Abstract
SWEETs (sugars will eventually be exported transporters) play a vital role in longer-distance sugar transportation, and thus control carbon flow and energy metabolism in plants. SWEET genes have been identified in various plant species, but their functions in fruit development remain uncharacterized. Here, we isolated 15 putative PsSWEETs from the Prunus salicina genome. For further analysis, comprehensive bioinformatics methods were applied to determine the gene structure, chromosome distribution, phylogeny, cis-acting regulatory elements, and expression profiles of PsSWEETs. qRT-PCR analysis suggested that these SWEETs might have diverse functions in the development of plum fruit. The relative expression levels of PsSWEET1 and PsSWEET9 were obviously higher in ripened fruit than the ones in other developmental stages, suggesting their possible roles in the transport and accumulation of sugars in plum fruit. Positive correlations were found between the expression level of PsSWEET3/10/13 and the content of sucrose, and the expression level of PsSWEET2 and the content of fructose, respectively, during the development of ‘Furongli’ fruit, suggesting their possible roles in the accumulation of sucrose and fructose. The current study investigated the initial genomic characterization and expression patterns of the SWEET gene family in plum, which could provide a foundation for the further understanding of the functional analysis of the SWEET gene family.
1. Introduction
Sugars, the principal products of photosynthesis in higher plants, can be used as a significant source of carbon skeletons for the synthesis of various cellular compounds, osmolytes, signaling molecules, and transport molecules, as well as a temporary energy carrier [1,2,3]. In addition to providing energy to plants, sugars also serve as crucial components of metabolic intermediates that play important roles not just in plant growth, but also in abiotic and biotic stress responses [4,5,6]. The cellular exchange of sugars from cells or subcellular compartments requires the involvement of multiple transporters, such as monosaccharide transporters (MSTs), sucrose transporters (SUTs), and SWEETs [7,8,9,10]. Among them, SWEET transporter proteins are primarily pH-insensitive and facilitate bidirectional transmembrane sugar transportation along the concentration gradient [11]. SWEET transporter proteins are capable of selectively transporting monosaccharides or disaccharides across the plasma membrane or within cells, and are widely distributed in prokaryotes, plants, humans, and other animals [12,13].
It is now widely acknowledged that many SWEET genes have crucial functions in regulating plant growth, encompassing phloem loading, nectar secretion, pollen development, seed filling, and biotic and abiotic stress responses [8,14]. In Arabidopsis thaliana, the initially identified SWEET gene was AtSWEET1, which acted as a single glucose transporter and contributed to flower development by providing nutrients to the gametophyte [2,15]. Compared with the wild type, the overexpression strains of AtSWEET4 have a higher plant height, while the mutant strain exhibits a shorter height, and lower contents of fructose and glucose in the leaves. It also has been reported that the overexpression of AtSWEET4 enhanced tolerance to freezing and drought stresses [16]. Previously, AtSWEET11 and AtSWEET12, which are localized at the plasma membrane (PM) of phloem parenchyma cells, were responsible for sucrose phloem loading in pollen development. Mutations in AtSWEET11 and AtSWEET12 significantly retarded embryonic development [1]. The identification of ZmSWEET4c in maize and its ortholog OsSWEET4 in rice as crucial genes for grain filling has been reported. Mutants of these two genes resulted in an impaired seed filling [17]. These findings highlighted the significance of SWEET genes in seed development. VvSWEET4, a glucose transporter situated on the plasma membrane in Vitis vinifera, participated in the host interaction with Botrytis cinerea [18]. Several reports indicated that the SWEET gene family also plays a vital role in fruit development. For example, SlSWEET12C and SlSWEET14 exhibit a relatively high expression in ripening the tomato fruit [19]. In banana, it was discovered that over 80% of the MaSWEETs genes were expressed throughout five developmental and ripening stages. Additionally, more than 27% of these genes exhibited high expression levels (FPKM > 10) at each stage [20]. The transcript levels of ten MdSWEETs were detected in apple leaves and fruit. The expression of MdSWEET1.1/2 was twice as high in the fruit compared to the leaves and shoot tips, implying that its expression level might not be influenced by the concentrations of glucose, fructose, and sucrose in apple cells [21]. Taken together, the SWEET gene family has been found to participate in multiple vital biological processes in plants, including the growth, development of seeds, fruit, and pollen, environmental adaptation, and host–pathogen interactions.
Plum, a perennial deciduous fruit tree belonging to the Prunus genus in the Rosaceae family, is cultivated globally and considered an important traditional fruit tree [22]. Plum has a long history of cultivation and possesses strong adaptability [23]. It has been cultivated worldwide and has abundant genetic resources. To date, there are 30–40 species of Prunus plants in the world [24]. Over 800 varieties were found in China [25]. Among them, the ‘Furongli’ plum (P. salicina Lindl.), which has a red peel and red pulp, has been grown for over 700 years in Fujian Province, China [26]. It is considered to be an economically important fruit tree in China due to its beautiful appearance, beneficial antioxidant potential, and bioactive compounds [27]. Despite detailed investigations of the sugar, organic acid, and secondary compound contents in ‘Furongli’ plum through previous studies [28], there is still a lack of full understanding regarding sugar transport during the development of the ‘Furongli’ plum. In the current study, we conducted a comprehensive analysis of gene structures, conserved domains, expression patterns, and phylogenetic relationships of the SWEET gene family based on a whole-genome-wide level in plum. This comprehensive study serves to facilitate our understanding of the evolutionary patterns and the expression of PsSWEETs in plum fruit development.
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of PsSWEET Genes
The genome sequences of P. salicina was downloaded from the Rosaceae Genome Database (https://www.rosaceae.org/, downloaded on 22 December 2022) [29]. AtSWEET genome sequences were downloaded from TAIR (https://www.arabidopsis.org/, downloaded on 22 December 2022). Firstly, the Arabidopsis AtSWEET genes was employed as a query sequence to perform a Blast search against the plum genome using the BLASTP tool with an E-value threshold of 1.0. Secondly, the HMMER3 software suite (version 3.3.2) was utilized to obtain the seed alignment file for the MtN3/saliva domain (PF03083) from the Pfam database (https://pfam.xfam.org/, downloaded on 25 December 2022) [30]. Subsequently, the HMMER3 software suite (version 3.3.2) was used to conduct HMM searches against the local protein databases of plum with an E-value < 1.0 [31]. Finally, the sequences of PsSWEET proteins were employed for the computation of basic biochemical characteristics using the ExPASy tool (http://www.expasy.org/tools, downloaded on 28 December 2022) [32], including molecular weight, theoretical isoelectric point, and hydrophilic mean. MEGA-X (v10.2.6) was used to investigate the phylogenetic and molecular evolutionary genetics of PsSWEET proteins with the default settings [33]. The neighbor-joining (NJ) approach was employed to construct the evolutionary tree with a bootstrap of 1000. The tree was further edited using iTOL (https://itol.embl.de, accessed on 29 December 2022) [34].
2.2. Analysis of Gene Structure, Protein Motif, and Promoter Region
The gene structures of PsSWEETs were virtualized using the Gene Structure Display Service (available online: http://gsds.cbi.pku.edu.cn, accessed on 20 February 2023) [35]. In addition, the conserved motifs were displayed by using the online tool MEME Suite 5.1.1 (http://meme-suite.org/, visited on 20 February 2023) [36]. The cis-regulatory elements from gene promoters (1500 bp before ATG) were analyzed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/, visited on 25 February 2023).
2.3. Gene Duplication, Synteny, and Chromosomal Locations Analysis of PsSWEET Genes
Synteny analysis among the SWEET family members was carried out using MCScanX and Tbtools [37,38]. Additionally, the synteny analysis of plum with Arabidopsis, apple, pear, and prune was also constructed using the same tools. The positions of the SWEET genes on chromosomes were drawn using MapInspect software (version 1.0).
2.4. Total RNA Isolation and Quantitative Real-Time PCR Analysis (qRT-PCR)
Plant material was collected as previously described [39]. Briefly, the fruits of the ‘Furongli’ plum were harvested from 6-year-old field-grown trees in Fuda Village, Fujian Province, China. Fruit samples were picked 23, 43, 70, 98, 112, 127, and 157 days after flowering (DAF), respectively. All the trees received the standard horticultural practices and insect prevention. The fruit samples were stored at −80 °C until use. After that, they were peeled and sliced into appropriate pieces and frozen in liquid nitrogen immediately. For qRT-PCR analysis, the total RNA was isolated using the RNA prep Pure Plant Total RNA Extraction Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. First-strand cDNA was synthesized for qPCR using the PrimeScript RT reagent kit with gDNA Eraser (Takara, Dalian, China). The qRT-PCR reactions were conducted in 20 µL volumes, utilizing 1 µL of cDNA and 2 × SYBR Premix Ex TaqTM II (Tli RNaseH Plus, TaKaRa), based on the Eppendorf RealPlex4 system (Hamburg, Germany). The experiment procedures of qRT-PCR were 40 cycles of 95 °C for 15 s and then 68 °C for 30 s. The actin gene was utilized as the control standard, and the 2−ΔΔCT method was employed to analyze the relative expression levels of the genes [40]. Three biological and three technical replicates were performed. The primer sequences are provided in Table S1.
2.5. Statistical Analysis
The statistical analysis of the qRT-PCR data was performed via one-way ANOVA using SPSS19 software. The means were separated using Duncan’s new multiple-range test p < 0.05. Different letters were used to indicate significant differences (p < 0.05). The ggcor package was used to perform the correlation test in the R circumstance [41].
3. Results
3.1. Identification and Phylogenetic Analysis of PsSWEET Family Members
After confirming the SWEET domain using the CDD and PFAM (PF03083) databases, a total of 15 PsSWEETs were discovered and named based on their corresponding orthologous genes in A. thaliana (Table 1). The molecular characteristics of these proteins are described in this study. These proteins varied in sizes ranging from 134 (in the case of PsSWEET6) to 291 (in the case of PsSWEET9), and also varied in isoelectric points ranging from 6.27 to 10.44 (Table 1). The PsSWEET proteins displayed a high degree of variability in their structure, implying that these proteins may have different roles in various biological processes or under different growth conditions.

Table 1.
Detailed information on 15 PsSWEET genes in plum.
To investigate the evolutionary events of the SWEET gene family in P. salicina, we constructed a phylogenetic tree based on A. thaliana, Oryza sativa, and Prunus avium. All PsSWEETs proteins were successfully divided into four clades (I–IV) according to the phylogenetic tree (Figure 1). Among the four clades (I–IV), clade III was the largest clade with 21 members, containing four PsSWEET, six AtSWEET, five OsSWEETs, and six PaSWEETs members. Clade I had the same PsSWEET members (Figure 1). Clade IV was the smallest clade with seven members, including two PsSWEETs, two AtSWEETs, one OsSWEETs, and two PaSWEETs members. The distribution of PsSWEETs, AtSWEETs, OsSWEETs, and PaSWEETs proteins encompassed all clades. Notably, PsSWEETs exhibited a closer relationship with PaSWEETs compared to AtSWEETs and OsSWEETs (Figure 1).
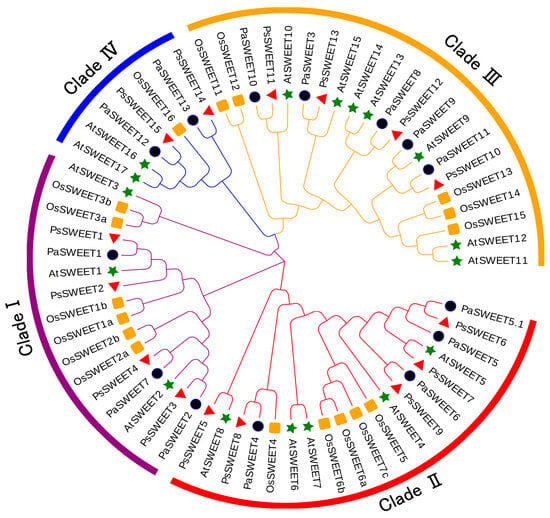
Figure 1.
Phylogenetic analysis of SWEET proteins from Prunus salicina, Prunus avium, Oryza sativa, and Arabidopsis thaliana. The SWEETs from P. salicina, P. avium, O. sativa, and A. thaliana were marked with a red triangle, purple circle, yellow square, and green star, respectively. The phylogenetic tree of the SWEET family protein sequences was conducted using MEGA 6.0 via the neighbor-joining method with 1000 bootstrap replicates. The gene family was divided into four SWEET subfamilies, clade I, II, III, and IV.
3.2. Analysis of Gene Structure, Motif, and Cis-Element in Promoter Regions of PsSWEETs
Generally speaking, the 15 PsSWEET proteins were clustered into three categories (Figure 2A). The 20 conserved motifs (motif 1–motif 20) of the PsSWEET gene family members were predicted using the MEME program. Motifs 1, 2, 5, and 19 were found in all PsSWEETs (Figure 2B). PsSWEET5 had the largest number of motifs, containing 16 motifs, while PsSWEET2 had the fewest, only containing 2 motifs. These results suggest that the majority of PsSWEETs grouped together share similar conserved motifs, which provides additional support for the phylogenetic results. The exon/intron structures of the PsSWEET gene family members were analyzed. Among the SWEET genes in plum, five introns were found in most PsSWEETs (Figure 2C). PsSWEET5 has the most prominent intron numbers, containing 12 introns. Compared to other PsSWEET genes, PsSWWET2 possesses the lowest number of introns, containing three introns (Figure 2C). The intron numbers of PsSWEETs ranged from three to twelve, exhibiting considerable variation. Remarkably, the PsSWEETs contained 5 exons in Clades III and IV, 3–8 exons in Clade I, and 4–12 exons in Clade II (Figure 2C). The exon–intron structure of the most of PsSWEETs within the same groups displayed a similar exon–intron structure, suggesting a high level of conservation.
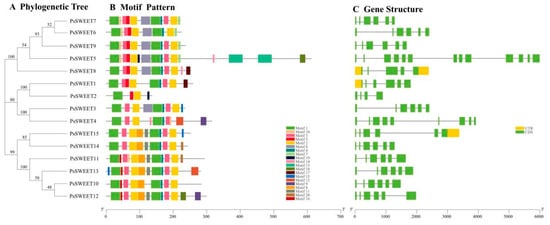
Figure 2.
Phylogeny, motifs, and exon–intron structures of PsSWEET proteins or genes. (A) Neighbor-joining phylogenetic tree of PsSWEETs. (B) Distribution of the conserved motifs in PsSWEET proteins. Twenty conserved motifs are marked with different colored boxes. (C) Exon-intron structure of PsSWEETs genes. Green boxes indicate exons, yellow boxes indicate UTR, and black lines indicate introns, respectively.
To enhance the comprehension of PsSWEETs’ transcriptional regulation and potential functions, 5′ regulatory elements in the 1500 bp sequences located upstream of their translation start sites were analyzed using the PlantCARE server. Twelve different types of putative cis-elements responsive to biotic stresses, fruit development, and hormones, including 3-AF1, AAGAA-motif, ABRE, AE-box, ARE, AT-rich, AT1-motif, AT~TATA-box, AuxRR-core, were found in the 1500 bp promoters of PsSWEET genes (Figure 3).
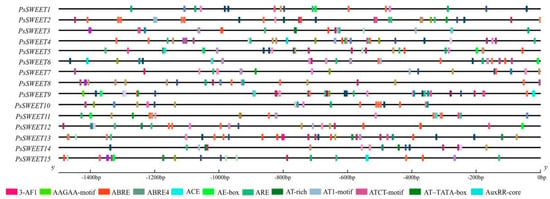
Figure 3.
Predicted cis elements in the promoter regions of PsSWEET genes, which were presented as colored rectangles. The numbers on the top indicate the relative positions to the start codon.
3.3. Duplication and Synteny Analysis of PsSWEET Family Members
Previous studies have shown that gene duplication, including tandem and segmental duplication, plays a significant role in the evolution of plant genomes. Three segmental duplication events (PsSWEET7/8, PsSWEET1/12, and PsSWEET5/9) and two tandem duplication events (PsSWEET9/10 and PsSWEET13/14) were identified in the present study (Figure 4). These results indicated that some PsSWEET genes were probably generated via gene segment or tandem duplication.
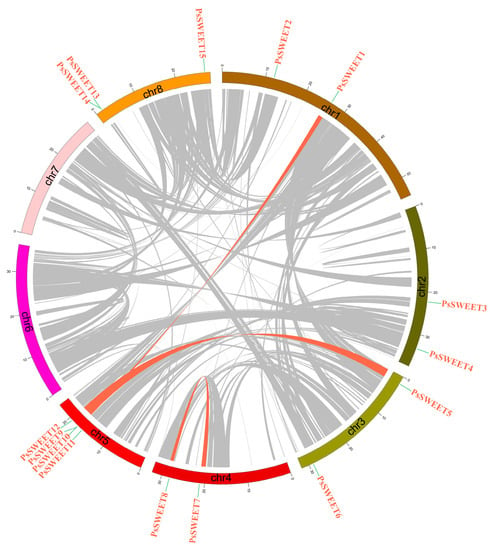
Figure 4.
Gene duplication analysis of PsSWEET genes. Pink lines connect the syntenic regions between plum PsSWEET genes.
To explore the potential evolutionary clues of the PsSWEET gene family, a series of comparative syntenic maps of P. avium associated with the four Rosaceae plant species, including Malus domestica, Prunus persica, Pyrus communis and Prunus mume, were constructed in this study. The syntenic analysis of PsSWEET family members exhibited that 15 PsSWEET genes were in synteny with M. domestica (8), P. persica (10), P. communis (8), and P. mume (10) (Figure 5).
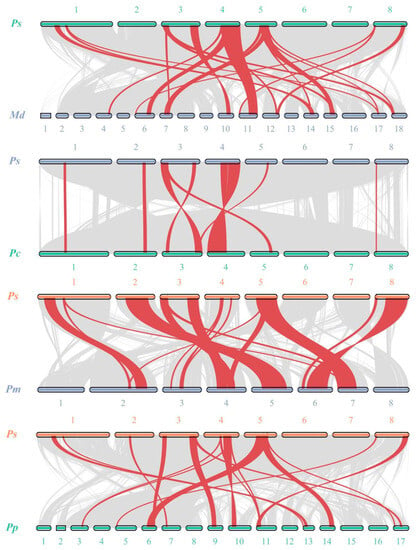
Figure 5.
Synteny analysis of PsSWEET genes between P. avium and Malus domestica, Prunus persica, Pyrus communis, Prunus mume.
3.4. Chromosomal Localization Analysis of PsSWEETs
As shown in Figure 6, the distribution of all PsSWEET genes was uneven across the six LGs in the plum genome. Among these genes, there was no detection of PsSWEET genes on chromosomes 6 and 7. The PsSWEET1/2 genes are observed on Chromosome 1, PsSWEET3/4 are on Chromosome 2, PsSWEET5/6 are on Chromosome 3, PsSWEET7/8 are on Chromosome 4, and PsSWEET9/10/11/12 are on Chromosome 8. Chromosome 5 has the largest number of PsSWEET9 genes (PsSWEET9/10/11/12).
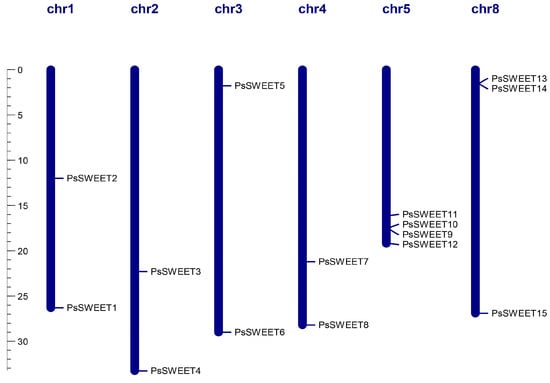
Figure 6.
Schematic representations of the chromosomal location of the PsSWEET genes. The chromosome number is marked on each chromosome.
3.5. Verification of Key PsSWEET Family Members during Fruit Development via qRT-PCR
The regulation of sugar transport from source to sink cells is a key factor in plant fruit development, and SWEETs are integral to this process by controlling sugar efflux transport [2]. In order to better understand the functions of SWEETs in ‘Furongli’ plums during fruit development and ripening, the expression levels of ten detectable PsSWEETs were determined using qRT-PCR (Figure 7). PsSWEET1/9 showed relatively lower levels of expression at the early stages of fruit development, while it sharply increased at the later stages. Noteworthy is that the expression patterns of PsSWEET3, PsSWEET4, PsSWEET7, and PsSWEET13 were similar during fruit development, and were the highest at 70 DAF. PsSWEET5 had the highest expression level at 70 DAF and 112 DAF, and PsSWEET14 had the highest expression level at 70 DAF. The expression levels of two other genes (PsSWEET2 and PsSWEET12) peaked at 94 DAF, then decreased gradually. It is worth noting that the most of PsSWEET genes displayed higher expression at the early stage of fruit development compared to the late stage (Figure 7).
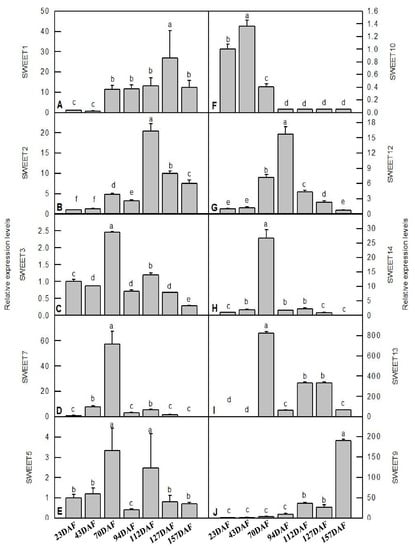
Figure 7.
qRT-PCR analysis of key PsSWEETs during fruit development and ripening of ‘Furongli’ plum. (A) SWEET1; (B) SWEET2; (C) SWEET3; (D) SWEET7; (E) SWEET5; (F) SWEET10; (G) SWEET12; (H) SWEET14; (I) SWEET13; (J) SWEET9. Data are the means ± SD of three biological replications (n = 3). Different letters above the bars indicate a significant difference at p < 0.05.
3.6. Expression Patterns Analysis of PsSWEET Family Members during Fruit Development
A correlation analysis between the expression levels of the five key PsSWEETs and soluble sugar contents (sucrose, fructose, and glucose) and related enzyme activity (PGI, HK, NI and AI) during plum development was conducted [39]. The results showed that among the five key PsSWEET genes, except for PsSWEET1, the expression levels of each of the other genes were significantly positively correlated with soluble sugar contents, and related to enzyme activity in the sugar metabolic pathway (Figure 8). A significant positive correlation was observed between PsSWEET9 and the content of glucose during the development of the ‘Furongli’ fruit. Furthermore, a significant positive correlation was found between PsSWEET3/10/13 and the content of sucrose, respectively. A significant positive correlation was also found between PsSWEET2 and PGI or the content of fructose, respectively (Mantel’s 0.01 < p < 0.05). Such a result might implicate the possible role of PsSWEET genes in the regulation of soluble sugar contents during the development of plum fruit.
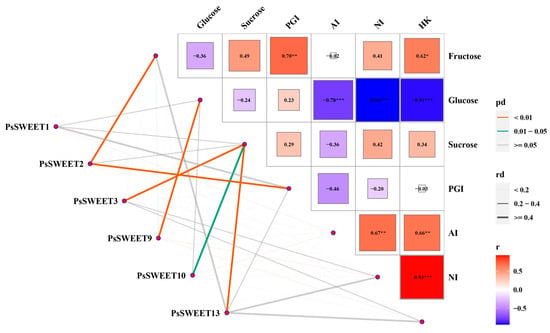
Figure 8.
The correlations among the expression levels of the six key PsSWEET genes and soluble sugar contents (sucrose, fructose, and glucose), and related enzyme activity (PGI, HK, and AI). Data of soluble sugar contents and enzyme activities used for the correlation analysis were extracted from our previous reported literature [39]. The lines correspond to the expression correlations between the six key PsSWEET genes and the soluble sugar contents, and related enzyme activity. The colors of the lines represent the Mantel’s pd, and the thicknesses of the lines represent the percentage of the Mantel’s rd. The intensity of the color from blue to red and size of the box indicate the degree of the expression correlation among the soluble sugar metabolic. “*”, “**”, and “***” mean a significant difference at p < 0.05, 0.01, and 0.001, respectively.
4. Discussion
The plum, as one of the most essential fruits, has been cultivated on a global scale. It is rich in bioactive substances such as vitamin C, carotenoids, polyphenols, and anthocyanins [28,42,43,44]. Except for bioactive substances, sugars play a crucial role in determining the quality of fruits and also contribute to their caloric value [45]. Sucrose and hexoses (mostly glucose and fructose), which are commonly found in fruits, are the primary sugars. Sucrose is transported by the apoplastic pathway and unloaded from the phloem into the loquat and cucumber fruits. A previous study indicated that hexose absorbed from the fruit apoplasm into the fruit storage parenchyma cells was facilitated by the CsHT3, which acts as a hexose transporter [46]. Still, the process remains unclear. Recently, the SWEET family, a novel type of sugar transporter, may play important roles in regulating the unloading process of sugars. SWEET proteins have been found in many plant species, such as mulberry [47], wheat [48], Arabidopsis [49], tea [50], Sorghum [51], and physic nut [52]. However, only a few species have been used to elucidate the genomics and function of SWEET genes until now. From this point, we investigated the main molecular characteristics and expression profiles of PsSWEET gene family members in the current study.
Here, 15 PsSWEET genes were identified based on the genome sequence of P. salicina. Their phylogenetic evolutionary relationship showed that these genes were classified into four subgroups (Figure 1). This classification is consistent with previous reports in banana [20], strawberry [53], tomato [54], soybean, watermelon [55], cabbage [56], and cucumber [57]. The majority of the PsSWEET genes grouped together share similar conserved motifs and gene structure, which provides additional support for the phylogenetic results (Figure 2). Similar results have been reported in other Rosaceae species, such as apple [58], pear [59], and loquat [47]. In terms of the phylogenetic relationship, gene structure, and conserved motifs, we found that the PsSWEET gene family might be relatively conservative in the process of evolution. It is widely recognized that gene duplication plays a significant role in the driving force for the expansion of gene families and greatly increases functional diversity [60]. The segmental and tandem gene duplications are considered as two key factors responsible for the generation and maintenance of gene families [61]. Three segmental duplication events (PsSWEET7/8, PsSWEET1/12, and PsSWEET5/9) and two tandem duplication events (PsSWEET9/10 and PsSWEET13/14) were identified in the present study (Figure 4). Interestingly, the tandem duplication events (PsSWEET9/10 and PsSWEET13/14) have divergent expression patterns during the development of plum fruits, which might be due to the different cis elements identified in these SWEET genes (Figure 3 and Figure 4). Therefore, the further details of transcriptional regulation need to be studied in the future. Only one segmental duplication event (LcSWEET4 and LcSWEET5) and three tandem duplication events (LcSWEET3a/3b, LcSWEET9a/9b, and LcSEET10/12) were found in Litchi chinensis [62]. The varied number of SWEET genes in different plant species could be a result of evolutionary mechanisms, such as tandem or segmental duplication. Together, it is proposed that the expansion of PsSWEETs might play a considerable role in the various gene functions of SWEET.
In recent years, growing evidence indicates that SWEET proteins exhibit a broad distribution across various plant species and participate in diverse physiological roles, including plant growth, fruit development, and stress responses. As previously mentioned, many researchers have found that the SWEET family is involved in the fruit development process in many plant species, such as M. domestica [58], Solanum lycopersicum [63], and V. vinifera [18]. However, it has still remained unclear whether PsSWEETs in plum also play a role in fruit development. Further, higher expression levels for the PsSWEET gene were detected during the development of plum fruit via qRT-PCR. The results indicated that the SWEET family in plum has participated in the fruit development process, which is similar to strawberry. Some PsSWEET genes (PsSWEET3/7/10/13/14) with a higher expression were observed during the early stage of fruit development (Figure 7). In prior studies, the sucrose content of ‘Furongli’ increased at the early stage of fruit development and then gradually decreased [39]. We found that there was positive correlation between PsSWEET3/10/13 gene expression and sucrose content during ‘Furongli’ development (Figure 8), which suggests that they play vital roles in the cellular transportation and unloading of sugars into fruits for sink storage. That is consistent with previous reports by Miao et al. [20]. Furthermore, AtSWEET16/17, a facilitative transporter, was demonstrated to mediate fructose transport across the tonoplast of roots and leaves in Arabidopsis [64,65,66,67]. Here, we also found that PsSWEET2 has a positive correlation with the fructose level during fruit development, and PsSWEET9 has a positive correlation with glucose levels during fruit development (Figure 8). These results indicate that PsSWEET genes have the potential to transport not just sucrose and glucose, but also fructose, which is supported by previous findings. Together, these data will supply abundant information for future investigations of SWEET gene functions for sugar transporters during fruit development.
5. Conclusions
It is well known that many SWEET genes have crucial functions in regulating sugar transportation during fruit development. Here, we successfully investigated the gene structures, conserved domains, expression patterns, and phylogenetic relationships of the SWEET gene family in plum. Fifteen PsSWEET genes were identified in P. salicina. Our qRT-PCR analyses have shown that PsSWEE2, PsSWEET3, PsSWEET4, PsSWEET5, PsSWEET7, PsSWEET13, and PsSWEET14 exhibited a relatively higher expression at the early stage of fruit development than at the later developmental stages. However, PsSWEET1/9 showed relatively lower levels of expression at the early stage of fruit development, while it sharply increased in the late stages. The divergent expression patterns of some tandem duplication events, such as PsSWEET9/10 and PsSWEET13/14 during the development of plum fruits, might be due to the different cis elements identified in these SWEET genes, respectively. Positive correlations were found between the expression level of PsSWEET3/10/13 and the content of sucrose, the expression level of PsSWEET2 and the content of fructose, and the expression level of PsSWEET9 and the content of glucose, respectively, during the development of the ‘Furongli’ fruit. Therefore, we proposed that PsSWEETs might have the potential to transport not just sucrose and glucose, but also fructose during ‘Furongli’ fruit development.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes14091679/s1, Table S1: Primers sequences used for RT-qPCR analysis in this study.
Author Contributions
Conceptualization, formal analysis, C.J.; writing—original draft preparation, C.J. and X.W.; methodology, S.Z.; software, J.Y. and X.W.; validation, J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funds from the Natural Science Foundation of Fujian Province (2020J011363), the Yong Talent Program of Fujian Academy of Agricultural Sciences (YC20210037), and the Basic Scientific Research Funds of Public Welfare Scientific Research Institutes of Fujian Province (2020R1028008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used in this study are available from the corresponding authors upon reasonable request.
Conflicts of Interest
The authors have declared no conflict of interest.
References
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Lin, I.W.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londono, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 2015, 67, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014, 201, 1150–1155. [Google Scholar] [CrossRef]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef]
- Ayre, B.G. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 2011, 4, 377–394. [Google Scholar] [CrossRef]
- Kuhn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 288–298. [Google Scholar] [CrossRef]
- Poudel, K.; Luo, X.; Chen, L.; Jing, D.; Xia, X.; Tang, L.; Li, H.; Cao, S. Identification of the SUT Gene Family in Pomegranate (Punica granatum L.) and Functional Analysis of PgL0145810.1. Int. J. Mol. Sci. 2020, 21, 6608. [Google Scholar] [CrossRef]
- Slewinski, T.L. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ren, Z.; Wang, Z.; Sun, K.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Song, C.; Zhou, X.; et al. Evolution and Stress Responses of Gossypium hirsutum SWEET Genes. Int. J. Mol. Sci. 2018, 19, 769. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wang, S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET Family of Sugar Transporters: Structure, Evolution and Biological Functions. Biomolecules 2022, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Anjali, A.; Fatima, U.; Manu, M.S.; Ramasamy, S.; Senthil-Kumar, M. Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 2020, 156, 1–6. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Chong, J.; Piron, M.C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef]
- Ru, L.; He, Y.; Zhu, Z.; Patrick, J.W.; Ruan, Y.L. Integrating Sugar Metabolism With Transport: Elevation of Endogenous Cell Wall Invertase Activity Up-Regulates SlHT2 and SlSWEET12c Expression for Early Fruit Development in Tomato. Front. Genet. 2020, 11, 592596. [Google Scholar] [CrossRef]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W.; Zhang, J.; Wang, J.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 3536. [Google Scholar] [CrossRef]
- Wei, X.; Liu, F.; Chen, C.; Ma, F.; Li, M. The Malus domestica sugar transporter gene family: Identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front. Plant Sci. 2014, 5, 569. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Mula, H.M.; Zapata, P.J.; Guillén, F.; Castillo, S.; Martínez-Romero, D.; Valero, D.; Serrano, M. Changes in physicochemical and nutritive parameters and bioactive compounds during development and on-tree ripening of eight plum cultivars: A comparative study. J. Sci. Food Agric. 2008, 88, 2499–2507. [Google Scholar] [CrossRef]
- Guerra, M.E.; Rodrigo, J. Japanese plum pollination: A review. Sci. Hortic. 2015, 197, 674–686. [Google Scholar] [CrossRef]
- Treutter, D.; Wang, D.; Farag, M.A.; Baires, G.D.; Ruhmann, S.; Neumuller, M. Diversity of phenolic profiles in the fruit skin of Prunus domestica plums and related species. J. Agric. Food Chem. 2012, 60, 12011–12019. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.S.; Liu, D.C.; Feng, C.J.; Zhang, A.M.; Li, S.H. Genetic diversity and phylogenetic relationships in plum germplasm resources revealed by RAPD markers. J. Hortic. Sci. Biotechnol. 2015, 81, 242–250. [Google Scholar] [CrossRef]
- Fang, Z.Z.; Zhou, D.R.; Ye, X.F.; Jiang, C.C.; Pan, S.L. Identification of candidate anthocyanin-related genes by transcriptomic analysis of ‘Furongli’ plum (Prunus salicina Lindl.) during Fruit Ripening Using RNA-Seq. Front. Plant Sci. 2016, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chang, X.X.; Wang, H.; Brennan, C.S.; Guo, X.B. Phytochemicals Accumulation in Sanhua Plum (Prunus salicina L.) during Fruit Development and Their Potential Use as Antioxidants. J. Agric. Food Chem. 2019, 67, 2459–2466. [Google Scholar] [CrossRef]
- Valero, D.; Díaz-Mula, H.M.; Zapata, P.J.; Guillén, F.; Martínez-Romero, D.; Castillo, S.; Serrano, M. Effects of alginate edible coating on preserving fruit quality in four plum cultivars during postharvest storage. Postharvest Biol. Technol. 2013, 77, 1–6. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Jiang, C.C.; Fang, Z.Z.; Zhou, D.R.; Pan, S.L.; Ye, X.F. Changes in secondary metabolites, organic acids and soluble sugars during the development of plum fruit cv. ‘Furongli’ (Prunus salicina Lindl). J. Sci. Food Agric. 2019, 99, 1010–1019. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Yang, X.; Wei, C.; Changqing, X.; Zhang, X. Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress. Genes 2021, 12, 326. [Google Scholar] [CrossRef]
- Jing, Q.; Hou, H.; Meng, X.; Chen, A.; Wang, L.; Zhu, H.; Zheng, S.; Lv, Z.; Zhu, X. Transcriptome analysis reveals the proline metabolic pathway and its potential regulation TF-hub genes in salt-stressed potato. Front. Plant Sci. 2022, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.D.; Lucas, E.A.; Hooshmand, S.; Campbell, S.; Akhter, M.P.; Arjmandi, B.H. Addition of fructooligosaccharides and dried plum to soy-based diets reverses bone loss in the ovariectomized rat. Evid. Based Complement. Altern. Med. 2011, 2011, 836267. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lillehoj, H.S.; Cho, S.M.; Chun, H.K.; Park, H.J.; Lim, C.I.; Lillehoj, E.P. Immunostimulatory effects of oriental plum (Prunus salicina Lindl.). Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Shukitt-Hale, B.; Kalt, W.; Carey, A.N.; Vinqvist-Tymchuk, M.; McDonald, J.; Joseph, J.A. Plum juice, but not dried plum powder, is effective in mitigating cognitive deficits in aged rats. Nutrition 2009, 25, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Ornelas-Paz Jde, J.; Yahia, E.M.; Ramirez-Bustamante, N.; Perez-Martinez, J.D.; Escalante-Minakata Mdel, P.; Ibarra-Junquera, V.; Acosta-Muniz, C.; Guerrero-Prieto, V.; Ochoa-Reyes, E. Physical attributes and chemical composition of organic strawberry fruit (Fragaria x ananassa Duch, Cv. Albion) at six stages of ripening. Food Chem. 2013, 138, 372–381. [Google Scholar] [CrossRef]
- Cheng, J.; Wen, S.; Bie, Z. Overexpression of hexose transporter CsHT3 increases cellulose content in cucumber fruit peduncle. Plant Physiol. Biochem. 2019, 145, 107–113. [Google Scholar] [CrossRef]
- Li, B.; Ali, M.M.; Guo, T.; Alam, S.M.; Gull, S.; Iftikhar, J.; Yousef, A.F.; Mosa, W.F.A.; Chen, F. Genome-wide identification, in silico analysis and expression profiling of SWEET gene family in loquat (Eriobotrya japonica Lindl.). Agriculture 2022, 12, 1312. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, P.; Zhu, X.F.; Li, T.Y.; Jia, B.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, P.; Xu, S.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. Genome-wide identification, expression patterns and sugar transport of the physic nut SWEET gene family and a functional analysis of JcSWEET16 in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 5391. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Lyu, W.Y.; Tian, S.H.; Zou, X.H.; Zhang, L.Q.; Gao, Q.H.; Ni, D.A.; Duan, K. The SWEET family genes in strawberry: Identification and expression profiling during fruit development. S. Afr. J. Bot. 2019, 125, 176–187. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Lan, G.; Si, F.; Zeng, Z.; Wang, C.; Yadav, V.; Wei, C.; Zhang, X. Systematic genome-wide study and expression analysis of SWEET gene family: Sugar transporter family contributes to biotic and abiotic stimuli in watermelon. Int. J. Mol. Sci. 2021, 22, 8407. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.; Li, J. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef]
- Hu, L.-P.; Zhang, F.; Song, S.-H.; Tang, X.-W.; Xu, H.; Liu, G.-M.; Wang, Y.; He, H.-J. Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J. Integr. Agric. 2017, 16, 1486–1501. [Google Scholar] [CrossRef]
- Zhen, Q.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hort. Res. 2018, 5, 14. [Google Scholar] [CrossRef]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A New Insight into the Evolution and Functional Divergence of SWEET Transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Wang, D.; Qin, Y.; Ma, A.; Fu, J.; Qin, Y.; Hu, G.; Zhao, J. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499. [Google Scholar] [CrossRef]
- Shammai, A.; Petreikov, M.; Yeselson, Y.; Faigenboim, A.; Moy-Komemi, M.; Cohen, S.; Cohen, D.; Besaulov, E.; Efrati, A.; Houminer, N.; et al. Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 2018, 96, 343–357. [Google Scholar] [CrossRef]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.; Spinner, L.; Clement, G.; Chietera, G.; Leran, S.; Ferrand, M.; Lacombe, B.; et al. Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.; Aubry, E.; Marmagne, A.; Dinant, S.; Chardon, F.; Le Hir, R. Impairment of sugar transport in the vascular system acts on nitrogen remobilization and nitrogen use efficiency in Arabidopsis. Physiol. Plant 2022, 174, e13830. [Google Scholar] [CrossRef] [PubMed]
- Aubry, E.; Hoffmann, B.; Vilaine, F.; Gilard, F.; Klemens, P.A.W.; Guérard, F.; Gakière, B.; Neuhaus, H.E.; Bellini, C.; Dinant, S.; et al. A vacuolar hexose transport is required for xylem development in the inflorescence stem. Plant Physiol. 2022, 188, 1229–1247. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).