Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identifification of the WRKY Gene Family in A. offificinalis
2.2. Phylogenetic Analysis and Classification of AoWRKY Genes
2.3. Gene Structure, Cis-Acting Regulatory and Conserved Motif Analysis of AoWRKY Genes
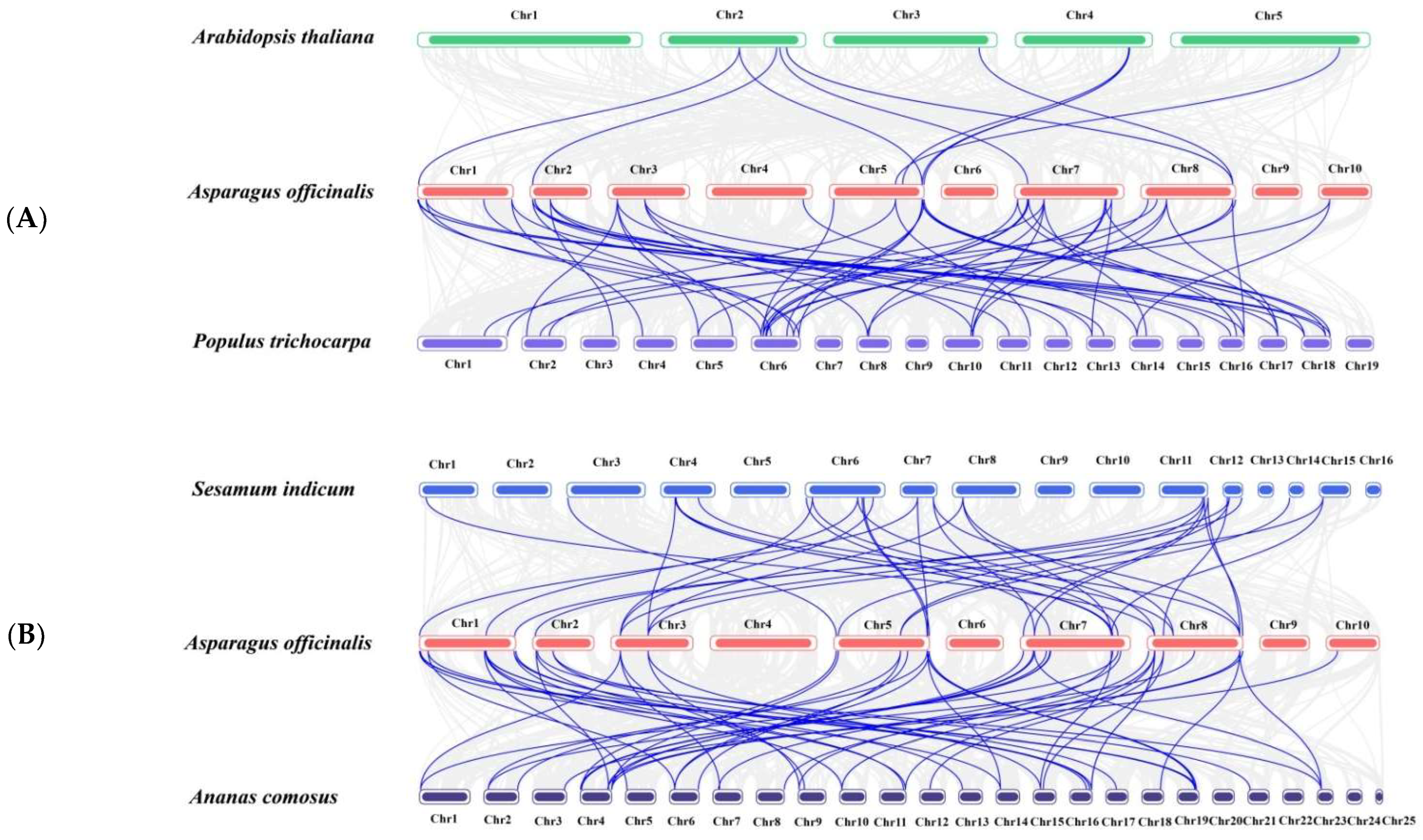
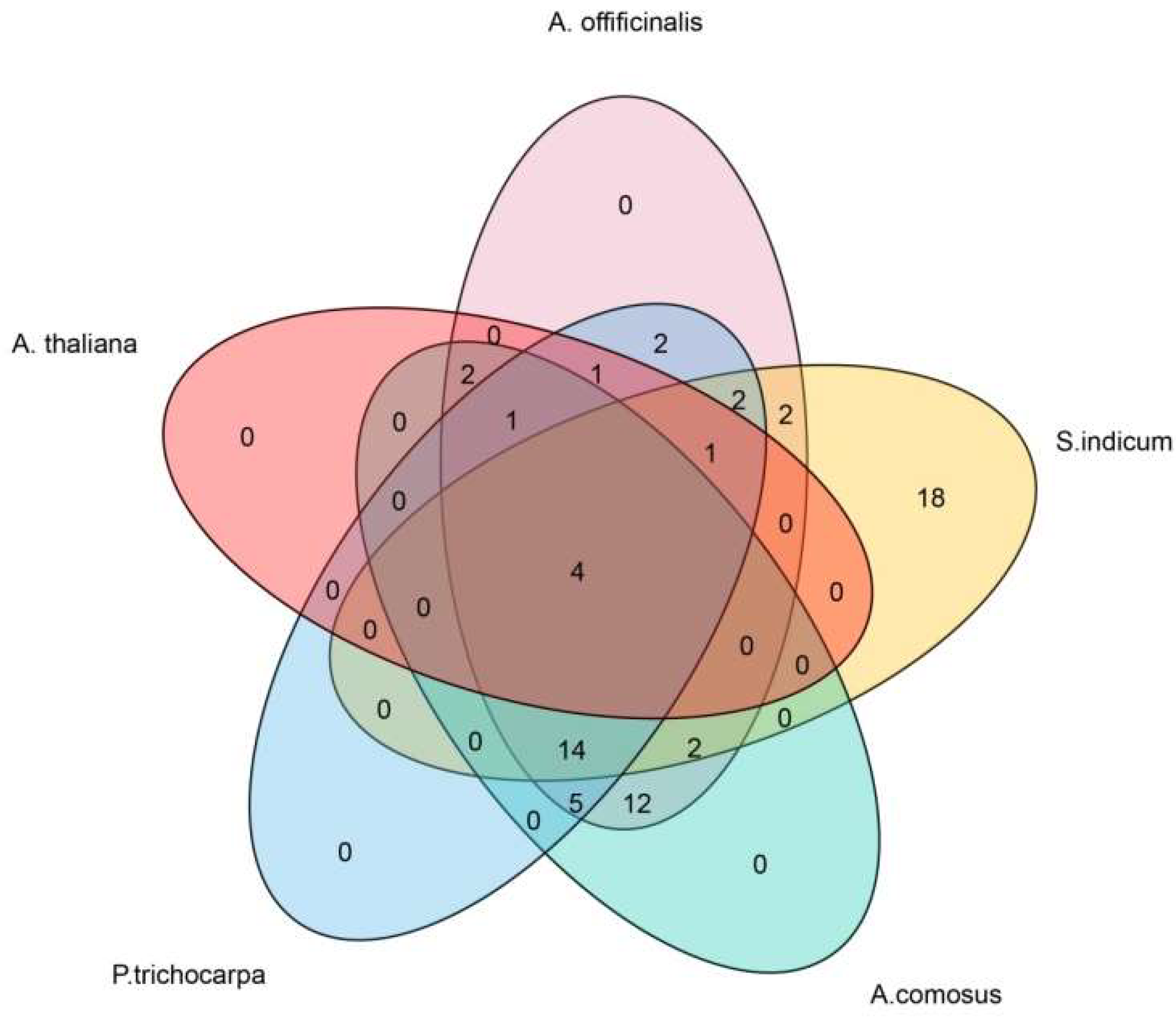
2.4. Chromosome Localization, Duplication and Synteny Analysis
2.5. Analysis of AoWRKY Gene Expression Profiles under Different Conditions
3. Results
3.1. Identification of AoWRKY Genes in A. offificinalis
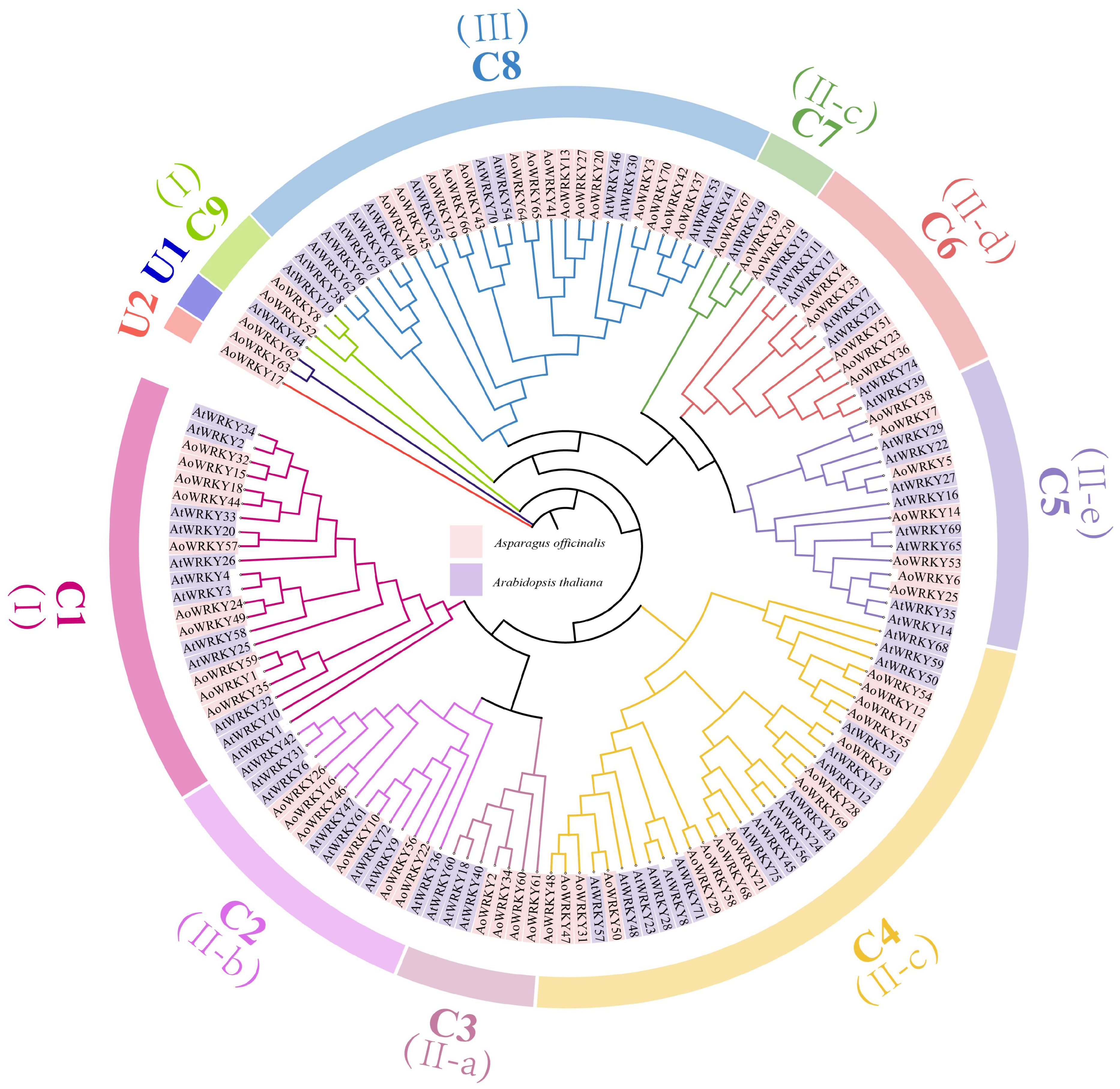
3.2. Systematic Classification Analysis of AoWRKY and AtWRKY
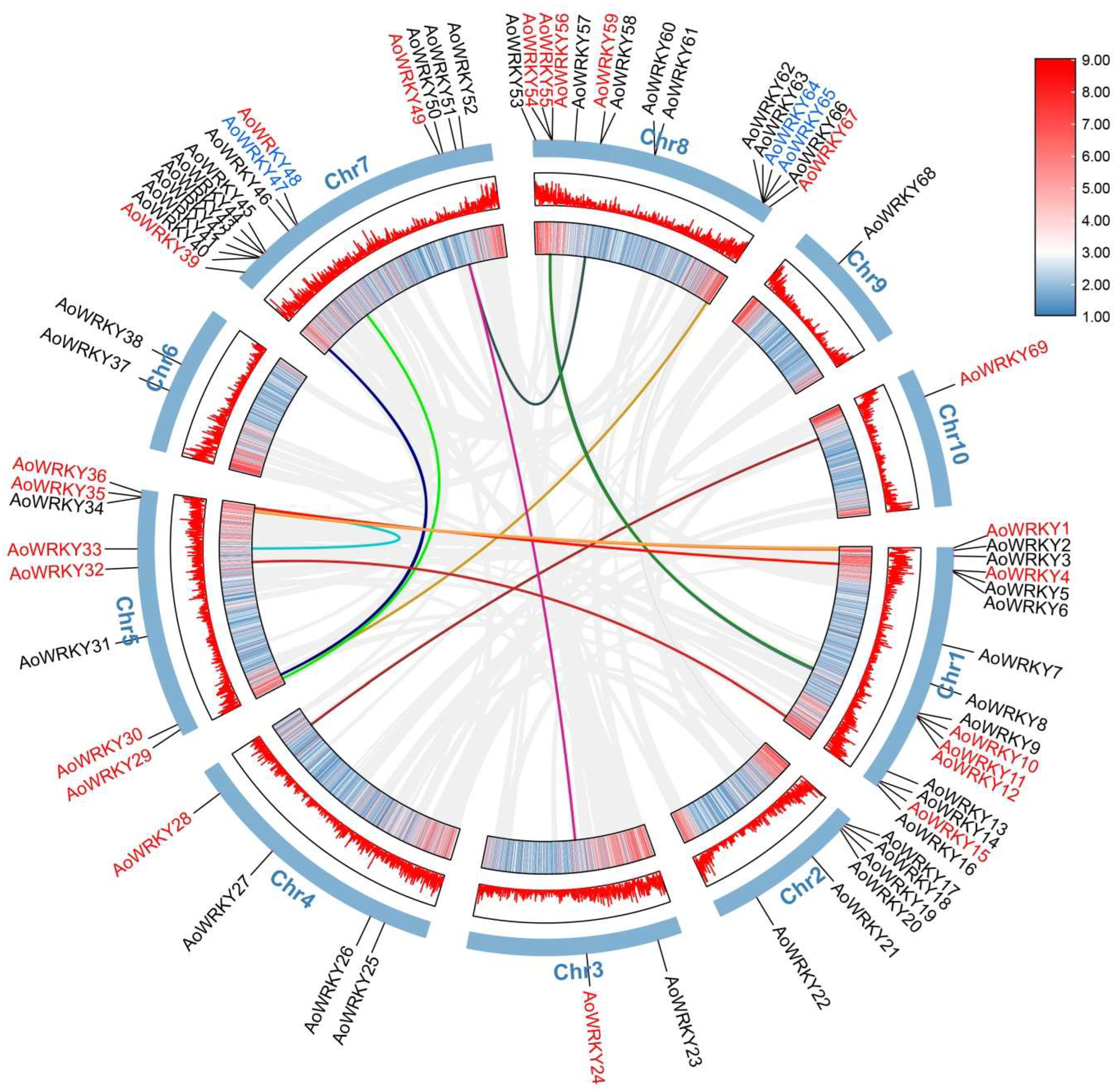
3.3. Chromosome Localization and Cis-Acting Regulatory Analysis of AoWRKY
3.4. Gene Structure and Conserved Motif Analysis of AoWRKY Genes
3.5. Tandem Gene Duplication and Segmental Gene Duplication of AoWRKY Genes
3.6. Analysis of AoWRKY Gene Expression under Different Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, L.; Li, W.; Chen, X. Transcription factor is not just a transcription factor. Trends Plant Sci. 2022, 27, 1087–1089. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, J.; Li, G.; Sun, Z.; Hu, S.; Chen, Y.; Guo, W.; Hou, H. Genome-wide identification and comparative analysis of the WRKY gene family in aquatic plants and their response to abiotic stresses in giant duckweed (Spirodela polyrhiza). Genomics 2021, 113, 1761–1777. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Watanabe, S.; Inoue, M.; Yamasaki, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J. Biol. Chem. 2012, 287, 7683–7691. [Google Scholar] [CrossRef] [PubMed]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. MGG 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Berri, S.; Abbruscato, P.; Faivre-Rampant, O.; Brasileiro, A.C.M.; Fumasoni, I.; Satoh, K.; Kikuchi, S.; Mizzi, L.; Morandini, P.; Pè, M.E.; et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009, 9, 120–142. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. MGG 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015, 34, 1295–1306. [Google Scholar]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef]
- Lei, R.; Li, X.; Ma, Z.; Lv, Y.; Hu, Y.; Yu, D. Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant Mol. Biol. 2017, 91, 962–976. [Google Scholar] [CrossRef]
- Cheng, Y.; JalalAhammed, G.; Yu, J.; Yao, Z.; Ruan, M.; Ye, Q.; Li, Z.; Wang, R.; Feng, K.; Zhou, G.; et al. Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Sci. Rep. 2016, 6, 39000. [Google Scholar] [CrossRef] [PubMed]
- Roscoe, T.J.; Vaissayre, V.; Paszkiewicz, G.; Clavijo, F.; Kelemen, Z.; Michaud, C.; Lepiniec, L.; Dubreucq, B.; Zhou, D.-X.; Devic, M. Regulation of FUSCA3 Expression During Seed Development in Arabidopsis. Plant Cell Physiol. 2019, 60, 476–487. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. Transparent Testa GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Li, Q.; Yin, M.; Li, Y.; Fan, C.; Yang, Q.; Wu, J.; Zhang, C.; Wang, H.; Zhou, Y. Expression of Brassica napus TTG2, a regulator of trichome development, increases plant sensitivity to salt stress by suppressing the expression of auxin biosynthesis genes. J. Exp. Bot. 2015, 66, 5821–5836. [Google Scholar] [CrossRef]
- Hu, X.; Kong, X.; Wang, C.; Ma, L.; Zhao, J.; Wei, J.; Zhang, X.; Loake, G.J.; Zhang, T.; Huang, J.; et al. Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 2014, 26, 4763–4781. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.-Y.; Shih, Y.-H.; Lin, P.-Y.; Feng, Y.-R.; Li, C.; Wu, K. WRKY63 transcriptional activation of COOLAIR and COLDAIR regulates vernalization-induced flowering. Plant Physiol. 2022, 190, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.P.; Dai, P.; Dai, S.Y. Effects of Salt Stress on the Growth of Asparagus officinalis L. Seedlings and on Na+, K+ and Ca2+ distribution in them. J. Southwest 2014, 36, 31–36. [Google Scholar]
- Nijveldt, R.J.; Van Nood, E.L.S.; Van Hoorn, D.E.; Boelens, P.G.; Van Norren, K.; Van Leeuwen, P.A. Flavonoids: A review of probable mechanisms of action and potential applications. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-alventosa, J.M.; Rodriguez, G.; Cermeno, P. Identification of Flavonoid Diglycosides in Several Genotypes of Asparagus from the Huetor-Tajar Population Variety. J. Agr. Food Chem. 2007, 55, 10028–10035. [Google Scholar] [CrossRef]
- Guardia, T.; Rotelli, A.E.; Juarez, A.O.; Pelzer, L.E. Anti-inflammatory properties of plant flavonoids. Effects of rutin, quercetin and hesperidin on adjuvant arthritis in rat. Farmaco 2001, 56, 683–687. [Google Scholar] [CrossRef]
- Narasimhanaidu, K.; Mainzen, P.P.S. Antihyperglycaemic and antioxidant effect of rutin, a polyphenolic flavonoid, in streptozotocin-induced diabetic wistar rats. Basic Clin. Pharmacol. 2006, 98, 97–103. [Google Scholar]
- Janbaz, K.H.; Saeed, S.A.; Gilani, A.H. Protective effect of rutin on paracetamol- and CCl4-induced hepatotoxicity in rodents. Fitoterapia 2002, 73, 557–563. [Google Scholar] [CrossRef]
- Schwarz, J.; Mathijs, E. Globalization and the sustainable exploitation of scarce groundwater in coastal Peru. J. Clean. Prod. 2017, 231–241. [Google Scholar] [CrossRef]
- Zafiriou, P.; Mamolos, A.P.; Menexes, G.C.; Siomos, A.S.; Tsatsarelis, C.A.; Kalburtji, K.L. Analysis of energy flow and greenhouse gas emissions in organic, integrated and conventional cultivation of white asparagus by PCA and HCA: Cases in Greece. J. Clean. Prod. 2012, 29, 20–27. [Google Scholar] [CrossRef]
- Global Production of Vegetables in 2021, by Type. 2023. Available online: https://www.statista.com/statistics/264065/globalproduction-of-vegetables-by-type (accessed on 20 August 2023).
- Guo, Q.; Wang, N.; Liu, H.; Li, Z.; Wang, C. The bioactive compounds and biological functions of Asparagus officinalis L.—A review. J. Funct. Foods 2019, 65, 103727. [Google Scholar] [CrossRef]
- Ashraf, M.; Munns, R. Evolution of Approaches to increase the salt tolerance of crops. Crit. Rev. Plant Sci. 2022, 41, 128–160. [Google Scholar] [CrossRef]
- Jia, Y.; Wu, J.; Cheng, M.; Xia, X. Global transfer of salinization on irrigated land: Complex network and endogenous structure. J. Environ. Manag. 2023, 336, 117592. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, S.; Ge, W.; Wang, T.; Fan, Z.; Wang, Y. Genome-wide identification and expression analysis of the WRKY gene family in cabbage (Brassica oleracea var. capitata L.). Biotechnol. Biotechnol. Equip. 2022, 36, 759–772. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. NAR 2004, 32, D142–D144. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Michael, L.; Christina, K.; Koichiro, T. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Castrense, S.; Luigi, M.P.; Piero, F.; Giuseppe, P.; Rita, C. BUSCA: An integrative web server to predict subcellular localization of proteins. NAR 2018, 46, W459–W466. [Google Scholar]
- Zhang, X.; Han, C.; Gao, H.; Cao, Y. Comparative transcriptome analysis of the garden asparagus (Asparagus officinalis L.) reveals the molecular mechanism for growth with arbuscular mycorrhizal fungi under salinity stress. Plant Physiol. Biochem 2019, 141, 20–29. [Google Scholar] [CrossRef]
- Andrews, S. FastQC:a quality control tool for high throughput sequence data. BMC Bioinform. 2014, 532, 1. [Google Scholar]
- Philip, E.; Måns, M.; Sverker, L.; Max, K. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Ryan, M.; Lance, C.; Davidson, L.C.; Osier, M.V. Comparison of Short-Read Sequence Aligners Indicates Strengths and Weaknesses for Biologists to Consider. Front. Plant Sci. 2021, 12, 657240. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–571. [Google Scholar] [CrossRef]
- Li, C.; Zhang, J.; Zhang, Q.; Dong, A.; Wu, Q.; Zhu, X.; Zhu, X. Genome-Wide identification and analysis of the NAC transcriptionfactor gene family in Garden Asparagus (Asparagus officinalis). Genes 2022, 13, 976. [Google Scholar] [CrossRef]
- Guo, X.; Ahmad, N.; Zhao, S.; Zhao, C.; Zhong, W.; Wang, X.; Li, G. Effect of salt Stress on growth and physiological properties of asparagus seedlings. Plants 2022, 11, 2836. [Google Scholar] [CrossRef]
- Chen, L.; Yang, Y.; Liu, C.; Zheng, Y.; Xu, M.; Wu, N.; Sheng, J.; Shen, L. Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem. Biophys. Res. Commun. 2015, 464, 962–968. [Google Scholar] [CrossRef]
- Pan, L.J.; Jiang, L. Identification and expression of the WRKY transcription factors of Carica papaya in response to abiotic and biotic stresses. Mol. Biol. Rep. 2014, 41, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genom. MGG 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Zhu, H.; Li, P.; Jiang, M.; Mao, W.; Ong, C.; Chu, Z. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 2014, 21, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Niu, E.; Du, H.; Zhao, L.; Feng, Y.; Guo, W. Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop J. 2014, 2, 87–101. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, D.; Shao, Y.; Liu, P.; Jiang, L.; Li, C. Genome-Wide Analysis of the WRKY Transcription Factors in Aegilops tauschii. Cytogenet. Genome Res. 2015, 144, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Chen, J.; Jiang, Y.; Lin, L.; Cao, Y.; Wang, M.; Zhang, Y.; Rong, J.; Ye, W. Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Genet. Genom. MGG 2015, 290, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.J.; Li, X.H.; Liu, Z.W.; Li, H.; Wang, Y.X.; Zhuang, J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol. Genet. Genom. MGG 2016, 291, 255–269. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, W.; Fang, L.; Sun, X.; Su, L.; Liang, Z.; Wang, N.; Londo, J.P.; Li, S.; Xin, H. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014, 14, 103–117. [Google Scholar] [CrossRef]
- Initiative, T.A.G.; Copenhaver, G.P. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 6814. [Google Scholar]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 2016, 7, 25–43. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. FPB 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Li, Q.; Tian, Q.; Zhang, Y.; Niu, M.; Yu, X.; Lian, C.; Liu, C.; Wang, H.-L.; Yin, W. Increased abscisic acid sensitivity and drought tolerance of Arabidopsis by overexpression of poplar abscisic acid receptors. PCTOC 2022, 148, 231–245. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Bijalwan, P.; Jeddi, K.; Saini, I.; Sharma, M.; Kaushik, P.; Hessini, K. Mitigation of saline conditions in watermelon with mycorrhiza and silicon application. Saudi. J. Biol. Sci. 2021, 28, 3678–3684. [Google Scholar] [CrossRef]



| Gene Symbol | pI | MW (Da) | Length (aa) | Instability Index | Aliphatic Index | Subcellular Localization | ORF |
|---|---|---|---|---|---|---|---|
| AoWRKY1 | 6.48 | 46,154.82 | 428 | 55.77 | 53.18 | Nucleus | 1287 |
| AoWRKY2 | 9.15 | 32,384.89 | 293 | 59.03 | 62.32 | Nucleus | 882 |
| AoWRKY3 | 8.75 | 28,313.14 | 254 | 44.03 | 64.53 | Extracellular | 765 |
| AoWRKY4 | undefined | undefined | 241 | 52.57 | 65.19 | Nucleus | 726 |
| AoWRKY5 | 5.24 | 29,765.87 | 264 | 46.71 | 57.99 | Nucleus | 795 |
| AoWRKY6 | 9.44 | 31,856.07 | 290 | 50.58 | 59.83 | Nucleus | 873 |
| AoWRKY7 | 5.41 | 31,262.65 | 285 | 49.21 | 56.18 | Nucleus | 858 |
| AoWRKY8 | 6.04 | 46,486.38 | 413 | 73.69 | 73.20 | Extracellular | 1242 |
| AoWRKY9 | 9.39 | 25,939.09 | 227 | 53.18 | 57.49 | Nucleus | 684 |
| AoWRKY10 | 9.07 | 46,949.56 | 419 | 62.02 | 72.12 | Nucleus | 1260 |
| AoWRKY11 | 8.97 | 13,655.35 | 122 | 33.04 | 51.89 | Nucleus | 369 |
| AoWRKY12 | 7.67 | 20,371.41 | 177 | 56.61 | 49.49 | Nucleus | 534 |
| AoWRKY13 | 5.61 | 27,669.79 | 244 | 51.91 | 63.52 | Extracellular | 735 |
| AoWRKY14 | 8.38 | 21,604.16 | 199 | 55.32 | 55.88 | Nucleus | 600 |
| AoWRKY15 | 6.21 | 63,743.09 | 585 | 49.54 | 56.02 | Nucleus | 1758 |
| AoWRKY16 | 9.04 | 45,956.35 | 420 | 49.61 | 62.50 | Nucleus | 1263 |
| AoWRKY17 | 8.53 | 44,932.82 | 399 | 57.93 | 58.87 | Nucleus | 1200 |
| AoWRKY18 | 8.73 | 55,651.20 | 505 | 61.60 | 49.49 | Nucleus | 1518 |
| AoWRKY19 | 8.81 | 21,958.55 | 196 | 60.57 | 51.79 | Extracellular | 591 |
| AoWRKY20 | 5.65 | 30,925.36 | 272 | 50.22 | 57.79 | Extracellular | 819 |
| AoWRKY21 | 9.54 | 20,022.42 | 174 | 40.10 | 58.16 | Nucleus | 525 |
| AoWRKY22 | 6.92 | 40,959.89 | 360 | 52.21 | 59.36 | Nucleus | 1083 |
| AoWRKY23 | 9.84 | 40,178.56 | 356 | 47.34 | 59.94 | Nucleus | 1071 |
| AoWRKY24 | 7.67 | 38,127.89 | 345 | 57.86 | 52.87 | Nucleus | 1038 |
| AoWRKY25 | 9.44 | 31,856.07 | 290 | 50.58 | 59.83 | Nucleus | 873 |
| AoWRKY26 | 8.59 | 56,176.11 | 517 | 51.30 | 56.52 | Nucleus | 1554 |
| AoWRKY27 | 5.12 | 28,238.23 | 253 | 52.89 | 61.66 | Extracellular | 762 |
| AoWRKY28 | 8.77 | 24,308.45 | 209 | 48.08 | 60.05 | Nucleus | 630 |
| AoWRKY29 | 4.66 | 19,682.85 | 181 | 49.78 | 51.22 | Nucleus | 546 |
| AoWRKY30 | 5.18 | 34,275.83 | 300 | 76.79 | 65.97 | Extracellular | 903 |
| AoWRKY31 | 8.34 | 26,410.43 | 230 | 58.47 | 61.43 | Nucleus | 693 |
| AoWRKY32 | 5.57 | 72,655.57 | 674 | 61.01 | 53.25 | Nucleus | 2025 |
| AoWRKY33 | 10.02 | 29,933.48 | 267 | 45.29 | 59.63 | Nucleus | 804 |
| AoWRKY34 | undefined | undefined | 259 | 51.14 | 65.52 | Nucleus | 780 |
| AoWRKY35 | 5.99 | 46,045.61 | 422 | 52.88 | 53.39 | Nucleus | 1269 |
| AoWRKY36 | 10.1 | 17,298.73 | 156 | 56.66 | 52.63 | Nucleus | 471 |
| AoWRKY37 | 5.54 | 32,599.17 | 298 | 59.24 | 49.43 | Extracellular | 897 |
| AoWRKY38 | 10.24 | 24,637.92 | 222 | 58.85 | 65.45 | Extracellular | 669 |
| AoWRKY39 | 5.06 | 31,499.98 | 278 | 58.6 | 73.31 | Nucleus | 837 |
| AoWRKY40 | 4.89 | 32,499.36 | 293 | 41.65 | 66.93 | Extracellular | 882 |
| AoWRKY41 | 8.56 | 25,121.05 | 219 | 43.79 | 55.25 | Extracellular | 660 |
| AoWRKY42 | 5.54 | 32,599.17 | 298 | 59.24 | 49.43 | Extracellular | 897 |
| AoWRKY43 | 5.22 | 35,686.71 | 327 | 60.99 | 68.01 | Extracellular | 984 |
| AoWRKY44 | 6.74 | 54,215.42 | 487 | 54.77 | 49.47 | Chloroplast | 1464 |
| AoWRKY45 | 5.36 | 34,350.08 | 313 | 54.95 | 59.87 | Nucleus | 942 |
| AoWRKY46 | 9.62 | 47,107.05 | 428 | 61.23 | 59.77 | Extracellular | 1287 |
| AoWRKY47 | 7.72 | 34,634.99 | 315 | 65.79 | 63.14 | Nucleus | 948 |
| AoWRKY48 | 9.28 | 31,193.24 | 281 | 64.28 | 61 | Nucleus | 846 |
| AoWRKY49 | 8.32 | 46,637.34 | 423 | 56.38 | 50.45 | Nucleus | 1272 |
| AoWRKY50 | 6.31 | 30,633.89 | 277 | 70.53 | 46.14 | Nucleus | 834 |
| AoWRKY51 | 9.9 | 36,191.16 | 318 | 43.8 | 60.13 | Nucleus | 957 |
| AoWRKY52 | 7.78 | 12,996.32 | 114 | 42.92 | 39.3 | Nucleus | 345 |
| AoWRKY53 | 5.49 | 32,548.79 | 299 | 43.69 | 52.51 | Nucleus | 900 |
| AoWRKY54 | 9.17 | 19,282.63 | 169 | 51.94 | 62.31 | Nucleus | 510 |
| AoWRKY55 | 7.05 | 22,006.13 | 200 | 53.76 | 56.55 | Nucleus | 603 |
| AoWRKY56 | 6.2 | 57,636.71 | 529 | 52.79 | 54.97 | Nucleus | 1590 |
| AoWRKY57 | 5.85 | 59,592.16 | 541 | 50.51 | 58.15 | Nucleus | 1626 |
| AoWRKY58 | 9.38 | 14,978.79 | 136 | 76.11 | 53.82 | Nucleus | 411 |
| AoWRKY59 | 6.82 | 14,042.8 | 122 | 70.52 | 58.36 | Nucleus | 369 |
| AoWRKY60 | 6.55 | 27,418.73 | 242 | 44.53 | 64.88 | Nucleus | 729 |
| AoWRKY61 | 6.11 | 25,787.13 | 232 | 60.7 | 75.26 | Extracellular | 699 |
| AoWRKY62 | 4.83 | 32,010.46 | 283 | 57.63 | 60.99 | Extracellular | 852 |
| AoWRKY63 | 6.35 | 27,294.78 | 241 | 52.99 | 68.84 | Extracellular | 726 |
| AoWRKY64 | 6.76 | 30,279.76 | 266 | 60.72 | 61.62 | Extracellular | 801 |
| AoWRKY65 | 5.88 | 31,046.72 | 277 | 57.25 | 78.19 | Extracellular | 834 |
| AoWRKY66 | 6.37 | 26,946.25 | 238 | 54.49 | 61.89 | Extracellular | 717 |
| AoWRKY67 | 5.29 | 40,200.29 | 356 | 43.22 | 78.99 | Extracellular | 1071 |
| AoWRKY68 | 9.17 | 18,202.33 | 154 | 47.22 | 54.35 | Nucleus | 465 |
| AoWRKY69 | 8.97 | 23,501.23 | 204 | 44.12 | 48.24 | Nucleus | 615 |
| AoWRKY70 | 6.26 | 33,056.91 | 297 | 53.52 | 60.77 | Extracellular | 894 |
| Tandem-Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Chromosome | Subfamily |
|---|---|---|---|---|---|
| AoWRKY47 & AoWRKY48 | 0.194258151 | 0.418275774 | 0.464426015 | Chr7 | C4 |
| AoWRKY64 & AoWRKY65 | 0.272881601 | 0.909314382 | 0.300095992 | Chr8 | C8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Hou, S.; Zhang, Q.; Meng, J.; Zhang, Y.; Du, J.; Wang, C.; Liang, D.; Guo, Y. Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis. Genes 2023, 14, 1704. https://doi.org/10.3390/genes14091704
Chen J, Hou S, Zhang Q, Meng J, Zhang Y, Du J, Wang C, Liang D, Guo Y. Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis. Genes. 2023; 14(9):1704. https://doi.org/10.3390/genes14091704
Chicago/Turabian StyleChen, Jing, Sijia Hou, Qianqian Zhang, Jianqiao Meng, Yingying Zhang, Junhong Du, Cong Wang, Dan Liang, and Yunqian Guo. 2023. "Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis" Genes 14, no. 9: 1704. https://doi.org/10.3390/genes14091704
APA StyleChen, J., Hou, S., Zhang, Q., Meng, J., Zhang, Y., Du, J., Wang, C., Liang, D., & Guo, Y. (2023). Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis. Genes, 14(9), 1704. https://doi.org/10.3390/genes14091704




