Abstract
Purpose: Triple-negative breast cancer (TNBC) is a molecularly complex and heterogeneous breast cancer subtype with distinct biological features and clinical behavior. Although TNBC is associated with an increased risk of metastasis and recurrence, the molecular mechanisms underlying TNBC metastasis remain unclear. We performed whole-exome sequencing (WES) analysis of primary TNBC and paired recurrent tumors to investigate the genetic profile of TNBC. Methods: Genomic DNA extracted from 35 formalin-fixed paraffin-embedded tissue samples from 26 TNBC patients was subjected to WES. Of these, 15 were primary tumors that did not have recurrence, and 11 were primary tumors that had recurrence (nine paired primary and recurrent tumors). Tumors were analyzed for single-nucleotide variants and insertions/deletions. Results: The tumor mutational burden (TMB) was 7.6 variants/megabase in primary tumors that recurred (n = 9); 8.2 variants/megabase in corresponding recurrent tumors (n = 9); and 7.3 variants/megabase in primary tumors that did not recur (n = 15). MUC3A was the most frequently mutated gene in all groups. Mutations in MAP3K1 and MUC16 were more common in our dataset. No alterations in PI3KCA were detected in our dataset. Conclusions: We found similar mutational profiles between primary and paired recurrent tumors, suggesting that genomic features may be retained during local recurrence.
1. Introduction
Triple-negative breast cancer (TNBC), a highly aggressive breast cancer subtype that lacks the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), accounts for 10–20% of all breast cancers [1,2]. Most TNBCs are high-grade, poorly differentiated carcinomas with high proliferation rates. TNBCs are challenging to treat due to their highly heterogeneous nature, rapid proliferative capabilities, chemoresistance, and high rates of metastasis to distant organs and tumor recurrence [3,4]. Although only 5% of TNBC patients are diagnosed with de novo metastatic disease, most of the patients typically experience relapse following treatment with curative intent [5,6]. TNBCs typically metastasize to the brain, liver, and lungs [7]. The prognosis of patients with metastatic TNBC is poor, and the majority of deaths among TNBC patients are due to progressive metastatic disease. Unlike other breast cancer subtypes, TNBC lacks clinically available tests to predict the risk of distant metastasis or disease relapse. Additionally, there are no metastasis-specific biomarkers to identify and treat patients at high risk of metastasis. The identification of molecules and pathways that drive TNBC metastasis may uncover distant metastasis-specific biomarkers. Hence, an in-depth understanding of the biology of metastatic dissemination may improve survival outcomes in patients with TNBC.
High-throughput next-generation sequencing techniques such as whole-exome sequencing (WES) have provided enormous insights into the genomic landscapes of several tumor types, leading to the identification of new druggable targets and the definition of new tumor subtypes and shedding light on the heterogeneity of many tumors [8,9]. In particular, through target enrichment, WES represents a cost-effective strategy for the identification of mutations in the protein-coding exons of the human genome. Essentially, knowledge of alterations in the coding regions of all genes in the genome may guide immediate treatment choices and strengthen therapeutic discovery efforts. Furthermore, the reduced costs and increased practical availability of tumor genomic profiling has generated ample opportunities to test “the precision medicine” hypothesis in clinical oncology. However, the approach suffers from a number of challenges that limit its application for widespread clinical WES implementation. The foremost is the rapid generation of high-quality WES data from archival FFPE tissue. Next is the ability to clinically interpret WES data for prospective use, which could maximize clinical and biological explorations. Overcoming these limitations would allow the rigorous assessment of the value of WES to guide clinical decision making [10].
The clinical management of metastatic cancer often relies on actionable molecular targets derived from primary tumors [11]. Potential genomic discordances in the molecular profiles of primary tumors and metastatic lesions are therapeutically relevant. Discordance of actionable molecular targets between primary tumors and metastatic recurrence can result in the non-optimal treatment of metastatic disease or cause unnecessary side effects [12,13,14]. Although the mutational landscape of primary breast tumors has been extensively analyzed, the mutational profiles of metastatic or recurrent breast tumors remain elusive [15,16]. Paired analyses of primary and metastatic tumors are pivotal for the optimal management of metastatic disease because (i) spatial and temporal differences might exist between primary tumors and matched metastatic lesions [17]; (ii) disseminating metastatic cells from primary tumors can activate specific transcriptomic programs to colonize and adapt to new tissue microenvironments; and (iii) additional molecular changes may be acquired by metastatic tumors due to therapeutic interventions, such as adjuvant chemotherapy [18]. Hence, the molecular profiling of primary tumors and matched metastatic lesions could facilitate the identification of actionable metastasis-specific targets. In this study, we performed whole-exome sequencing (WES) of nine matched primary and recurrent TNBC tumors to compare their mutational profiles and identify molecular alterations associated with metastatic progression in TNBC. We also performed WES of 15 primary tumors that remained recurrence-free to compare the mutational landscapes of primary TNBC tumors that recurred to those that did not recur.
2. Materials and Methods
2.1. Patients and Patient Samples
Formalin-fixed paraffin-embedded (FFPE) primary and matched recurrent tumor tissue (n = 35 samples) from 26 patients with TNBC was obtained from the University Hospital Zurich, Switzerland (Table 1). Fifteen of the 35 samples were tissue from primary tumors that did not recur, and 11 samples were primary tumors that recurred (9 paired primary and recurrent tumors). Of the 9 recurrent tumors, 7 (78%) were lymph node recurrences, and 2 (22%) were soft tissue and intramammary recurrences.

Table 1.
Descriptive statistics for the clinicopathological characteristics of patients with TNBC.
All 9 patients with paired primary and recurrent samples were diagnosed with recurrences (lymph node or other sites) after treatment for early or advanced breast cancer. The median age at diagnosis was 55 years. The median follow-up time was 3 years for patients with recurrence and 2.4 years for patients without recurrence. The clinicopathological characteristics of the patients are summarized in Table 1. The status of ER, PR, and HER2 was evaluated using immunohistochemistry (IHC). Because of the retrospective study design, we did not have access to blood samples or matched non-tumor tissue for these patients. Therefore, we performed variant filtering by frequency in a healthy population to exclude potential germline mutations. Previous comparisons of germline variants between unrelated individuals have shown that germline variants can be used as an effective filter, obviating the need for sequence-matched tumors and normal tissue [19,20]. The study protocol was approved by the Institutional Review Board and was in compliance with material transfer guidelines and data use agreements between Georgia State University and the University Hospital Zurich, Switzerland. The study was conducted in accordance with International Ethical Guidelines for Biomedical Research involving human subjects. Written informed consent was obtained from all the participants.
2.2. WES and Variant Calling
Hematoxylin and eosin (H&E) slides were prepared for all samples, and the tumor content was assessed by a pathologist. Genomic DNA (gDNA) was extracted using the NucleoSpin DNA FFPE Kit (Macherey-Nagel, Düren, Germany), and its concentration and purity were measured using NanoDrop and Qubit (ThermoScientific, Waltham, MA, USA). DNA electrophoresis using 2200 Tapestation (Agilent Technologies, Inc., Santa Clara, CA, USA) was conducted to confirm gDNA purity and concentration. One of the challenges of working with FFPE samples is that DNA extracted from this tissue is often of limited quantity. DNA yields from FFPE tissue samples might be insufficient for standard next-generation sequencing protocols. Despite this limitation, several studies have successfully sequenced samples starting with inputs as low as 10 ng [21,22]. The DNA available as input in our study ranged from 0.06 µg to 5.6 µg. Additionally, the majority of the samples sequenced (34/35) had a DNA integrity number ≥ 3 (Supplementary Table S1). The average coverage depth was 100×.
WES libraries were prepared using the SureSelect V6 exome kit from Agilent (Santa Clara, CA, USA), following the manufacturer’s instructions. The resulting libraries were sequenced on a NextSq 500 system (Illumina, San Diego, CA, USA) according to the standard operation protocol. The sequence quality of the resulting paired-end 150-nucleotide reads was assessed using FastQC [23].
Reads were trimmed using Trim-Galore to remove adapter sequences and low-quality sequences. Trimmed reads were mapped to the human reference genome GRCh38 using the BWA software [24]. After alignment, all samples were preprocessed according to the Genomics Analysis Toolkit (GATK) germline variant calling best practice workflow [25]. Germline variant calling was performed on all the samples using GATK HaplotypeCaller followed by VariantRecalibrator and ApplyVQSR.
GATK output files (VCF files) were screened for high-quality variants using SnpEff [26]. Mutants with a ‘PASS’ filter or no filter and a quality score of 30 or more were considered for downstream analysis. VCF files related to the same tumor type were merged using BCFtools, and merged VCF files were annotated using ANNOVAR [27]. ANNOVAR was used to filter out mutations with a minor allele frequency (MAF) of 0.01 in the “exac03”, “esp6500siv2”, and “gnomad_exom” germline databases. Thereafter, ANNOVAR was used to annotate variants and screen each variant using the LJB* databases. All single-nucleotide variants (SNVs) were scored using the SIFT, PolyPhen2 HDIC, LRT, MutationTaster, MutationAssesor, and FATHMM tools to predict the effect of mutation on each corresponding protein. SNVs that were found to have deleterious effects by at least 3 tools were selected for further analysis. This process ensured that common germline mutations and mutations with no adverse effect on the protein were excluded. Using the maftools R package, we analyzed the mutational landscapes of primary tumors and metastatic tumors separately and performed a comparative mutational analysis between tumor groups [28].
3. Results
3.1. Mutational Landscape of TNBC
The WES of 35 samples from 26 patients with TNBC revealed a total of 33,853 variants. Among the nine paired primary and recurrent tumor samples, the median number of variants per sample was 732 for primary tumors and 781 for the corresponding recurrent tumors. The median number of SNVs was 431 in primary tumors and 456 in matched recurrent tumors. We detected a total of 219–445 indels in primary tumors and 228–2089 indels in the corresponding recurrent tumors (Supplementary Table S2). We used variant data to calculate the tumor mutational burden (TMB) for each sample, which was defined as the number of mutations per megabase of the human genome sequenced. The median TMB in the nine primary tumors was 7.6 variants per megabase, and the median TMB in matched recurrent tumors was 8.2 variants per megabase. The TMB observed in our dataset is similar to the previously reported average TMB of 7.3 variants per megabase in the Thai TNBC dataset [29].
We found a significant overlap in the genes mutated in the nine primary and recurrent tumors; however, a few genes were mutated only in primary or recurrent tumors (Figure 1). Mutations in ATXN3, CAMKK2, L00134391, MAML2, NCOR2, RPL14, TRAK1, and ZAN were enriched only in primary tumors. On the other hand, mutations in COL17A1, LAMC3, and MMP27 were restricted to recurrent tumors (Figure 1). Among the genes mutated only in recurrent tumors, COL17A1 has been shown to prevent breast cancer cell invasion and migration [30]. Based on the number of mutations found in each gene, we identified the top 10 most mutated genes in both primary and matched recurrent tumors. MUC3A was the most frequently mutated gene in both groups (Supplementary Figure S1A).
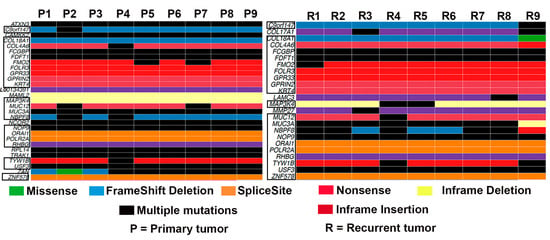
Figure 1.
Oncoplot showing the frequency of gene mutation in all the primary tumors and matched recurrent tumors (n = 9). Genes that are mutated in both groups are shown in the boxes.
We also compared the mutational profiles of primary tumors with recurrence (n = 11) to those of primary tumors without recurrence (n = 15). The median variant number per sample was 696 for primary tumors without recurrence and 764 for primary tumors that recurred. In total, 219–1171 indels were detected in primary tumors that recurred, and 208–389 indels were detected in primary tumors that did not recur. The median number of SNVs was 431 in primary tumors that recurred and 398 in primary tumors that remained recurrence-free (Supplementary Table S3). We found that the TMB in recurrent-free primary tumors was 7.3 variants per megabase.
Next, we compared the genes that were altered in all the samples in both groups. We found that the genes C9orf147, FMO2, FOLR3, GPR33, L00134391, MUC3A, MUC12, POLR2A, RPL14, and USF3 were mutated in both the primary tumors that recurred and in those that did not recur. MUC3A remained the most mutated gene in primary tumors without recurrence (Supplementary Figure S1B).
3.2. Frequency of Recurrent Gene Mutations in TNBC
cBioportal was used to identify breast-cancer-specific genes; PI3KCA, TP53, PTEN, GATA3, SYNE1, MAP3K1, MUC16, and CDH1 were the genes with the highest mutational frequencies in the TCGA and METABRIC cohorts. Additionally, we analyzed mutational alterations in TNBC-associated risk genes, including BRCA1 and BRCA2. We compared the frequency of recurrent gene mutations in matched primary and recurrent tumor pairs (Figure 2). PTEN is a negative regulator of the PI3K pathway, and mutations in PI3KCA and PTEN are often mutually exclusive [31,32,33]. Although PI3KCA was not mutated in any of the primary or recurrent tumors, three of nine primary and recurrent tumors and one primary–recurrent tumor pair (P4-R4) harbored a mutation in PTEN.
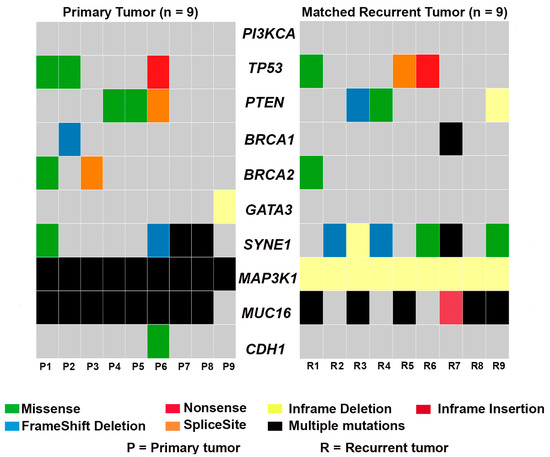
Figure 2.
Oncoplot showing the frequency of recurrent gene mutations in paired primary and recurrent tumors.
MAP3K1 mutations are more frequent in hormone-receptor-positive (HR+) breast cancer than in TNBC [34]. However, in our dataset, MAP3K1 was mutated in all primary TNBC tumors and their matched recurrent tumors. Although primary tumors had multiple mutations in MAP3K1, all recurrent tumors had in-frame MAP3K1 deletions. Interestingly, mutations in CDH1 and GATA3 were observed in only two primary TNBC tumors (P6 and P9); similar alterations in these genes were missing in their paired recurrent samples. In addition, no mutations in CDH1 and GATA3 were observed in any of the other recurrent tumors. Although 8/9 primary tumors and 6/9 recurrent tumors harbored an alteration in MUC16, five out of nine matched primary–recurrent pairs (P1-R1, P3-R3, P5-R5, P7-R7, and P8-R8) had MUC16 alterations. Similarly, SYNE1 was mutated in around 6/9 recurrent tumors, 4/9 primary tumors, and only two matched primary–recurrent pairs (P6-R6 and P7-R7). TP53 was mutated in three primary and three recurrent tumors; however, only two matched primary–recurrent pairs (P1-R1 and P6-R6) shared a TP53 mutation. Although BRCA2 was altered in the primary tumor P3, this mutation was not observed in the corresponding recurrent tumor. Only one primary–recurrent tumor pair (P1-R1) exhibited BRCA2 mutations. No pair-wise alterations were observed in BRCA1; one primary tumor (P2) and one recurrent tumor (R7) harbored a frameshift deletion and multiple BRCA1 mutations, respectively.
Next, we assessed the frequency of recurrent gene mutations in primary tumors with and without recurrence (Figure 3). We found that mutations in PI3KCA and PTEN were mutually exclusive in the four primary tumors that recurred. Primary tumors without a recurrence (except for P10′) had no mutations in PTEN or PI3KCA. Intriguingly, MAP3K1 was mutated in all primary tumors except for one (P15′). No alterations in CDH1 and GATA3 were observed in the primary tumors that remained recurrence-free, although both of these genes were mutated in at least one of the primary tumors that later had a recurrence. TP53 was altered in 9/15 primary tumors that did not recur and only 4/11 primary tumors that recurred. MUC16 mutations were detected in most primary tumors; 10/11 primary tumors that recurred and 9/15 primary tumors in those that remained recurrence-free harbored MUC16 mutations. Only 2 and 3 out of 11 primary tumors that recurred exhibited BRCA1 and BRCA2 mutations, respectively; BRCA1 and BRCA2 were mutated in 2/15 primary tumors that did not recur.
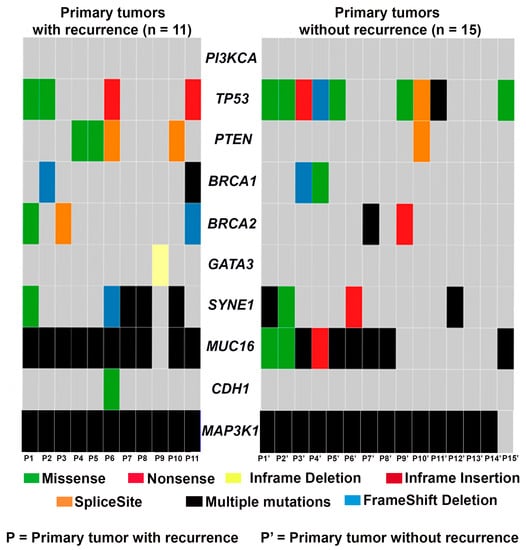
Figure 3.
Oncoplot showing the frequency of recurrent gene mutations in primary tumors that recurred (n = 11) and those that remained recurrence-free (n = 15).
4. Discussion
In this study, we investigated the mutational landscapes of primary TNBC tumors and their matched recurrent tumors and compared the mutational profiles of primary tumors that had recurrence to those that remained recurrence-free. Mutational analysis in TNBC patients remains fertile ground for the search for actionable targets owing to the inherent heterogeneous nature of the disease and the varied outcomes seen in the patients. Notably, genomic alterations were observed in all the samples investigated. Primary TNBC tumors and matched recurrent tumors showed similar mutational profiles, evidenced by the similar TMB and number of variants per sample. There was also an overlap in gene mutations between primary and matched recurrent tumors. Discordance in gene alterations between primary and matched recurrent TNBC tumors can be attributed to temporal and spatial differences between primary and recurrent lesions and neoadjuvant chemotherapy received by four of the nine (~44%) patients with matched primary and recurrent tumors. The neoadjuvant chemotherapy may have led to the acquisition of mutations in recurrent tumors that were initially absent in the corresponding primary tumors.
The high concordance in the mutational landscapes observed in our dataset is in line with previous studies showing similar genomic alterations in primary tumors and metastatic or recurrent tumor samples [15,35]. For instance, a study by Moreno et al. found that 85.5% of variants in primary tumors were also present in metastatic tissue [36]. Similarly, a study by Roy-Chowdhuri et al. also found 77% concordance between matched primary and metastatic breast tumors [37]. Varying degrees of genomic concordance between primary tumors and their corresponding metastatic tumors have been previously reported, suggesting that primary tumors harbor genetic alterations essential for successful metastatic dissemination [15,35]. These findings also suggest that the mutational signatures of primary tumors can serve as a proxy for cells that ultimately participate in metastatic dissemination and are responsible for tumor recurrence and disease relapse.
The identification of consistent mutational profiles between primary and corresponding recurrent tumors can aid in the identification and development of novel diagnostic and therapeutic targets in TNBC. The largely similar profiles of primary and recurrent tumors may prove useful in clinical research investigating global mutational changes during tumor progression or treatment responses, and for the clinical management of TNBC patients. For instance, the detection of mutations in immune response genes in TNBC allows for the prediction of the efficiency of response to immune checkpoint inhibitors or anti-PDL1 drugs [38]. Similarly, mutations in the genes that were altered in both primary and recurrent tumors (Figure 1) may allow for the tailoring of the targeted therapy and prediction of the response to treatment. The high recurrence risk is one of the main problems in the clinical management of TNBC. A comparison of the mutational profiles between the primary tumors that recurred compared to the ones that did not (Figure 3) could lead to the development of mutational signatures that can help to predict recurrence. From this point of view, the unique mutational signatures that can prognosticate patients into high- and low-recurrence groups could be valuable.
Non-recurrent primary tumors also harbored a similar number of variants as the primary tumors that metastasized. MUC3A was the most frequently mutated gene both in primary tumors with recurrence and in those that did not have recurrence. The differences in the frequency of gene mutations between the primary tumors that recurred and those that did not have recurrence could be attributed to differences in follow-up times between the comparison groups. MUC3A was also the top mutated gene both in primary TNBC tumors and in recurrent tumors. MUC3A encodes mucin 3A, a protein that belongs to the family of mucins, which are large glycoproteins expressed in various epithelial and malignant cells. The abnormal expression or glycosylation of mucins results in alterations in cell growth, differentiation, adhesion, and invasion and has been implicated in the development of neoplasms, including breast cancer [39,40]. Rakha et al. found that MUC3 was expressed in 91% of invasive breast cancer samples and that its expression was significantly associated with the lymph node stage, a poor Nottingham prognostic index (NPI), a high grade, and an increased risk of local recurrence [41]. MUC16, a gene encoding another mucin family member, was also mutated in most tumors in our dataset. Interestingly, previous findings suggest a role for MUC16 in promoting metastasis, therapy resistance, and disease progression in multiple malignancies, including breast cancer [42,43,44,45,46].
Previous next-generation sequencing studies have revealed the distinct mutational spectrum of TNBC. TP53 is the most commonly mutated gene (up to 80%) [47], whereas PI3KCA has the lowest mutational frequency in TNBC (approximately 9%) [48,49]. Consistently, we found no PI3KCA mutations in primary TNBC tumors (irrespective of their recurrent status) and matched recurrent tumors. The frequency of PI3KCA mutations is substantially higher in hormone-receptor-positive breast cancer compared with the TNBCs as a whole. Reports have suggested an association between TNBC molecular subtypes and alterations in the PI3K pathway. Specifically, it has been reported that PI3KCA mutations are more common in luminal TNBC and found in up to 40% of androgen-receptor-positive TNBCs [50,51,52]. Since we did not have information about the molecular subtypes of the TNBCs used for the study, it is possible that the luminal subtype was under-represented or absent in this cohort owing to the small sample size. Nonetheless, the dominance of PI3KCA mutations in specific TNBC subsets suggest the potential for targeted therapy for PI3KCA-mutant TNBCs. The frequency of TP53 mutations varied between primary and recurrent tumors in our dataset. A recent study showed that patients with TNBC harboring mutant TP53 and wild-type PI3KCA could benefit from immune checkpoint inhibitors (ICIs) [53].
MAP3K1 (or MEKK1) encodes a serine/threonine kinase that regulates the activity of various kinases regulating cell proliferation, migration, survival, and death [34,54]. Comprehensive genomic analyses revealed multiple alterations in MAP3K1 in different cancer types, including ER+ breast cancer. Except for TP53 and PI3KCA, most of the significantly mutated genes in non-TNBCs are rarely mutated in TNBC [34]. However, in our dataset, primarily composed of TNBC samples, we found a striking mutation pattern for MAP3K1, which was altered in all the samples except for one primary tumor (P15′). In addition to commonly mutated genes, infrequently mutated genes contribute to the mutational landscape of TNBC and may serve as actionable targets.
BRCA1 and BRCA2 are tumor suppressor genes involved in DNA repair and genome integrity. Somatic or germline BRCA1 or BRCA2 mutations are found in 10% to 40% of patients with TNBC. Often, high-grade breast cancer and TNBCs show somatic mutations or abnormal BRCA1/BRCA2 expression [55,56,57,58]. The varied prevalence of BRCA1/BRCA2 mutations in this study could be attributed to a number of factors, including the age at diagnosis, menopausal status, ethnicity, and therapy. Patients harboring mutations in genes involved in DNA damage repair, including BRCA1 and BRCA2, may benefit from platinum-based chemotherapy or poly(ADP-ribose) polymerase (PARP) inhibitors [59,60].
Our study had several limitations. First, the cohort size was small; however, this dataset represented a unique resource, as it included matched primary and recurrent tumor samples from the same patient. It is challenging to obtain matched sets of primary and recurrent tumors because biopsy samples of recurrent tumors are rare. Second, cases were chosen based on sample availability, inadvertently introducing selection bias. Third, although the incidence of TNBC is higher in young patients than in older individuals, more than half of our samples were obtained from older (≥50 years) patients. Nevertheless, the comparison of TMB data from our dataset with previously published literature could have been limited by differences in cohort, study design, and data analysis methods. Moreover, the median follow-up time of 3 years was not long enough to identify clinically relevant relationships between genomic alterations and clinical outcomes. Additionally, we used whole tumor samples instead of laser-capture microdissection samples [61]. Although great care was taken to reduce contamination from stromal and immune components, tumor microenvironment components that remained may have influenced our findings [62].
Next-generation sequencing technologies enable the characterization of the mutational and transcriptomic profiles of primary and metastatic tumors. Genome-wide comparisons of gene expression profiles in paired samples can aid in the detection of mutations that drive malignancy and perturbed genes and pathways that promote metastasis. Massively parallel sequencing techniques enable the comparison of the global gene expression profiles of matched primary and metastatic/recurrent TNBC tumors. Additionally, quantitative and spatial proteomics approaches can aid in strengthening the next-generation sequencing study findings. Ongoing studies in our laboratory aim to recruit large cohorts of patients with metastatic disease and integrate various omics platforms to map the complex genomic and proteomic profiles of primary and recurrent and/or metastatic tumors from the same patient. We envision that an integrative approach will provide a holistic view of the complexity associated with the metastatic dissemination of cancer cells. This in turn will ultimately lead to the refinement of our understanding of metastatic TNBC and the identification of metastasis-specific biomarkers and improve outcomes in TNBC patients by elucidating the multi-level alterations during metastatic disease progression.
5. Conclusions
Metastatic TNBCs are particularly aggressive, and the lack of actionable molecular alterations makes metastatic TNBC a challenging disease. Although the mutational landscapes of primary TNBC have been extensively characterized, analyses of paired metastatic lesions are scarce. In this study, we found that the primary tumors and metastatic lesions had similar mutational landscapes, suggesting that the primary tumor could serve as a surrogate for the detection of disseminated cancer cells and locoregional recurrences. However, we also found that mutations in COL17A1, LAMC3, and MMP27 were enriched only in recurrent tumors. This finding indicates that the TNBCs in our cohort followed a mixed model of tumor evolution and that recurrent tumors consisted of both clones that disseminated early and late from primary tumors during tumor evolution. MUC3A was the most frequently mutated gene in the dataset. This study led to the identification of previously unexplored, metastasis-specific, actionable targets (i.e., MUC3A, COL17A1, LAMC3, and MMP27) that can be further validated and developed for the management of metastatic TNBC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14091690/s1, Figure S1: MAF plot showing the top 10 most commonly mutated genes in primary (A) and matched recurrent tumors (n = 9) and (B) in primary tumors with (n = 11) and without recurrence (n = 15); Table S1: DNA amount and DNA integrity number for primary tumors that recurred (P; n = 11); recurrent tumors (R; n = 9) and primary tumors that did not recur (P’; n = 15); Table S2: Summary of SNVs and indels in paired primary (P; n = 9) and recurrent (R; n = 9) tumors; Table S3: Summary of SNVs and indels in primary tumors that recurred (P; n = 11) and primary tumors that remained recurrence-free (P’; n = 15).
Author Contributions
J.K. (Jaspreet Kaur) designed the study, interpreted the data, and wrote the manuscript. D.S.C. and K.G. processed and analyzed the data. Z.V. and B.S. contributed tumor tissue and associated clinical data. E.J. and U.K. helped in the scoring of tissue. E.J., J.K. (Jeanne Kowalski), and S.V. revised the manuscript and contributed to the discussion. R.A. conceived and designed the study and critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by a grant from the National Cancer Institute of Health (R01CA239120) to R.A. J. Kowalski received funds from CPRIT#RR160093, Department of Oncology, Dell Medical School, and the Glenn Breast Cancer Research Scholar Award.
Institutional Review Board Statement
The study protocol was approved by the Institutional Review Board and was in compliance with material transfer guidelines and data use agreements between Georgia State University and the University Hospital Zurich, Switzerland. The study was conducted in accordance with International Ethical Guidelines for Biomedical Research involving human subjects. This work obtained ethics approval from the Ethical Committee of the Canton Zurich, with ethics code KEK-2012-552 and approval date 8 May 2013. Written informed consent was obtained from all the participants involved in the study.
Informed Consent Statement
Written informed consent was obtained from all the participants involved in the study.
Data Availability Statement
The data presented in this study are available in this article (and Supplementary Material).
Acknowledgments
Editorial support for this manuscript was provided by Christos Evangelou.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.K.; Carey, L.A. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer 2009, 9 (Suppl. 2), S73–S81. [Google Scholar] [CrossRef]
- Rakha, E.A.; El-Sayed, M.E.; Green, A.R.; Lee, A.H.; Robertson, J.F.; Ellis, I.O. Prognostic markers in triple-negative breast cancer. Cancer 2007, 109, 25–32. [Google Scholar] [CrossRef]
- O’Reilly, D.; Sendi, M.A.; Kelly, C.M. Overview of recent advances in metastatic triple negative breast cancer. World J. Clin. Oncol. 2021, 12, 164–182. [Google Scholar] [CrossRef]
- Yao, Y.; Chu, Y.; Xu, B.; Hu, Q.; Song, Q. Risk factors for distant metastasis of patients with primary triple-negative breast cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Jin, J.; Gao, Y.; Zhang, J.; Wang, L.; Wang, B.; Cao, J.; Shao, Z.; Wang, Z. Incidence, pattern and prognosis of brain metastases in patients with metastatic triple negative breast cancer. BMC Cancer 2018, 18, 446. [Google Scholar] [CrossRef]
- Beltran, H.; Eng, K.; Mosquera, J.M.; Sigaras, A.; Romanel, A.; Rennert, H.; Kossai, M.; Pauli, C.; Faltas, B.; Fontugne, J.; et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol. 2015, 1, 466–474. [Google Scholar] [CrossRef]
- Rabbani, B.; Tekin, M.; Mahdieh, N. The promise of whole-exome sequencing in medical genetics. J. Hum. Genet. 2014, 59, 5–15. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Wagle, N.; Stojanov, P.; Perrin, D.L.; Cibulskis, K.; Marlow, S.; Jane-Valbuena, J.; Friedrich, D.C.; Kryukov, G.; Carter, S.L.; et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014, 20, 682–688. [Google Scholar] [CrossRef]
- Kroigard, A.B.; Larsen, M.J.; Thomassen, M.; Kruse, T.A. Molecular Concordance Between Primary Breast Cancer and Matched Metastases. Breast J. 2016, 22, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, T.; Niikura, N.; Ogiya, R.; Yasojima, H.; Watanabe, K.I.; Kanbayashi, C.; Tsuneizumi, M.; Matsui, A.; Fujisawa, T.; Iwasa, T.; et al. Distinct gene expression profiles between primary breast cancers and brain metastases from pair-matched samples. Sci. Rep. 2019, 9, 13343. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.E.; Kim, N.I.; Lee, J.S.; Park, M.H.; Kang, K. Differentially Expressed Genes in Matched Normal, Cancer, and Lymph Node Metastases Predict Clinical Outcomes in Patients With Breast Cancer. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Vareslija, D.; Priedigkeit, N.; Fagan, A.; Purcell, S.; Cosgrove, N.; O’Halloran, P.J.; Ward, E.; Cocchiglia, S.; Hartmaier, R.; Castro, C.A.; et al. Transcriptome Characterization of Matched Primary Breast and Brain Metastatic Tumors to Detect Novel Actionable Targets. J. Natl. Cancer Inst. 2019, 111, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.R.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Song, M.; Chouchane, L.; Ma, X. Functional Genomic Analysis of Breast Cancer Metastasis: Implications for Diagnosis and Therapy. Cancers 2021, 13, 3276. [Google Scholar] [CrossRef]
- Kim, K.P.; Kim, J.E.; Hong, Y.S.; Ahn, S.M.; Chun, S.M.; Hong, S.M.; Jang, S.J.; Yu, C.S.; Kim, J.C.; Kim, T.W. Paired Primary and Metastatic Tumor Analysis of Somatic Mutations in Synchronous and Metachronous Colorectal Cancer. Cancer Res. Treat. 2017, 49, 161–167. [Google Scholar] [CrossRef]
- Aftimos, P.; Oliveira, M.; Irrthum, A.; Fumagalli, D.; Sotiriou, C.; Gal-Yam, E.N.; Robson, M.E.; Ndozeng, J.; Di Leo, A.; Ciruelos, E.M.; et al. Genomic and Transcriptomic Analyses of Breast Cancer Primaries and Matched Metastases in AURORA, the Breast International Group (BIG) Molecular Screening Initiative. Cancer Discov. 2021, 11, 2796–2811. [Google Scholar] [CrossRef]
- Kumar, A.; White, T.A.; MacKenzie, A.P.; Clegg, N.; Lee, C.; Dumpit, R.F.; Coleman, I.; Ng, S.B.; Salipante, S.J.; Rieder, M.J.; et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc. Natl. Acad. Sci. USA 2011, 108, 17087–17092. [Google Scholar] [CrossRef]
- Snezhkina, A.V.; Lukyanova, E.N.; Kalinin, D.V.; Pokrovsky, A.V.; Dmitriev, A.A.; Koroban, N.V.; Pudova, E.A.; Fedorova, M.S.; Volchenko, N.N.; Stepanov, O.A.; et al. Exome analysis of carotid body tumor. BMC Med. Genom. 2018, 11, 17. [Google Scholar] [CrossRef]
- Bonin, S.; Hlubek, F.; Benhattar, J.; Denkert, C.; Dietel, M.; Fernandez, P.L.; Hofler, G.; Kothmaier, H.; Kruslin, B.; Mazzanti, C.M.; et al. Multicentre validation study of nucleic acids extraction from FFPE tissues. Virchows Arch. 2010, 457, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.M.; Belvedere, O.; Conway, C.; Daly, C.; Chalkley, R.; Bickerdike, M.; McKinley, C.; Egan, P.; Ross, L.; Hayward, B.; et al. Using next-generation sequencing for high resolution multiplex analysis of copy number variation from nanogram quantities of DNA from formalin-fixed paraffin-embedded specimens. Nucleic Acids Res. 2010, 38, e151. [Google Scholar] [CrossRef][Green Version]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Niyomnaitham, S.; Parinyanitikul, N.; Roothumnong, E.; Jinda, W.; Samarnthai, N.; Atikankul, T.; Suktitipat, B.; Thongnoppakhun, W.; Limwongse, C.; Pithukpakorn, M. Tumor mutational profile of triple negative breast cancer patients in Thailand revealed distinctive genetic alteration in chromatin remodeling gene. PeerJ 2019, 7, e6501. [Google Scholar] [CrossRef]
- Yodsurang, V.; Tanikawa, C.; Miyamoto, T.; Lo, P.H.Y.; Hirata, M.; Matsuda, K. Identification of a novel p53 target, COL17A1, that inhibits breast cancer cell migration and invasion. Oncotarget 2017, 8, 55790–55803. [Google Scholar] [CrossRef]
- Carracedo, A.; Pandolfi, P.P. The PTEN-PI3K pathway: Of feedbacks and cross-talks. Oncogene 2008, 27, 5527–5541. [Google Scholar] [CrossRef] [PubMed]
- Mukohara, T. PI3K mutations in breast cancer: Prognostic and therapeutic implications. Breast Cancer 2015, 7, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.T.; Angus, S.P.; Johnson, G.L. MAP3K1, Genomic Alterations in Cancer and Function in Promoting Cell Survival or Apoptosis. Genes Cancer 2013, 4, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Glas, A.M.; Wessels, L.F.; Witteveen, A.T.; Peterse, J.L.; van’t Veer, L.J. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc. Natl. Acad. Sci. USA 2003, 100, 15901–15905. [Google Scholar] [CrossRef] [PubMed]
- Moreno, F.; Gayarre, J.; Lopez-Tarruella, S.; Del Monte-Millan, M.; Picornell, A.C.; Alvarez, E.; Garcia-Saenz, J.A.; Jerez, Y.; Marquez-Rodas, I.; Echavarria, I.; et al. Concordance of Genomic Variants in Matched Primary Breast Cancer, Metastatic Tumor, and Circulating Tumor DNA: The MIRROR Study. JCO Precis Oncol. 2019, 3, 1–16. [Google Scholar] [CrossRef]
- Roy-Chowdhuri, S.; de Melo Gagliato, D.; Routbort, M.J.; Patel, K.P.; Singh, R.R.; Broaddus, R.; Lazar, A.J.; Sahin, A.; Alvarez, R.H.; Moulder, S.; et al. Multigene clinical mutational profiling of breast carcinoma using next-generation sequencing. Am. J. Clin. Pathol. 2015, 144, 713–721. [Google Scholar] [CrossRef]
- Lei, Q.; Wang, D.; Sun, K.; Wang, L.; Zhang, Y. Resistance Mechanisms of Anti-PD1/PDL1 Therapy in Solid Tumors. Front. Cell Dev. Biol. 2020, 8, 672. [Google Scholar] [CrossRef]
- Hauselmann, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Chakraborty, S.; Ponnusamy, M.P.; Lakshmanan, I.; Jain, M.; Batra, S.K. Mucins in the pathogenesis of breast cancer: Implications in diagnosis, prognosis and therapy. Biochim. Biophys. Acta 2011, 1815, 224–240. [Google Scholar] [CrossRef]
- Rakha, E.A.; Boyce, R.W.; Abd El-Rehim, D.; Kurien, T.; Green, A.R.; Paish, E.C.; Robertson, J.F.; Ellis, I.O. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod. Pathol. 2005, 18, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Aithal, A.; Rauth, S.; Kshirsagar, P.; Shah, A.; Lakshmanan, I.; Junker, W.M.; Jain, M.; Ponnusamy, M.P.; Batra, S.K. MUC16 as a novel target for cancer therapy. Expert Opin. Ther. Targets 2018, 22, 675–686. [Google Scholar] [CrossRef]
- Haridas, D.; Chakraborty, S.; Ponnusamy, M.P.; Lakshmanan, I.; Rachagani, S.; Cruz, E.; Kumar, S.; Das, S.; Lele, S.M.; Anderson, J.M.; et al. Pathobiological implications of MUC16 expression in pancreatic cancer. PLoS ONE 2011, 6, e26839. [Google Scholar] [CrossRef]
- Klug, T.L.; Bast, R.C., Jr.; Niloff, J.M.; Knapp, R.C.; Zurawski, V.R., Jr. Monoclonal antibody immunoradiometric assay for an antigenic determinant (CA 125) associated with human epithelial ovarian carcinomas. Cancer Res. 1984, 44, 1048–1053. [Google Scholar]
- Lakshmanan, I.; Ponnusamy, M.P.; Das, S.; Chakraborty, S.; Haridas, D.; Mukhopadhyay, P.; Lele, S.M.; Batra, S.K. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene 2012, 31, 805–817. [Google Scholar] [CrossRef]
- Lakshmanan, I.; Salfity, S.; Seshacharyulu, P.; Rachagani, S.; Thomas, A.; Das, S.; Majhi, P.D.; Nimmakayala, R.K.; Vengoji, R.; Lele, S.M.; et al. MUC16 Regulates TSPYL5 for Lung Cancer Cell Growth and Chemoresistance by Suppressing p53. Clin. Cancer Res. 2017, 23, 3906–3917. [Google Scholar] [CrossRef]
- Wang, X.; Guda, C. Integrative exploration of genomic profiles for triple negative breast cancer identifies potential drug targets. Medicine 2016, 95, e4321. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, L.; Qi, Y.; Stemke-Hale, K.; Wang, B.; Young, E.J.; Booser, D.J.; Holmes, F.A.; O’Shaughnessy, J.; Hellerstedt, B.; Pippen, J.; et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res. Treat. 2012, 134, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Jin, J.; Ji, W.; Guan, X. Therapeutic landscape in mutational triple negative breast cancer. Mol. Cancer 2018, 17, 99. [Google Scholar] [CrossRef] [PubMed]
- Bareche, Y.; Venet, D.; Ignatiadis, M.; Aftimos, P.; Piccart, M.; Rothe, F.; Sotiriou, C. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 2018, 29, 895–902. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Schafer, J.M.; Pendleton, C.S.; Tang, L.; Johnson, K.C.; Chen, X.; Balko, J.M.; Gomez, H.; Arteaga, C.L.; et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 2014, 16, 406. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Turner, N.C. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann. Oncol. 2019, 30, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Ding, X.; Xu, S.; Zhu, B.; Jia, Q. Gene expression profiling identified TP53(Mut)PIK3CA(Wild) as a potential biomarker for patients with triple-negative breast cancer treated with immune checkpoint inhibitors. Oncol. Lett. 2020, 19, 2817–2824. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Atchley, D.P.; Albarracin, C.T.; Lopez, A.; Valero, V.; Amos, C.I.; Gonzalez-Angulo, A.M.; Hortobagyi, G.N.; Arun, B.K. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J. Clin. Oncol. 2008, 26, 4282–4288. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Stefansson, I.M.; Chappuis, P.O.; Begin, L.R.; Goffin, J.R.; Wong, N.; Trudel, M.; Akslen, L.A. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 2003, 95, 1482–1485. [Google Scholar] [CrossRef]
- Hartman, A.R.; Kaldate, R.R.; Sailer, L.M.; Painter, L.; Grier, C.E.; Endsley, R.R.; Griffin, M.; Hamilton, S.A.; Frye, C.A.; Silberman, M.A.; et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 2012, 118, 2787–2795. [Google Scholar] [CrossRef]
- von Wahlde, M.K.; Timms, K.M.; Chagpar, A.; Wali, V.B.; Jiang, T.; Bossuyt, V.; Saglam, O.; Reid, J.; Gutin, A.; Neff, C.; et al. Intratumor Heterogeneity of Homologous Recombination Deficiency in Primary Breast Cancer. Clin. Cancer Res. 2017, 23, 1193–1199. [Google Scholar] [CrossRef]
- Papadimitriou, M.; Mountzios, G.; Papadimitriou, C.A. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat. Rev. 2018, 67, 34–44. [Google Scholar] [CrossRef]
- Vollebergh, M.A.; Lips, E.H.; Nederlof, P.M.; Wessels, L.F.; Wesseling, J.; Vd Vijver, M.J.; de Vries, E.G.; van Tinteren, H.; Jonkers, J.; Hauptmann, M.; et al. Genomic patterns resembling BRCA1- and BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res. 2014, 16, R47. [Google Scholar] [CrossRef]
- Grigoriadis, A.; Mackay, A.; Reis-Filho, J.S.; Steele, D.; Iseli, C.; Stevenson, B.J.; Jongeneel, C.V.; Valgeirsson, H.; Fenwick, K.; Iravani, M.; et al. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on MPSS and array expression data. Breast Cancer Res. 2006, 8, R56. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tarin, D. Gene expression profiling of human lymph node metastases and matched primary breast carcinomas: Clinical implications. Mol. Oncol. 2007, 1, 172–180. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).