Transcriptomic Profiling of Subcutaneous Backfat in Castrated and Intact Alentejano Pigs Finished Outdoors with Commercial and Fiber-Rich Diets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Tissue Sampling
2.2. Carcass Traits and Tissue Sampling
2.3. Muscle and Fat Quality Traits
2.4. RNA Extraction
2.5. Library Preparation
2.6. Bioinformatic Analysis
2.7. RT qPCR Validation Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Effect of Castration and Diet in Carcass Traits, Meat Quality Traits, and in Fatty Acid Composition
3.2. Sequencing and Mapping Statistics
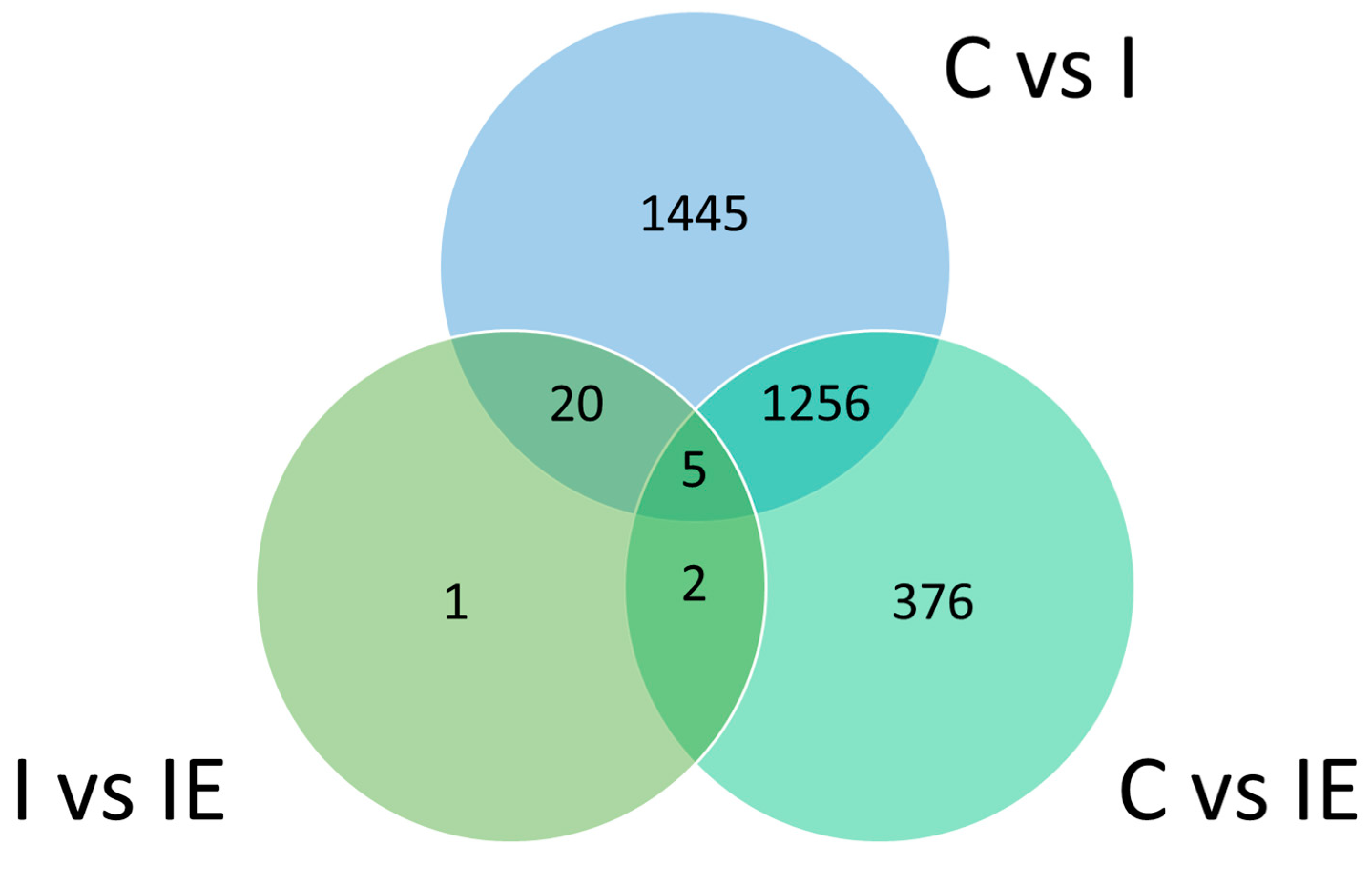
3.3. Differential Gene Expression Analysis
3.4. Validation Analysis
3.5. Functional Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muñoz, M.; Bozzi, R.; García, F.; Núñez, Y.; Geraci, C.; Crovetti, A.; García-Casco, J.; Alves, E.; Škrlep, M.; Charneca, R.; et al. Diversity across Major and Candidate Genes in European Local Pig Breeds. PLoS ONE 2018, 13, e0207475. [Google Scholar] [CrossRef] [PubMed]
- Gama, L.T.; Martínez, A.M.; Carolino, I.; Landi, V.; Delgado, J.V.; Vicente, A.A.; Vega-Pla, J.L.; Cortés, O.; Sousa, C.O. Genetic Structure, Relationships and Admixture with Wild Relatives in Native Pig Breeds from Iberia and Its Islands. Genet. Sel. Evol. 2013, 45, 18. [Google Scholar] [CrossRef]
- Charneca, R.; Martins, J.; Freitas, A.; Neves, J.; Nunes, J.; Paixim, H.; Bento, P.; Batorek-Lukač, N. Alentejano Pig. In European Local Pig Breeds-Diversity and Performance; Candek-Potokar, M., Linan, R.M.N., Eds.; IntechOpen: London, UK, 2019; pp. 13–36. [Google Scholar] [CrossRef]
- Lopez-Bote, C.J. Sustained Utilization of the Iberian Pig Breed. Meat Sci. 1998, 49, S17–S27. [Google Scholar] [CrossRef]
- Larzul, C. How to Improve Meat Quality and Welfare in Entire Male Pigs by Genetics. Animals 2021, 11, 699. [Google Scholar] [CrossRef] [PubMed]
- Rault, J.-L.; Lay, D.C.; Marchant-Forde, J.N. Castration Induced Pain in Pigs and Other Livestock. Appl. Anim. Behav. Sci. 2011, 135, 214–225. [Google Scholar] [CrossRef]
- Claus, R.; Lacorn, M.; Ostertag, C. An Improved Microtitre Enzyme Immunoassay to Measure the Boar Taint Steroid 5α-Androst-16-En-3-One in Blood Plasma of Pigs. Meat Sci. 2008, 80, 934–938. [Google Scholar] [CrossRef]
- Patterson, R.L.S. 5α-Androst-16-Ene-3-One: Compound Responsible for Taint in Boar Fat. J. Sci. Food Agric. 1968, 19, 31–38. [Google Scholar] [CrossRef]
- Lin, Z.; Lou, Y.; Squires, E.J. Functional Polymorphism in Porcine CYP2E1 Gene: Its Association with Skatole Levels. J. Steroid Biochem. Mol. Biol. 2006, 99, 231–237. [Google Scholar] [CrossRef]
- Vold, E. Fleischproduktionseigenschaften Bei Ebern Und Kastraten: III. Untersuchungen der Schlachtkörperzusammensetzung, sowie der Fleisch-und Speckqualität bei Ebern und Kastraten; Meldinger fra Norges Landbrukshøgskole, NLH: Vollebekk, Norway, 1969; Volume 48, pp. 1–25. [Google Scholar]
- Walstra, P.; Maarse, G. Onderzoek Gestachlengen van Mannelijke Mestvarkens. In Researchgroep voor Vlees en Vleeswaren TNO, IVO-Rapport C 147; TNO: Zeist, The Netherlands, 1970; Volume 2, pp. 1–30. [Google Scholar]
- Bonneau, M.; Kempster, A.J.; Claus, R.; Claudi-Magnussen, C.; Diestre, A.; Tornberg, E.; Walstra, P.; Chevillon, P.; Weiler, U.; Cook, G.L. An International Study on the Importance of Androstenone and Skatole for Boar Taint: I. Presentation of the Programme and Measurement of Boar Taint Compounds with Different Analytical Procedures. Meat Sci. 2000, 54, 251–259. [Google Scholar] [CrossRef]
- Jensen, B.; Jensen, M. Microbial Production of Skatole in the Digestive Tract of Entire Male Pigs. In Skatole and Boar Taint: Results from an Integrated National Research Project Investigating Causes of Boar Taint in Danish Pigs; Jensen, W.K., Ed.; Danish Meat Research Institute: Roskilde, Denmark, 1998; pp. 41–75. [Google Scholar]
- Jensen, B.B. Prevention of Boar Taint in Pig Production. Factors Affecting the Level of Skatole. Acta Vet. Scand. 2006, 48, S6. [Google Scholar] [CrossRef]
- Friis, C. Distribution, Metabolic Fate and Elimination of Skatole in the Pig. In Proceedings of the Colloques de l’INRA, Meeting of the EAAP Working Group Production and Utilization of Meat from Entire Male Pigs, Roskilde, Danemark, 12–14 October 1993; INRA Edition: Paris, France, 1993; pp. 113–115. [Google Scholar]
- Kristina Andersson, H.; Andersson, K.; Zamaratskaia, G.; Rydhmer, L.; Chen, G.; Lundström, K. Effect of Single-Sex or Mixed Rearing and Live Weight on Performance, Technological Meat Quality and Sexual Maturity in Entire Male and Female Pigs Fed Raw Potato Starch. Acta Agric. Scand. Sect.—Anim. Sci. 2005, 55, 80–90. [Google Scholar] [CrossRef]
- Øverland, M.; Kjos, N.K.; Fauske, A.K.; Teige, J.; Sørum, H. Easily Fermentable Carbohydrates Reduce Skatole Formation in the Distal Intestine of Entire Male Pigs. Livest. Sci. 2011, 140, 206–217. [Google Scholar] [CrossRef]
- Heyrman, E.; Millet, S.; Tuyttens, F.A.M.; Ampe, B.; Janssens, S.; Buys, N.; Wauters, J.; Vanhaecke, L.; Aluwé, M. On Farm Intervention Studies on Reduction of Boar Taint Prevalence: Feeding Strategies, Presence of Gilts and Time in Lairage. Res. Vet. Sci. 2018, 118, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.L.; Mejer, H.; Thamsborg, S.M.; Byrne, D.V.; Roepstorff, A.; Karlsson, A.H.; Hansen-Møller, J.; Jensen, M.T.; Tuomola, M. Influence of Chicory Roots (Cichorium intybus L.) on Boar Taint in Entire Male and Female Pigs. Anim. Sci. 2006, 82, 359–368. [Google Scholar] [CrossRef]
- Zamaratskaia, G.; Squires, E.J. Biochemical, Nutritional and Genetic Effects on Boar Taint in Entire Male Pigs. Anim. Int. J. Anim. Biosci. 2009, 3, 1508–1521. [Google Scholar] [CrossRef]
- Rasmussen, M.K.; Brunius, C.; Zamaratskaia, G.; Ekstrand, B. Feeding Dried Chicory Root to Pigs Decrease Androstenone Accumulation in Fat by Increasing Hepatic 3β Hydroxysteroid Dehydrogenase Expression. J. Steroid Biochem. Mol. Biol. 2012, 130, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B. Impact of Feed Composition and Feed Processing on the Gastrointestinal Ecosystem in Pigs. In Proceedings of the Nutrition and Gastrointestinal Physiology–Today and Tomorrow; TNO Nutrition and Food Research Institute: Wageningen, The Netherlands, 1999; pp. 43–56. [Google Scholar]
- Boyd, W.L.; Lichstein, H.C. The Effect of Carbohydrates on the Tryptophanase Activity of Bacteria. J. Bacteriol. 1955, 69, 584–589. [Google Scholar] [CrossRef]
- Van Oeckel, M.J.; Warnants, N.; De Paepe, M.; Casteels, M.; Boucqué, C.V. Effect of Fibre-Rich Diets on the Backfat Skatole Content of Entire Male Pigs. Livest. Prod. Sci. 1998, 56, 173–180. [Google Scholar] [CrossRef]
- Knarreborg, A.; Beck, J.; Jensen, M.T.; Laue, A.; Agergaard, N.; Jensen, B.B. Effect of Non-Starch Polysaccharides on Production and Absorption of Indolic Compounds in Entire Male Pigs. Anim. Sci. 2002, 74, 445–453. [Google Scholar] [CrossRef]
- Whittington, F.M.; Nute, G.R.; Hughes, S.I.; McGivan, J.D.; Lean, I.J.; Wood, J.D.; Doran, E. Relationships between Skatole and Androstenone Accumulation, and Cytochrome P4502E1 Expression in Meishan×Large White Pigs. Meat Sci. 2004, 67, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.M.; Varino, R.; Charneca, R.; Albuquerque, A.; Garrido, N.; Neves, J.; Freitas, A.; Costa, F.; Marmelo, C.; Ramos, A.; et al. Outdoor Finishing of Intact Male Portuguese Alentejano Pigs on a Sustainable High-Fiber Diet: Impacts on Blood, Growth, Carcass, Meat Quality and Boar Taint Compounds. Animals 2023, 13, 2221. [Google Scholar] [CrossRef] [PubMed]
- NP-2931; Swine Slaughtered for Direct Consumption-Cutting Half Carcass. Instituto Português da Qualidade: Lisboa, Portugal, 2006.
- ISO 1442:1997; Meat and Meat Products—Determination of Moisture Content (Reference Method). International Organization for Standardization: Geneva, Switzerland, 1997.
- AOAC. Official Methods of Analysis of AOAC International, 18th ed.; AOAC: Gaithersburg, MD, USA, 2006. [Google Scholar]
- Folch, J.; Lees, M.; Sloane, S.G.H.; Folch, J.; Lees, M.; Sloane-Stanley, G. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Alves, S.P.; Raundrup, K.; Cabo, Â.; Bessa, R.J.B.; Almeida, A.M. Fatty Acid Composition of Muscle, Adipose Tissue and Liver from Muskoxen (Ovibos moschatus) Living in West Greenland. PLoS ONE 2015, 10, e0145241. [Google Scholar] [CrossRef]
- ISO 2917:1999; Meat and Meat Products—Measurement of PH (Reference Method). International Organization for Standardization: Geneva, Switzerland, 1999.
- Hornsey, H.C. The Colour of Cooked Cured Pork. I.—Estimation of the Nitric Oxide-Haem Pigments. J. Sci. Food Agric. 1956, 7, 534–540. [Google Scholar] [CrossRef]
- Cava, R.; Estévez, M.; Ruiz, J.; Morcuende, D. Physicochemical Characteristics of Three Muscles from Free-Range Reared Iberian Pigs Slaughtered at 90 Kg Live Weight. Meat Sci. 2003, 63, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Woessner, J.F. The Determination of Hydroxyproline in Tissue and Protein Samples Containing Small Proportions of This Imino Acid. Arch. Biochem. Biophys. 1961, 93, 440–447. [Google Scholar] [CrossRef]
- Etherington, D.J.; Sims, T.J. Detection and Estimation of Collagen. J. Sci. Food Agric. 1981, 32, 539–546. [Google Scholar] [CrossRef]
- Martins, J.M.; Fialho, R.; Albuquerque, A.; Neves, J.; Freitas, A.; Nunes, J.T.; Charneca, R. Growth, Blood, Carcass and Meat Quality Traits from Local Pig Breeds and Their Crosses. Animal 2020, 14, 636–647. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinforma. Oxf. Engl. 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. Gprofiler2--an R Package for Gene List Functional Enrichment Analysis and Namespace Conversion Toolset g:Profiler. F1000Research 2020, 9, ELIXIR-709. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Miron, M.; Woody, O.Z.; Marcil, A.; Murie, C.; Sladek, R.; Nadon, R. A Methodology for Global Validation of Microarray Experiments. BMC Bioinform. 2006, 7, 333. [Google Scholar] [CrossRef]
- Zomeño, C.; Gispert, M.; Brun, A.; Carabús, A.; Soler, J.; Font-i-Furnols, M. Productive Performance and in Vivo Body Composition across the Growing and Finishing Period and Carcass Traits in Pigs of Four Sex Types. Meat Sci. 2022, 192, 108909. [Google Scholar] [CrossRef]
- Serrano, M.P. A Study of Factors That Influence Growth Performance and Carcass and Meat Quality of Iberian Pigs Reared under Intensive Management. Ph.D. Thesis, Universidad Politécnica de Madrid, Escuela Técnica Superior de Enginieros Agrónomos, Madrid, Spain, 2008. [Google Scholar]
- Christensen, K. In Vitro Studies on the Synthesis of Intramuscular Fat in the Longissimus Dorsh Muscle of Pigs. Livest. Prod. Sci. 1975, 2, 59–68. [Google Scholar] [CrossRef]
- Albuquerque, A.; Óvilo, C.; Núñez, Y.; Benítez, R.; López-Garcia, A.; García, F.; Félix, M.d.R.; Laranjo, M.; Charneca, R.; Martins, J.M. Comparative Transcriptomic Analysis of Subcutaneous Adipose Tissue from Local Pig Breeds. Genes 2020, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; García-Casco, J.M.; Caraballo, C.; Fernández-Barroso, M.Á.; Sánchez-Esquiliche, F.; Gómez, F.; Rodríguez, M.d.C.; Silió, L. Identification of Candidate Genes and Regulatory Factors Underlying Intramuscular Fat Content Through Longissimus Dorsi Transcriptome Analyses in Heavy Iberian Pigs. Front. Genet. 2018, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Houseknecht, K.L.; Baile, C.A.; Matteri, R.L.; Spurlock, M.E. The Biology of Leptin: A Review. J. Anim. Sci. 1998, 76, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Barb, C.R. The Brain-Pituitary-Adipocyte Axis: Role of Leptin in Modulating Neuroendocrine Function. J. Anim. Sci. 1999, 77, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, X.; Yu, S.; Zheng, R. The Leptin Resistance. Adv. Exp. Med. Biol. 2018, 1090, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Ovilo, C.; Fernández, A.; Fernández, A.I.; Folch, J.M.; Varona, L.; Benítez, R.; Nuñez, Y.; Rodríguez, C.; Silió, L. Hypothalamic Expression of Porcine Leptin Receptor (LEPR), Neuropeptide Y (NPY), and Cocaine- and Amphetamine-Regulated Transcript (CART) Genes Is Influenced by LEPR Genotype. Mamm. Genome 2010, 21, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Behre, H.M.; Simoni, M.; Nieschlag, E. Strong Association between Serum Levels of Leptin and Testosterone in Men. Clin. Endocrinol. 1997, 47, 237–240. [Google Scholar] [CrossRef]
- Lopez, S.; Bermudez, B.; Montserrat-De La Paz, S.; Jaramillo, S.; Varela, L.M.; Ortega-Gomez, A.; Abia, R.; Muriana, F.J.G. Membrane Composition and Dynamics: A Target of Bioactive Virgin Olive Oil Constituents. Biochim. Biophys. Acta-Biomembr. 2014, 1838, 1638–1656. [Google Scholar] [CrossRef]
- Doran, O.; Moule, S.K.; Teye, G.A.; Whittington, F.M.; Hallett, K.G.; Wood, J.D. A Reduced Protein Diet Induces Stearoyl-CoA Desaturase Protein Expression in Pig Muscle but Not in Subcutaneous Adipose Tissue: Relationship with Intramuscular Lipid Formation. Br. J. Nutr. 2006, 95, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Porter, V. Spain and Portugal. In Pigs: A Handbook to the Breeds of the World; Porter, V., Mountfield, T.J., Eds.; Cornell University Press: Ithaca, NY, USA, 1993; pp. 137–140. ISBN 1-873403-17-8. [Google Scholar]
- Bruininx, E.; van den Borne, J.; van Heugten, E.; van Milgen, J.; Verstegen, M.; Gerrits, W. Oxidation of Dietary Stearic, Oleic, and Linoleic Acids in Growing Pigs Follows a Biphasic Pattern. J. Nutr. 2011, 141, 1657–1663. [Google Scholar] [CrossRef]
- Garrido, N.; Izquierdo, M.; Hernández-García, F.I.; Núñez, Y.; García-Torres, S.; Benítez, R.; Padilla, J.Á.; Óvilo, C. Differences in Muscle Lipogenic Gene Expression, Carcass Traits and Fat Deposition among Three Iberian Pig Strains Finished in Two Different Feeding Systems. Animals 2023, 13, 1138. [Google Scholar] [CrossRef]
- Grzes, M.; Sadkowski, S.; Rzewuska, K.; Szydlowski, M.; Switonski, M. Pig Fatness in Relation to FASN and INSIG2 Genes Polymorphism and Their Transcript Level. Mol. Biol. Rep. 2016, 43, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Semenkovich, C.F. Regulation of Fatty Acid Synthase (FAS). Prog. Lipid Res. 1997, 36, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.C.; Xu, Z.R.; Han, X.Y.; Li, W.F. Effect of Dietary Betaine Supplementation on Lipogenic Enzyme Activities and Fatty Acid Synthase MRNA Expression in Finishing Pigs. Anim. Feed Sci. Technol. 2008, 140, 365–375. [Google Scholar] [CrossRef]
- Miao, Z.; Zhu, F.; Zhang, H.; Chang, X.; Xie, H.; Zhang, J.; Xu, Z. Developmental Patterns of FASN and LIPE MRNA Expression in Adipose Tissue of Growing Jinhua and Landrace Gilts. Czech J. Anim. Sci. 2010, 55, 557–564. [Google Scholar] [CrossRef]
- Albuquerque, A.; Neves, J.A.; Redondeiro, M.; Laranjo, M.; Félix, M.R.; Freitas, A.; Tirapicos, J.L.; Martins, J.M. Long Term Betaine Supplementation Regulates Genes Involved in Lipid and Cholesterol Metabolism of Two Muscles from an Obese Pig Breed. Meat Sci. 2017, 124, 25–33. [Google Scholar] [CrossRef]
- Moon, Y.-A.; Shah, N.A.; Mohapatra, S.; Warrington, J.A.; Horton, J.D. Identification of a Mammalian Long Chain Fatty Acyl Elongase Regulated by Sterol Regulatory Element-Binding Proteins. J. Biol. Chem. 2001, 276, 45358–45366. [Google Scholar] [CrossRef] [PubMed]
- Corominas, J.; Ramayo-Caldas, Y.; Puig-Oliveras, A.; Pérez-Montarelo, D.; Noguera, J.L.; Folch, J.M.; Ballester, M. Polymorphism in the ELOVL6 Gene Is Associated with a Major QTL Effect on Fatty Acid Composition in Pigs. PLoS ONE 2013, 8, e53687. [Google Scholar] [CrossRef] [PubMed]
- Corominas, J.; Marchesi, J.A.P.; Puig-Oliveras, A.; Revilla, M.; Estellé, J.; Alves, E.; Folch, J.M.; Ballester, M. Epigenetic Regulation of the ELOVL6 Gene Is Associated with a Major QTL Effect on Fatty Acid Composition in Pigs. Genet. Sel. Evol. GSE 2015, 47, 20. [Google Scholar] [CrossRef] [PubMed]
- Corominas, J.; Ramayo-Caldas, Y.; Puig-Oliveras, A.; Estellé, J.; Castelló, A.; Alves, E.; Pena, R.N.; Ballester, M.; Folch, J.M. Analysis of Porcine Adipose Tissue Transcriptome Reveals Differences in de Novo Fatty Acid Synthesis in Pigs with Divergent Muscle Fatty Acid Composition. BMC Genom. 2013, 14, 843. [Google Scholar] [CrossRef]
- Naomi, R.; Ridzuan, P.M.; Bahari, H. Current Insights into Collagen Type I. Polymers 2021, 13, 2642. [Google Scholar] [CrossRef]
- Akit, H.; Collins, C.; Fahri, F.; Hung, A.; D’Souza, D.; Leury, B.; Dunshea, F. Dietary Lecithin Decreases Skeletal Muscle COL1A1 and COL3A1 Gene Expression in Finisher Gilts. Animals 2016, 6, 38. [Google Scholar] [CrossRef]
- Akit, H.; Collins, C.L.; Fahri, F.T.; Hung, A.T.; D’Souza, D.N.; Leury, B.J.; Dunshea, F.R. Dietary Lecithin Improves Dressing Percentage and Decreases Chewiness in the Longissimus Muscle in Finisher Gilts. Meat Sci. 2014, 96, 1147–1151. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Myosin Isoforms in Mammalian Skeletal Muscle. J. Appl. Physiol. 1994, 77, 493–501. [Google Scholar] [CrossRef]
- Lefaucheur, L.; Ecolan, P.; Plantard, L.; Gueguen, N. New Insights into Muscle Fiber Types in the Pig. J. Histochem. Cytochem. 2002, 50, 719–730. [Google Scholar] [CrossRef]
- Hou, Y.; Jing, L.; Zhao, Y.; Wang, S.; Liu, A.; Zhao, S.; Li, X. Identification of Genome Regions and Important Variations Associated with Expression Level of MYH Genes and Its Correlation with Meat Quality Traits in Pigs. Front. Agric. Sci. Eng. 2015, 2, 237–241. [Google Scholar] [CrossRef]
- Ahn, J.S.; Kim, D.-H.; Park, H.-B.; Han, S.-H.; Hwang, S.; Cho, I.-C.; Lee, J.-W. Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice. Int. J. Mol. Sci. 2018, 19, 2959. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-D.; Jeong, J.-Y.; Hur, S.; Yang, H.-S.; Jeon, J.-T.; Joo, S.-T. The Relationship between Meat Color (CIE L* and A*), Myoglobin Content, and Their Influence on Muscle Fiber Characteristics and Pork Quality. Korean J. Food Sci. Anim. Resour. 2010, 30, 626–633. [Google Scholar] [CrossRef]
- Cheng, H.; Song, S.; Kim, G.-D. Frozen/Thawed Meat Quality Associated with Muscle Fiber Characteristics of Porcine Longissimus Thoracis et Lumborum, Psoas Major, Semimembranosus, and Semitendinosus Muscles. Sci. Rep. 2021, 11, 13354. [Google Scholar] [CrossRef]
- Kemp, J.D.; Shelley, J.M., Jr.; Ely, D.G.; Moody, W.G. Effects of Castration and Slaughter Weight on Fatness, Cooking Losses and Palatability of Lamb. J. Anim. Sci. 1972, 34, 560–562. [Google Scholar] [CrossRef]
- Teye, G.A.; Sheard, P.R.; Whittington, F.M.; Nute, G.R.; Stewart, A.; Wood, J.D. Influence of Dietary Oils and Protein Level on Pork Quality. 1. Effects on Muscle Fatty Acid Composition, Carcass, Meat and Eating Quality. Meat Sci. 2006, 73, 157–165. [Google Scholar] [CrossRef]
- Xu, Y.J.; Jin, M.L.; Wang, L.J.; Zhang, A.D.; Zuo, B.; Xu, D.Q.; Ren, Z.Q.; Lei, M.G.; Mo, X.Y.; Li, F.E.; et al. Differential Proteome Analysis of Porcine Skeletal Muscles between Meishan and Large White1. J. Anim. Sci. 2009, 87, 2519–2527. [Google Scholar] [CrossRef]
- Yang, H.; Xu, X.; Ma, H.; Jiang, J. Integrative Analysis of Transcriptomics and Proteomics of Skeletal Muscles of the Chinese Indigenous Shaziling Pig Compared with the Yorkshire Breed. BMC Genet. 2016, 17, 80. [Google Scholar] [CrossRef]
- Helferich, W.G.; Jump, D.B.; Anderson, D.B.; Skjaerlund, D.M.; Merkel, R.A.; Bergen, W.G. Skeletal Muscle α-Actin Synthesis Is Increased Pretranslationally in Pigs Fed the Phenethanolamine Ractopamine*. Endocrinology 1990, 126, 3096–3100. [Google Scholar] [CrossRef]
- Coma, J.; Zimmerman, D.R.; Carrion, D. Relationship of Rate of Lean Tissue Growth and Other Factors to Concentration of Urea in Plasma of Pigs. J. Anim. Sci. 1995, 73, 3649–3656. [Google Scholar] [CrossRef]
- Vidal, O.; Varona, L.; Oliver, M.A.; Noguera, J.L.; Sànchez, A.; Amills, M. Malic Enzyme 1 Genotype Is Associated with Backfat Thickness and Meat Quality Traits in Pigs. Anim. Genet. 2006, 37, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Michal, J.J.; Zhang, Z.W.; Gaskins, C.T.; Jiang, Z. The Bovine Fatty Acid Binding Protein 4 Gene Is Significantly Associated with Marbling and Subcutaneous Fat Depth in Wagyu x Limousin F2 Crosses. Anim. Genet. 2006, 37, 400–402. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Y.H.; Zhang, S.; Li, F.; Wang, S.; Dai, L.; Jiang, H.; Xiao, S.; Liu, D.; Sun, B.; et al. Association of A-FABP Gene Polymorphism in Intron 1 with Meat Quality Traits in Junmu No. 1 White Swine. Gene 2011, 487, 170–173. [Google Scholar] [CrossRef]
- Damon, M.; Louveau, I.; Lefaucheur, L.; Lebret, B.; Vincent, A.; Leroy, P.; Sanchez, M.P.; Herpin, P.; Gondret, F. Number of Intramuscular Adipocytes and Fatty Acid Binding Protein-4 Content Are Significant Indicators of Intramuscular Fat Level in Crossbred Large White × Duroc Pigs1. J. Anim. Sci. 2006, 84, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Gerbens, F.; Jansen, A.; Van Erp, A.J.M.; Harders, F.; Meuwissen, T.H.E.; Rettenberger, G.; Veerkamp, J.H.; Te Pas, M.F.W. The Adipocyte Fatty Acid-Binding Protein Locus: Characterization and Association with Intramuscular Fat Content in Pigs. Mamm. Genome 1998, 9, 1022–1026. [Google Scholar] [CrossRef]
- Wiercinska, P.; Lou, Y.; Squires, E.J. The Roles of Different Porcine Cytochrome P450 Enzymes and Cytochrome B5A in Skatole Metabolism. Animal 2012, 6, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Friis, C. Is Boar Taint Related to Sex Differences or Polymorphism of Skatole Metabolism? In Proceedings of the EAAP Working Group on Production and Utilization of Meat from Entire Male Pigs, Milton Keynes, UK, 27–29 September 1995; pp. 25–27. [Google Scholar]
- Skaanild, M.T.; Friis, C. Is Bupropion a More Specific Substrate for Porcine CYP2E than Chlorzoxazone and P-Nitrophenol? Basic Clin. Pharmacol. Toxicol. 2007, 101, 159–162. [Google Scholar] [CrossRef]
- Wiercinska, P.; Squires, E.J. Chlorzoxazone Metabolism by Porcine Cytochrome P450 Enzymes and the Effect of Cytochrome b5. Drug Metab. Dispos. 2010, 38, 857–862. [Google Scholar] [CrossRef]
- Kojima, M.; Sekimoto, M.; Degawa, M. A Novel Gender-Related Difference in the Constitutive Expression of Hepatic Cytochrome P4501A Subfamily Enzymes in Meishan Pigs. Biochem. Pharmacol. 2008, 75, 1076–1082. [Google Scholar] [CrossRef]
- Brunius, C.; Rasmussen, M.K.; Lacoutière, H.; Andersson, K.; Ekstrand, B.; Zamaratskaia, G. Expression and Activities of Hepatic Cytochrome P450 (CYP1A, CYP2A and CYP2E1) in Entire and Castrated Male Pigs. Animal 2012, 6, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway Enrichment Analysis and Visualization of Omics Data Using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
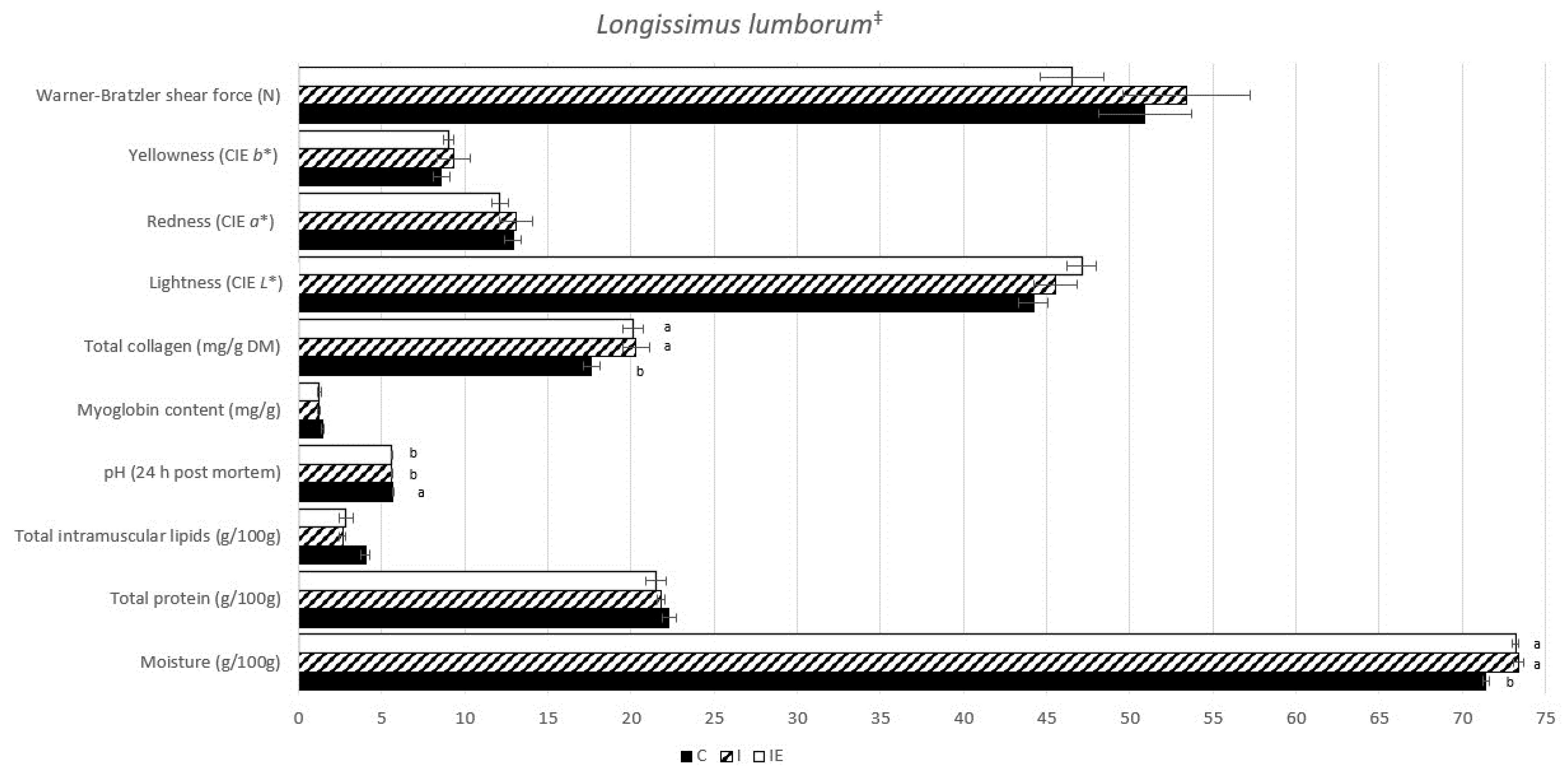
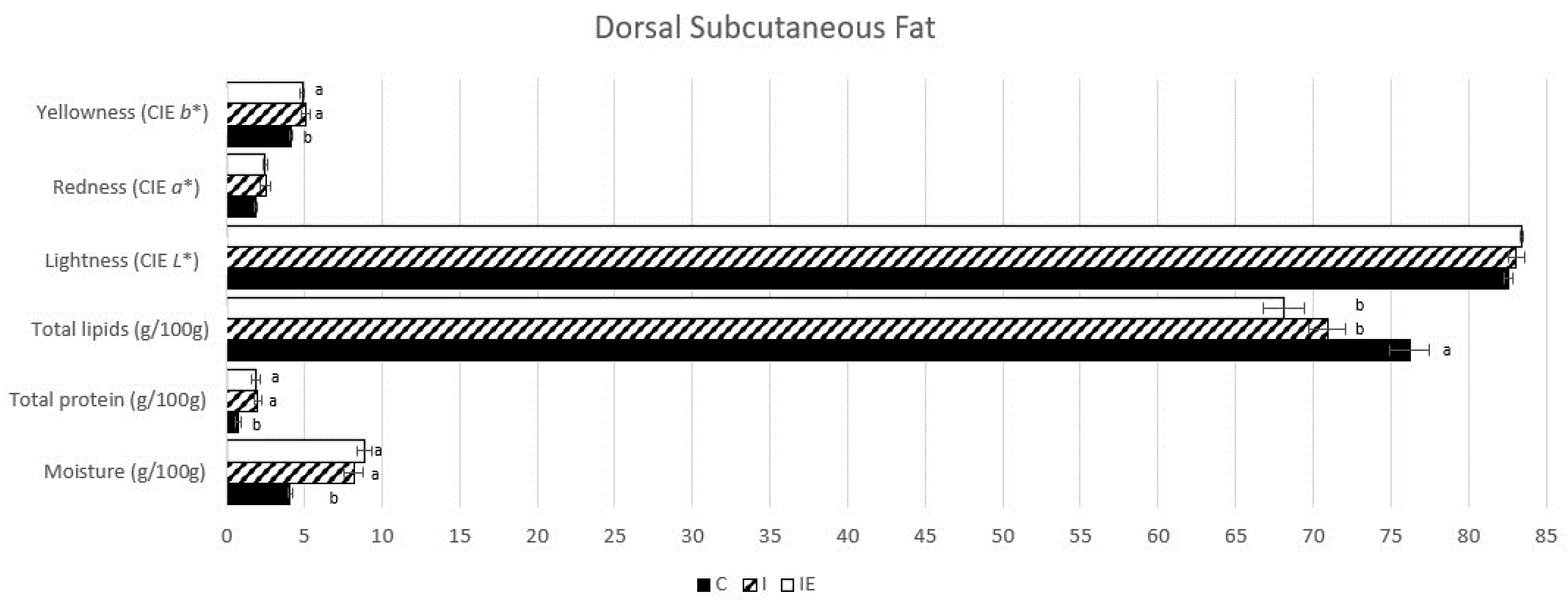
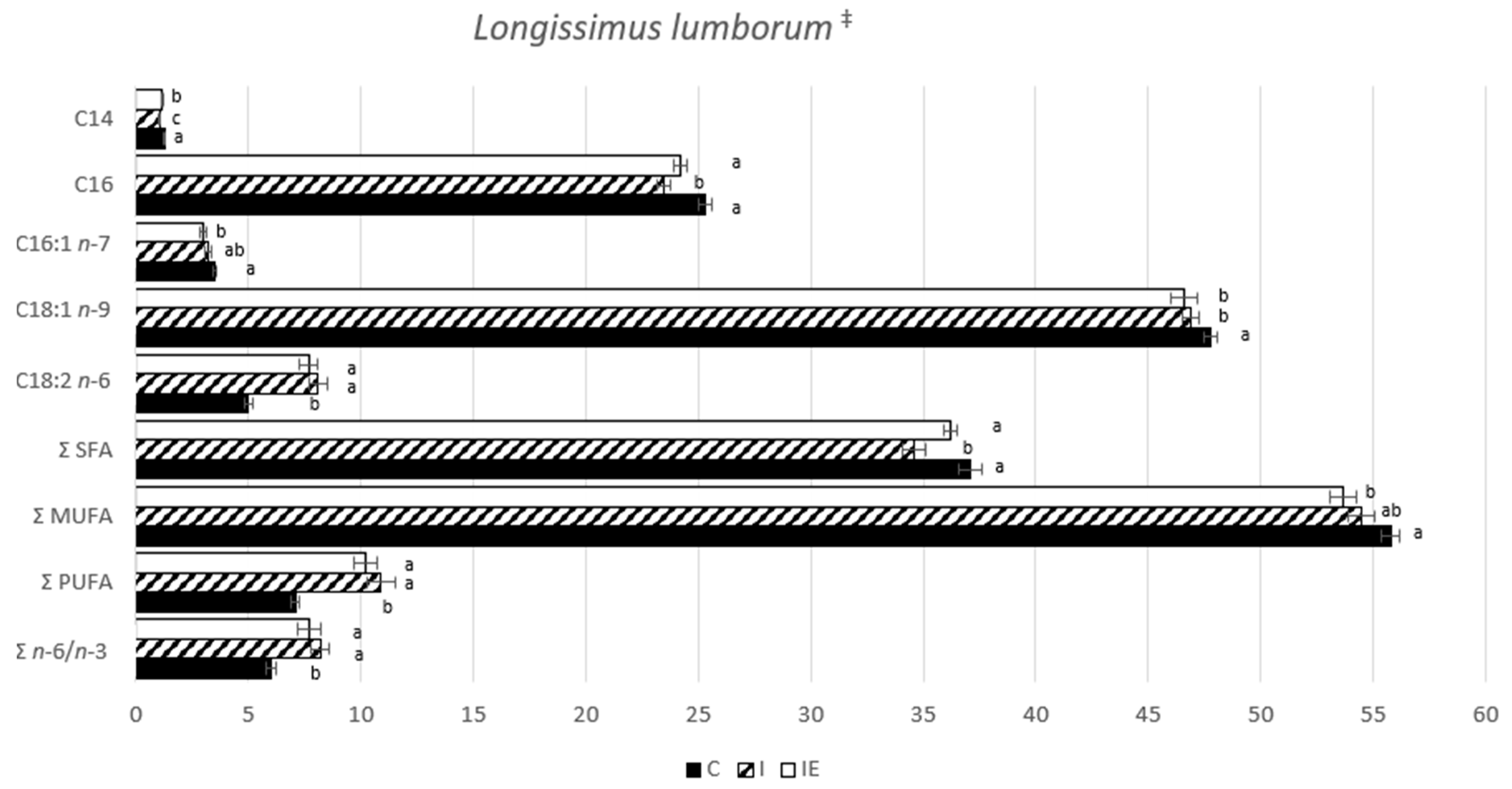
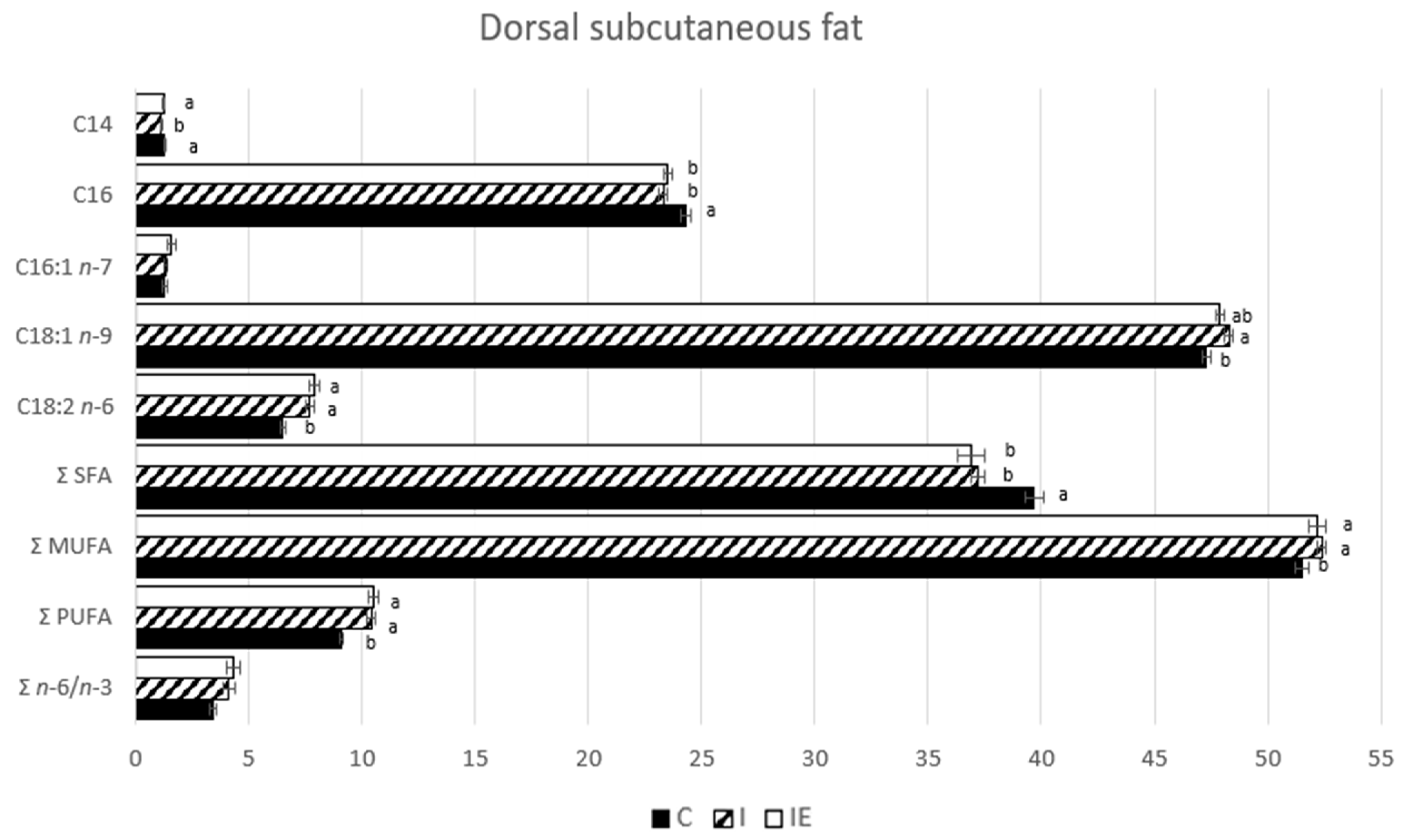
| Trait | C | I | IE | p-Value |
|---|---|---|---|---|
| Hot carcass weight (kg) | 125.0 | 120.3 | 124.6 | 0.163 |
| Carcass yield (%) | 78.8 a | 75.7 b | 77.8 a | <0.001 |
| Commercial yield (kg) 1 | 29.3 | 29.5 | 30.7 | 0.133 |
| Untrimmed shoulder (kg) | 10.6 c | 11.7 b | 12.6 a | 0.012 |
| Loin (kg) | 2.95 | 2.96 | 2.85 | 0.651 |
| Untrimmed ham (kg) | 15.5 a | 14.5 b | 15.0 ab | 0.029 |
| Tenderloin (g) | 315.4 | 342.3 | 312.9 | 0.281 |
| Fat cuts (kg) 2 | 17.9 a | 15.0 b | 15.3 b | 0.005 |
| Belly (kg) | 7.7 a | 5.9 b | 6.7 b | 0.014 |
| Backfat (kg) | 10.2 a | 9.0 b | 8.7 b | 0.019 |
| Lean to fat cuts ratio | 1.64 b | 1.98 a | 2.01 a | <0.001 |
| Average backfat thickness (mm) 3 | 69.4 a | 51.1 b | 52.4 b | <0.001 |
| Fatness: ZP fat depth (mm) 4 | 64.3 a | 47.9 b | 49.4 b | <0.001 |
| Average total feed consumption (kg) | 544.0 a | 486.1 b | 454.1 c | <0.001 |
| Average total feed rejection (kg) | 2.36 b | 18.05 a | 1.76 b | <0.001 |
| Gene | C vs. I | C vs. IE | I vs. IE | |||
|---|---|---|---|---|---|---|
| RNAseq | qPCR | RNAseq | qPCR | RNAseq | qPCR | |
| ACACA | 1.001 | 1.432 | 0.595 | 0.803 | −0.406 | −0.629 |
| ACLY | 0.837 | 1.203 | 0.816 | 0.964 | −0.021 | −0.239 |
| ADIPOQ | 1.076 | 0.834 | 0.209 | 0.198 | −0.868 | −0.636 |
| ELOVL6 | 1.987 | 2.120 | 1.033 | 0.872 | −0.953 | −1.248 |
| FASN | 2.021 | 1.033 | 1.092 | 0.637 | −0.929 | −0.395 |
| LEP | 2.486 | 2.487 | 1.817 | 1.455 | −0.669 | −1.032 |
| ME1 | 1.798 | 1.542 | 1.312 | 1.371 | −0.486 | −0.171 |
| SCD | 2.241 | 2.532 | 1.026 | 1.099 | −1.215 | −1.433 |
| FABP4 | 0.190 | −0.439 | 1.273 | −0.030 | 0.470 | 0.409 |
| IGF1 | −0.378 | 0.006 | 0.357 | 0.710 | −0.080 | −0.716 |
| Concordance Correlation Coefficient | ||||||
| C vs. I | C vs. IE | I vs. IE | ||||
| CCC | 0.876 | 0.714 | 0.767 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido, N.; Albuquerque, A.; Charneca, R.; Costa, F.; Marmelo, C.; Ramos, A.; Martin, L.; Martins, J.M. Transcriptomic Profiling of Subcutaneous Backfat in Castrated and Intact Alentejano Pigs Finished Outdoors with Commercial and Fiber-Rich Diets. Genes 2023, 14, 1722. https://doi.org/10.3390/genes14091722
Garrido N, Albuquerque A, Charneca R, Costa F, Marmelo C, Ramos A, Martin L, Martins JM. Transcriptomic Profiling of Subcutaneous Backfat in Castrated and Intact Alentejano Pigs Finished Outdoors with Commercial and Fiber-Rich Diets. Genes. 2023; 14(9):1722. https://doi.org/10.3390/genes14091722
Chicago/Turabian StyleGarrido, Nicolás, André Albuquerque, Rui Charneca, Filipa Costa, Carla Marmelo, Amélia Ramos, Luísa Martin, and José Manuel Martins. 2023. "Transcriptomic Profiling of Subcutaneous Backfat in Castrated and Intact Alentejano Pigs Finished Outdoors with Commercial and Fiber-Rich Diets" Genes 14, no. 9: 1722. https://doi.org/10.3390/genes14091722
APA StyleGarrido, N., Albuquerque, A., Charneca, R., Costa, F., Marmelo, C., Ramos, A., Martin, L., & Martins, J. M. (2023). Transcriptomic Profiling of Subcutaneous Backfat in Castrated and Intact Alentejano Pigs Finished Outdoors with Commercial and Fiber-Rich Diets. Genes, 14(9), 1722. https://doi.org/10.3390/genes14091722









