g001061 | 42,716,016 | 42,731,391 | 1212 | 3639 | 5.38 |
| CaPHD43 | Capana12g002774 | 227,622,114 | 227,626,663 | 691 | 2076 | 5.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, J.; Deng, M.; Zhao, K.; Zhou, H.; Wu, R.; Li, M.; Cheng, H.; Li, P.; Zhang, R.; Lv, J. Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper. Genes 2023, 14, 1737. https://doi.org/10.3390/genes14091737
Wen J, Deng M, Zhao K, Zhou H, Wu R, Li M, Cheng H, Li P, Zhang R, Lv J. Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper. Genes. 2023; 14(9):1737. https://doi.org/10.3390/genes14091737
Chicago/Turabian StyleWen, Jinfen, Minghua Deng, Kai Zhao, Huidan Zhou, Rui Wu, Mengjuan Li, Hong Cheng, Pingping Li, Ruihao Zhang, and Junheng Lv. 2023. "Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper" Genes 14, no. 9: 1737. https://doi.org/10.3390/genes14091737
APA StyleWen, J., Deng, M., Zhao, K., Zhou, H., Wu, R., Li, M., Cheng, H., Li, P., Zhang, R., & Lv, J. (2023). Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper. Genes, 14(9), 1737. https://doi.org/10.3390/genes14091737






