Exploring the Binding Affinity of the ARR2 GARP DNA Binding Domain via Comparative Methods
Abstract
1. Introduction
2. Results
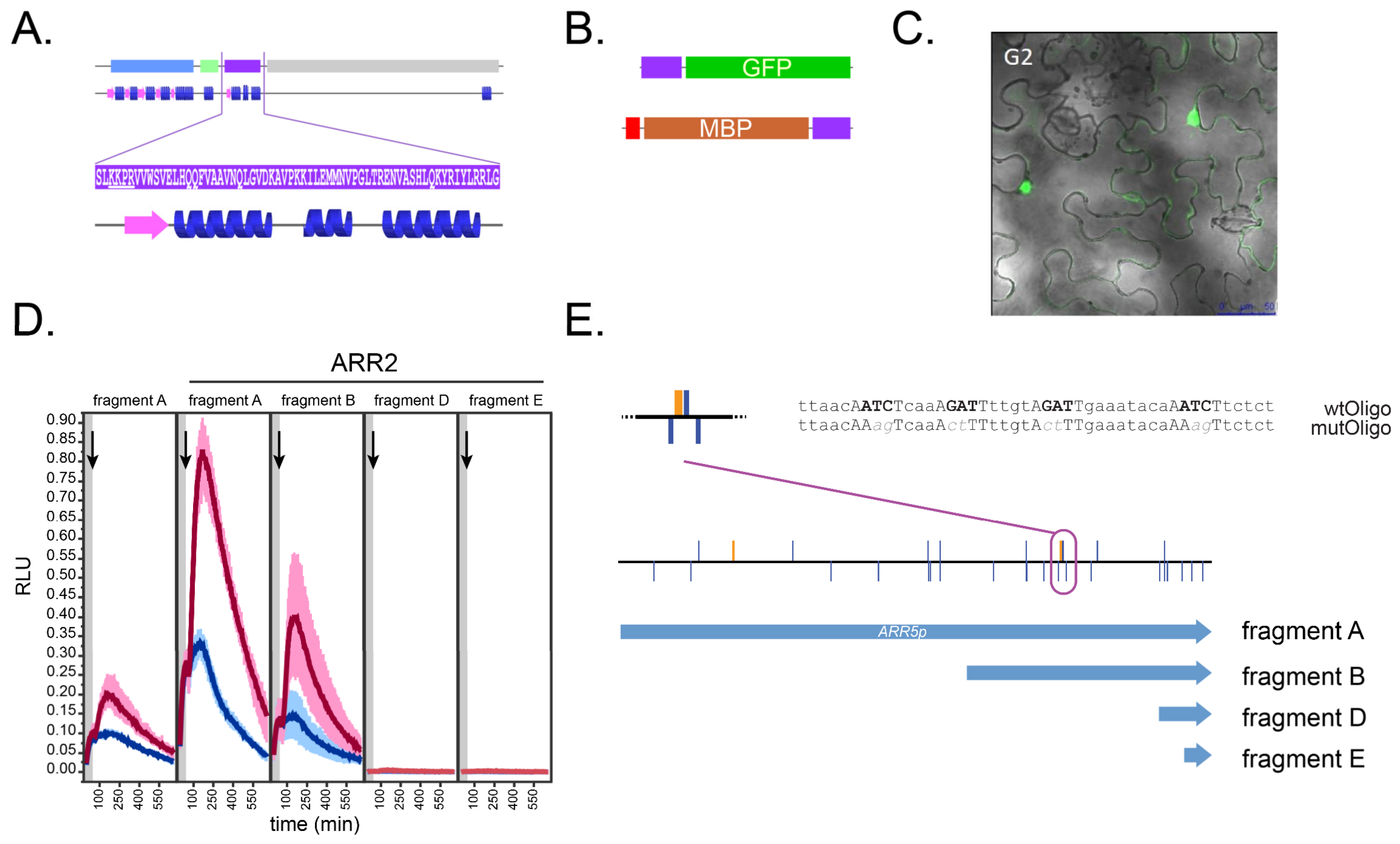
2.1. Generation of GARP2 Fusion Proteins and Its DNA Target Sequences within the ARR5 Promoter
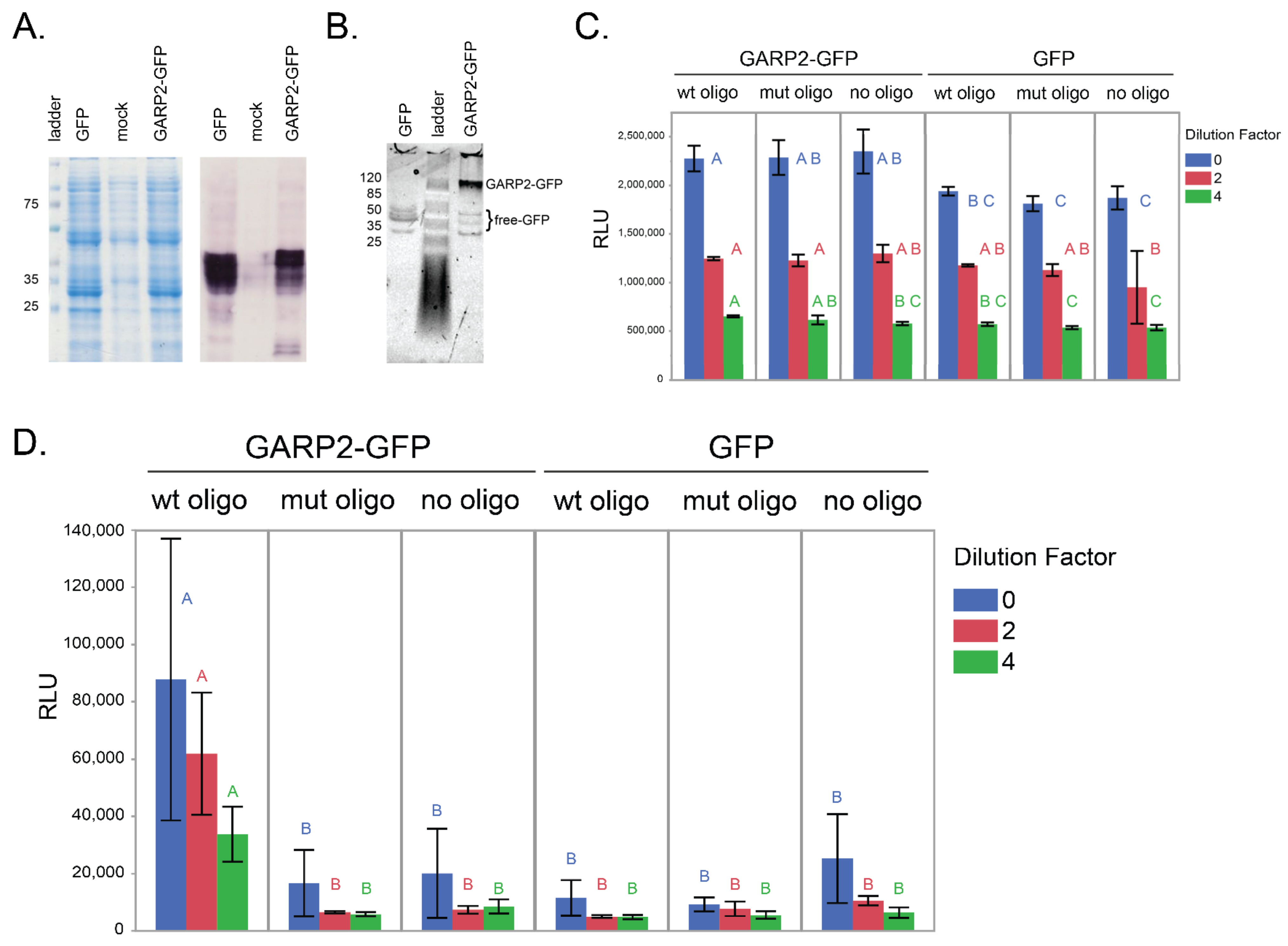
2.2. GARP2 Interaction with DNA Analyzed via qDPI-ELISA
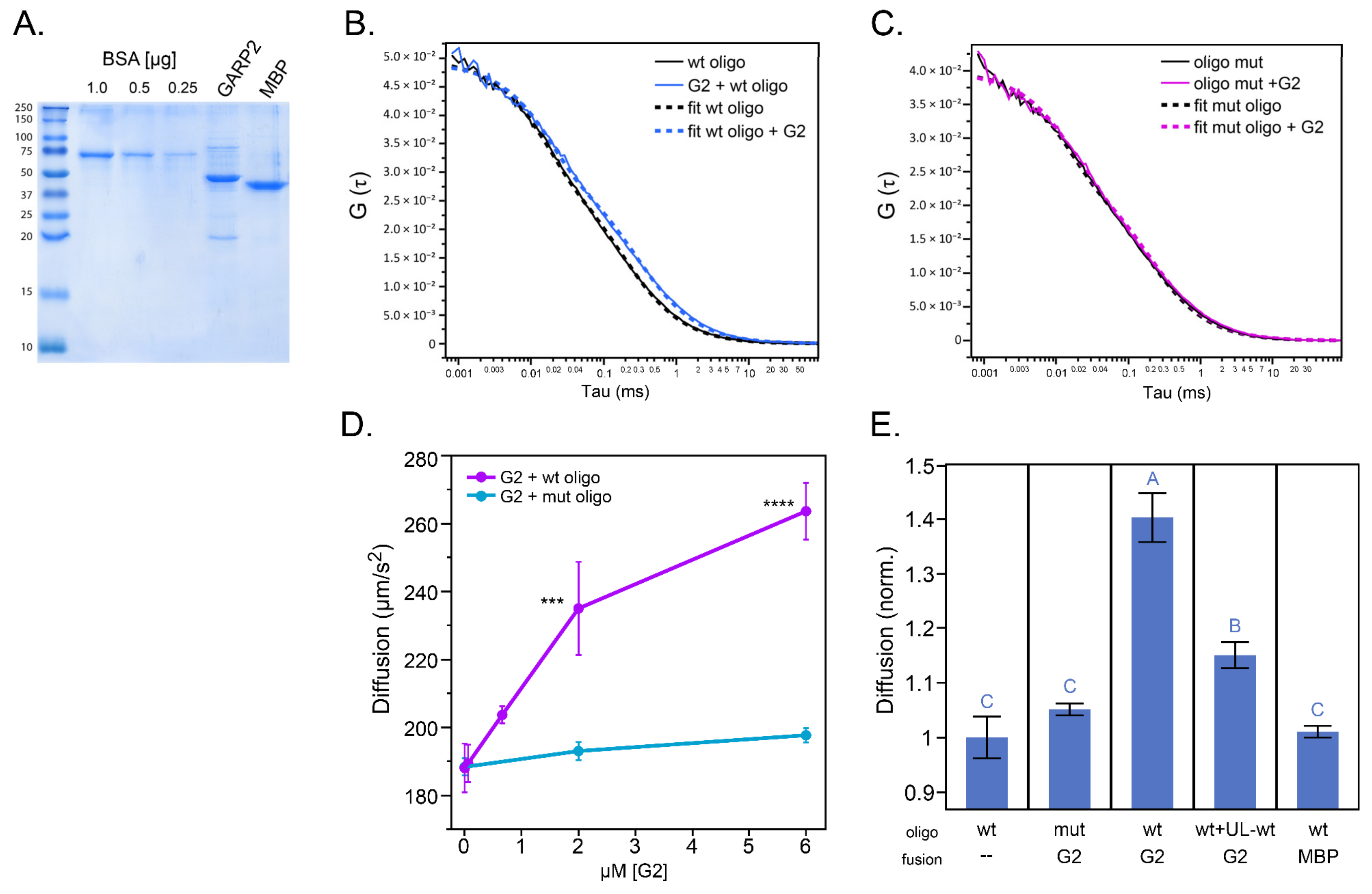
2.3. GARP2 Interaction with DNA Determined by Fluorescence Correlation Spectroscopy
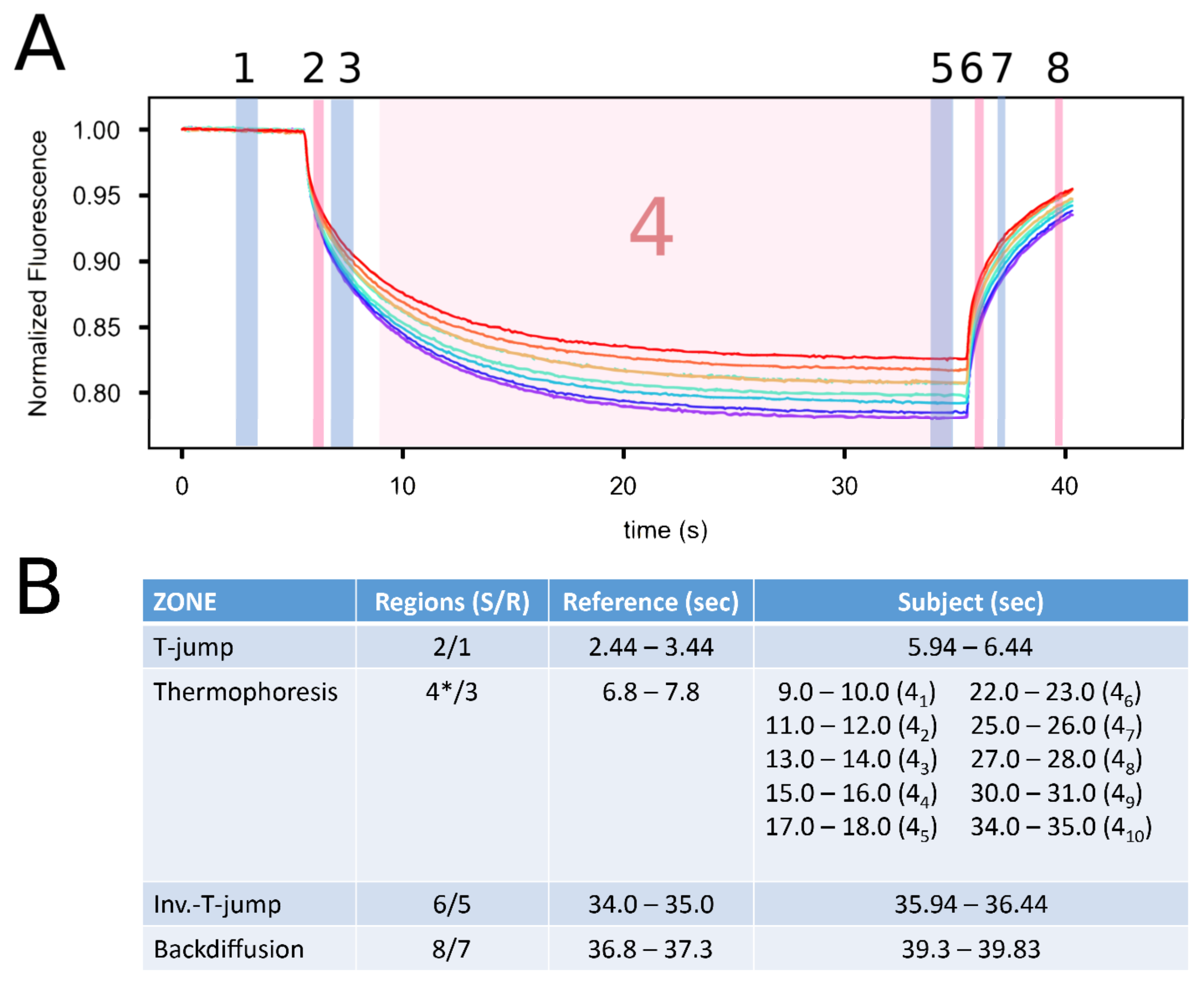
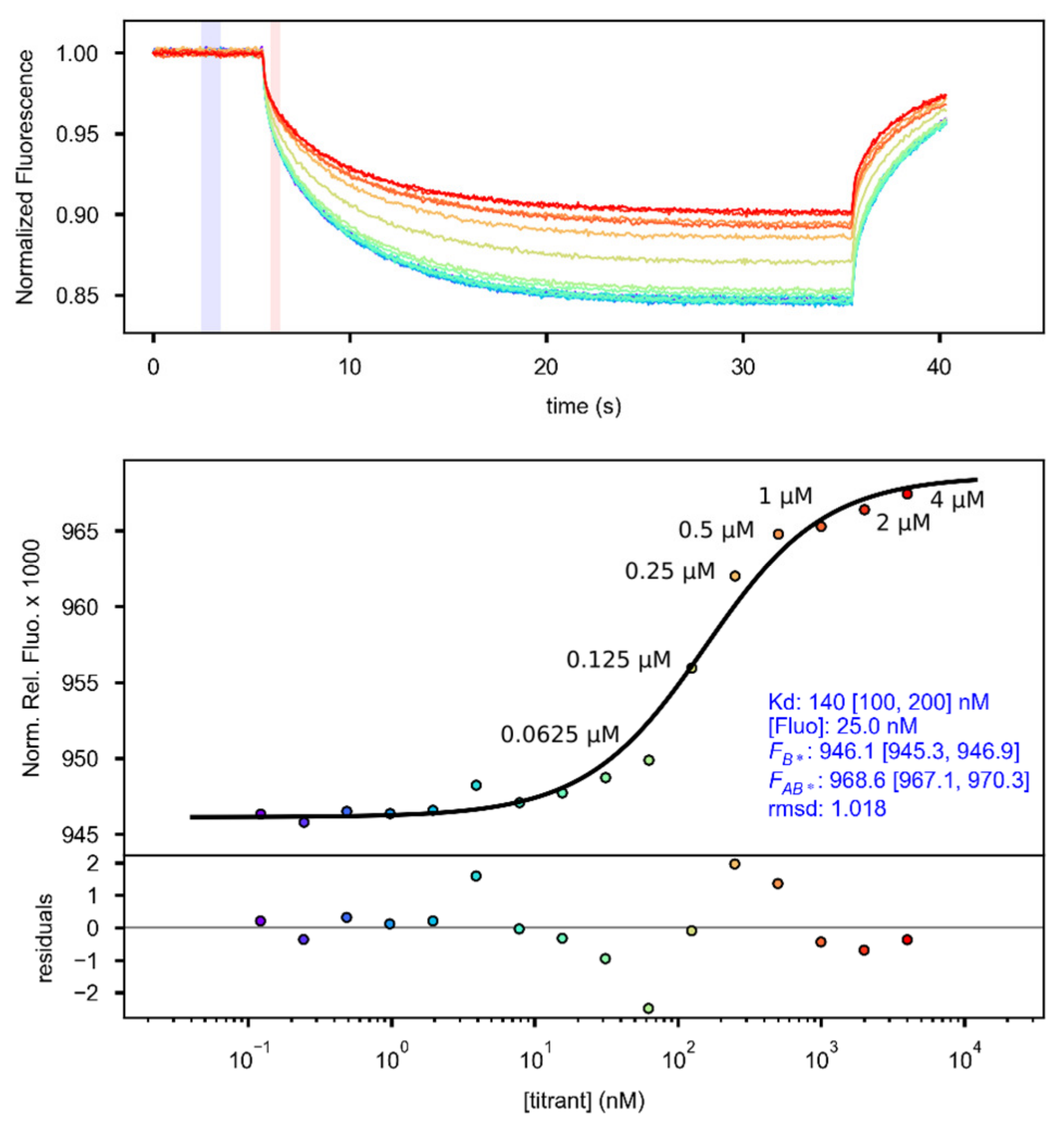
2.4. Quantification of the GARP2–DNA Binding Affinity via Microscale Thermophoresis
2.4.1. Establishing MST Limits in the Different MST Zones
2.4.2. Comparison of the KD from Different MST Zones
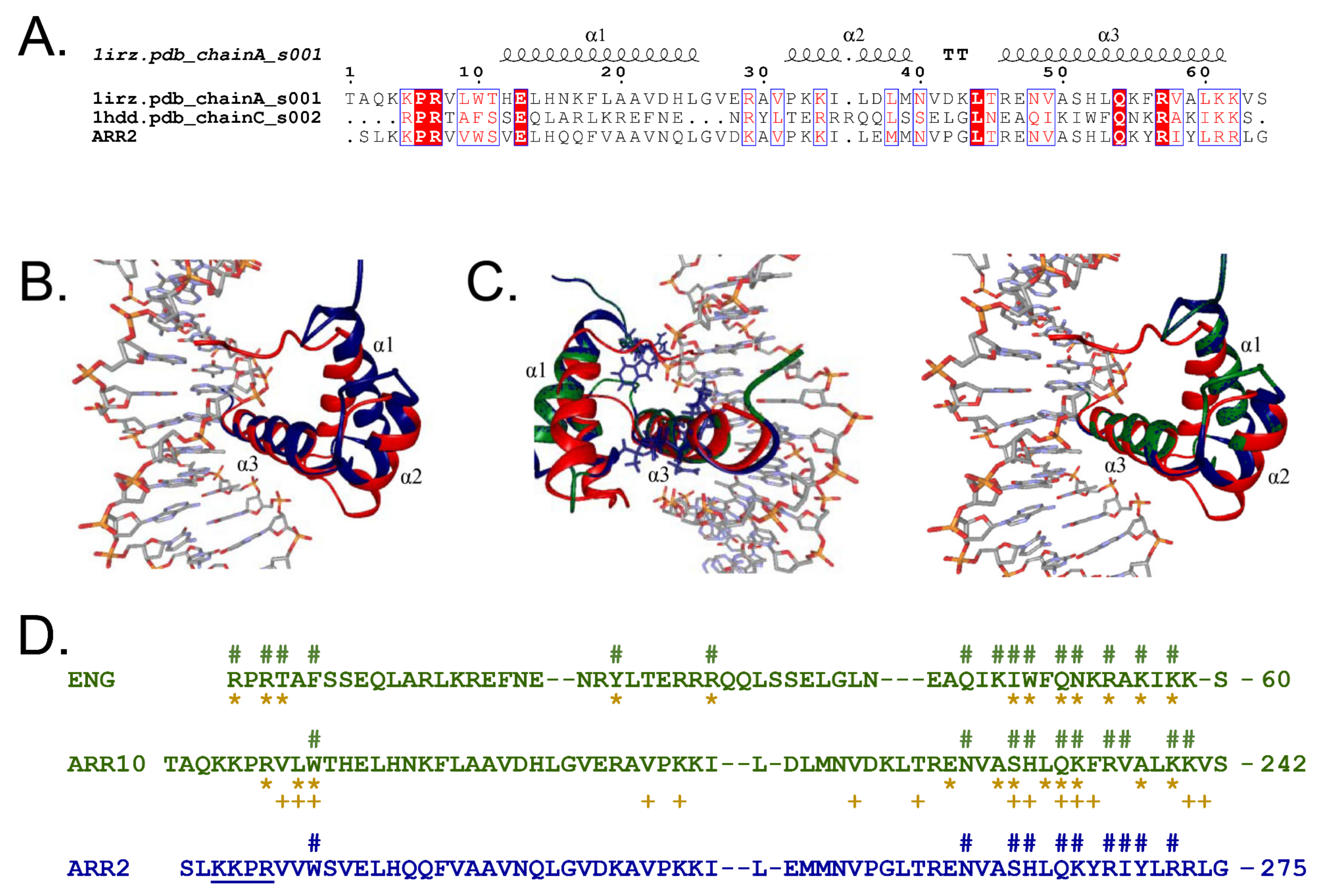
2.5. Structural Modeling of GARP2
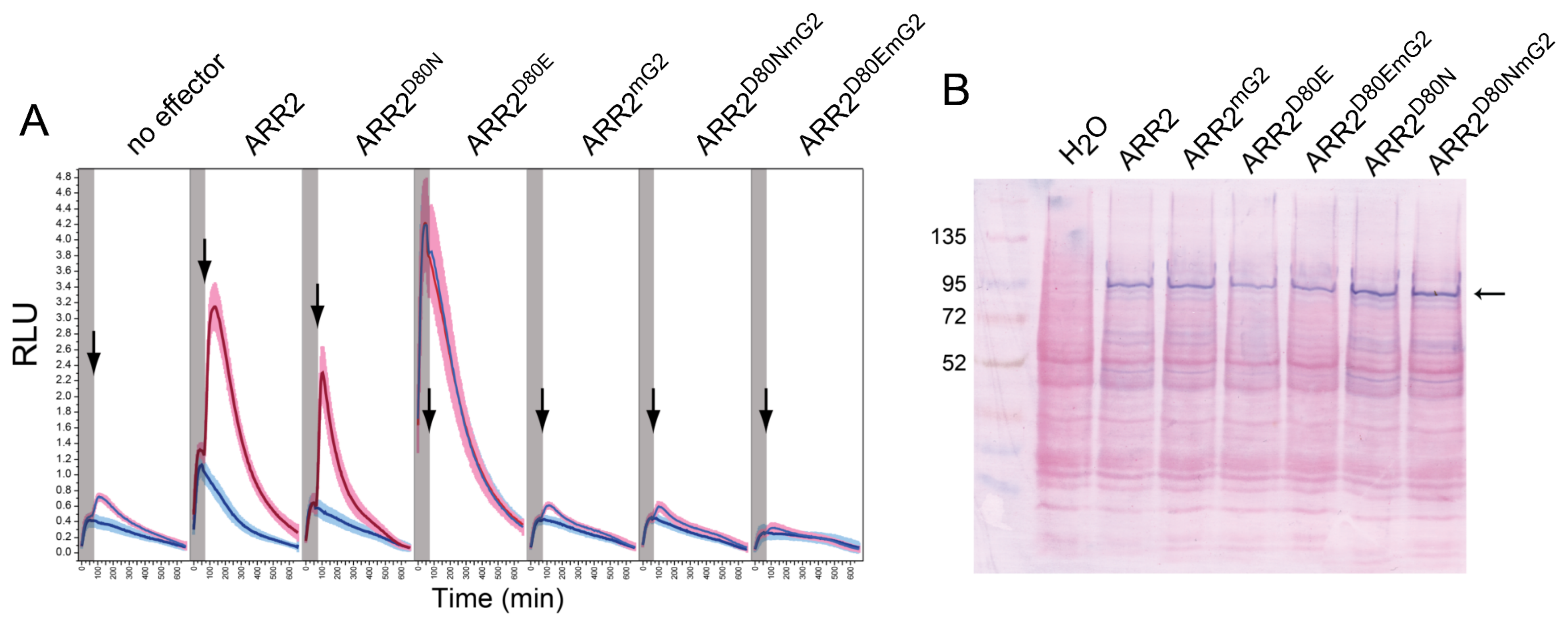
2.6. Mutations in the GARP2 Domain Affect ARR2’s Ability to Activate Transcription
3. Discussion
4. Methods
4.1. Cloning of GARPARR2
4.2. Target DNA
4.3. DPI ELISA
4.3.1. Bacterial Growth, GFP Induction and Extraction
4.3.2. Preparation of Ds Oligos
4.3.3. Immobilization of Ds Oligos to Plate
4.3.4. Protein Binding and Fluorescence Measurement
4.3.5. Quantification of GFP by Fluorescence
4.4. Reporter-Gene Transfection Assays
Immunoblot Analysis
4.5. Nickel-NTA Purification
4.5.1. Bacterial Growth and Protein Induction
4.5.2. Extraction
4.5.3. Protein Purification
4.5.4. Dialysis
4.5.5. Determining Protein Concentration
4.6. FCS
4.7. MST Preparation and Analysis
4.7.1. Annealing Ds Oligos
4.7.2. Identifying the Optimal Capillary Type
4.7.3. Affinity Test and Labeling
4.7.4. MST Acquisition
4.7.5. MST Analysis
4.8. Structural Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Janin, J. Protein-protein recognition. Prog. Biophys. Mol. Biol. 1995, 64, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Kastritis, P.L.; Bonvin, A.M. On the binding affinity of macromolecular interactions: Daring to ask why proteins interact. J. R. Soc. Interface 2013, 10, 20120835. [Google Scholar] [CrossRef] [PubMed]
- Vuignier, K.; Schappler, J.; Veuthey, J.L.; Carrupt, P.A.; Martel, S. Drug-protein binding: A critical review of analytical tools. Anal. Bioanal. Chem. 2010, 398, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, T.H.; Padrick, S.B.; Gardner, K.H.; Brautigam, C.A. On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 2016, 496, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; Andre, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Kim, J.S.; Chae, S.; Jun, K.M.; Pahk, Y.M.; Lee, T.H.; Chung, P.J.; Kim, Y.K.; Nahm, B.H. Genome-wide identification of grain filling genes regulated by the OsSMF1 transcription factor in rice. Rice 2017, 10, 16. [Google Scholar] [CrossRef]
- Brand, L.H.; Satbhai, S.B.; Kolukisaoglu, H.; Wanke, D. Limits and prospects of methods for the analysis of DNA-protein interaction. In The Analysis of Regulatory DNA: Current Developments, Knowledge and Applications Uncovering Gene Regulation; Bentham Science Publishers: Sharjah, United Arab Emirates, 2013; Volume 124. [Google Scholar]
- Rajeev, L.; Garber, M.E.; Mukhopadhyay, A. Tools to map target genes of bacterial two-component system response regulators. Environ. Microbiol. Rep. 2020, 12, 267–276. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Van Weemen, B.K.; Schuurs, A.H.W.M. Immunoassay using antigen—Enzyme conjugates. FEBS Lett. 1971, 15, 232–236. [Google Scholar] [CrossRef]
- Brand, L.H.; Kirchler, T.; Hummel, S.; Chaban, C.; Wanke, D. DPI-ELISA: A fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 2010, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.M.; Boser, A.; Hirsch, J.P.; Wanke, D. Quantitative Analysis of Protein-DNA Interaction by qDPI-ELISA. Methods Mol. Biol. 2016, 1482, 49–66. [Google Scholar] [CrossRef]
- Ries, J.; Schwille, P. Fluorescence correlation spectroscopy. Bioessays 2012, 34, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xing, J.; Qiu, Z.; He, Q.; Lin, J. Quantification of Membrane Protein Dynamics and Interactions in Plant Cells by Fluorescence Correlation Spectroscopy. Mol. Plant 2016, 9, 1229–1239. [Google Scholar] [CrossRef]
- Hess, S.T.; Huang, S.; Heikal, A.A.; Webb, W.W. Biological and chemical applications of fluorescence correlation spectroscopy: A review. Biochemistry 2002, 41, 697–705. [Google Scholar] [CrossRef]
- Habel, J.; Hansen, M.; Kynde, S.; Larsen, N.; Midtgaard, S.R.; Jensen, G.V.; Bomholt, J.; Ogbonna, A.; Almdal, K.; Schulz, A.; et al. Aquaporin-Based Biomimetic Polymeric Membranes: Approaches and Challenges. Membranes 2015, 5, 307–351. [Google Scholar] [CrossRef] [PubMed]
- Haustein, E.; Schwille, P. Ultrasensitive investigations of biological systems by fluorescence correlation spectroscopy. Methods 2003, 29, 153–166. [Google Scholar] [CrossRef]
- Tamura, T.; Endo, H.; Suzuki, A.; Sato, Y.; Kato, K.; Ohtani, M.; Yamaguchi, M.; Demura, T. Affinity-based high-resolution analysis of DNA binding by VASCULAR-RELATED NAC-DOMAIN7 via fluorescence correlation spectroscopy. Plant J. 2019, 100, 298–313. [Google Scholar] [CrossRef]
- Aker, J.; Hesselink, R.; Engel, R.; Karlova, R.; Borst, J.W.; Visser, A.J.; de Vries, S.C. In vivo hexamerization and characterization of the Arabidopsis AAA ATPase CDC48A complex using forster resonance energy transfer-fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. Plant Physiol. 2007, 145, 339–350. [Google Scholar] [CrossRef][Green Version]
- Leder, V.; Lummer, M.; Tegeler, K.; Humpert, F.; Lewinski, M.; Schuttpelz, M.; Staiger, D. Mutational definition of binding requirements of an hnRNP-like protein in Arabidopsis using fluorescence correlation spectroscopy. Biochem. Biophys. Res. Commun. 2014, 453, 69–74. [Google Scholar] [CrossRef]
- Cui, Y.; Choudhury, S.R.; Irudayaraj, J. Quantitative real-time kinetics of optogenetic proteins CRY2 and CIB1/N using single-molecule tools. Anal. Biochem. 2014, 458, 58–60. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muto, H.; Nagao, I.; Demura, T.; Fukuda, H.; Kinjo, M.; Yamamoto, K.T. Fluorescence cross-correlation analyses of the molecular interaction between an Aux/IAA protein, MSG2/IAA19, and protein-protein interaction domains of auxin response factors of arabidopsis expressed in HeLa cells. Plant Cell Physiol. 2006, 47, 1095–1101. [Google Scholar] [CrossRef]
- Clark, N.M.; Hinde, E.; Winter, C.M.; Fisher, A.P.; Crosti, G.; Blilou, I.; Gratton, E.; Benfey, P.N.; Sozzani, R. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 2016, 5, e14770. [Google Scholar] [CrossRef] [PubMed]
- Hink, M.A.; Shah, K.; Russinova, E.; de Vries, S.C.; Visser, A.J. Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys. J. 2008, 94, 1052–1062. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Seidel, S.A.; Dijkman, P.M.; Lea, W.A.; van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. [Google Scholar] [CrossRef]
- Perez, A.; Marchan, I.; Svozil, D.; Sponer, J.; Cheatham, T.E., 3rd; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef]
- Mira-Rodado, V. New Insights into Multistep-Phosphorelay (MSP)/Two-Component System (TCS) Regulation: Are Plants and Bacteria that Different? Plants 2019, 8, 590. [Google Scholar] [CrossRef]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arab. Book 2008, 6, e0112. [Google Scholar] [CrossRef]
- Hosoda, K.; Imamura, A.; Katoh, E.; Hatta, T.; Tachiki, M.; Yamada, H.; Mizuno, T.; Yamazaki, T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 2002, 14, 2015–2029. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Characterization of a nitrate-inducible transcriptional repressor NIGT1 provides new insights into DNA recognition by the GARP family proteins. Plant Signal. Behav. 2013, 8, e24447. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef]
- Kerstetter, R.A.; Bollman, K.; Taylor, R.A.; Bomblies, K.; Poethig, R.S. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef]
- Kissinger, C.R.; Liu, B.S.; Martin-Blanco, E.; Kornberg, T.B.; Pabo, C.O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: A framework for understanding homeodomain-DNA interactions. Cell 1990, 63, 579–590. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Bobola, N.; Merabet, S. Homeodomain proteins in action: Similar DNA binding preferences, highly variable connectivity. Curr. Opin. Genet. Dev. 2017, 43, 1–8. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Kiba, T.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999, 40, 733–742. [Google Scholar] [CrossRef]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Aoyama, T.; Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. Cell Mol. Biol. 2000, 24, 703–711. [Google Scholar] [CrossRef]
- Che, P.; Lall, S.; Howell, S.H. Acquiring competence for shoot development in Arabidopsis: ARR2 directly targets A-type ARR genes that are differentially activated by CIM preincubation. Plant Signal. Behav. 2008, 3, 99–101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Veerabagu, M.; Kirchler, T.; Elgass, K.; Stadelhofer, B.; Stahl, M.; Harter, K.; Mira-Rodado, V.; Chaban, C. The interaction of the Arabidopsis response regulator ARR18 with bZIP63 mediates the regulation of PROLINE DEHYDROGENASE expression. Mol. Plant 2014, 7, 1560–1577. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Sasaki, N.; Tsuge, T.; Aoyama, T.; Oka, A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007, 48, 263–277. [Google Scholar] [CrossRef]
- Zurcher, E.; Tavor-Deslex, D.; Lituiev, D.; Enkerli, K.; Tarr, P.T.; Muller, B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 2013, 161, 1066–1075. [Google Scholar] [CrossRef]
- Imamura, A.; Kiba, T.; Tajima, Y.; Yamashino, T.; Mizuno, T. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 122–131. [Google Scholar] [CrossRef]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 17, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Veerabagu, M.; Elgass, K.; Kirchler, T.; Huppenberger, P.; Harter, K.; Chaban, C.; Mira-Rodado, V. The Arabidopsis B-type response regulator 18 homomerizes and positively regulates cytokinin responses. Plant J. Cell Mol. Biol. 2012, 72, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Wallmeroth, N.; Anastasia, A.K.; Harter, K.; Berendzen, K.W.; Mira-Rodado, V. Arabidopsis response regulator 22 inhibits cytokinin-regulated gene transcription in vivo. Protoplasma 2017, 254, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Wallmeroth, N.; Jeschke, D.; Slane, D.; Nagele, J.; Veerabagu, M.; Mira-Rodado, V.; Berendzen, K.W. ARR22 overexpression can suppress plant Two-Component Regulatory Systems. PLoS ONE 2019, 14, e0212056. [Google Scholar] [CrossRef] [PubMed]
- Hass, C.; Lohrmann, J.; Albrecht, V.; Sweere, U.; Hummel, F.; Yoo, S.D.; Hwang, I.; Zhu, T.; Schafer, E.; Kudla, J.; et al. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 2004, 23, 3290–3302. [Google Scholar] [CrossRef]
- Lohrmann, J.; Sweere, U.; Zabaleta, E.; Baurle, I.; Keitel, C.; Kozma-Bognar, L.; Brennicke, A.; Schafer, E.; Kudla, J.; Harter, K. The response regulator ARR2: A pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Genet. Genom. MGG 2001, 265, 2–13. [Google Scholar]
- Tajima, Y.; Imamura, A.; Kiba, T.; Amano, Y.; Yamashino, T.; Mizuno, T. Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ryu, H.; Hong, S.H.; Woo, H.R.; Lim, P.O.; Lee, I.C.; Sheen, J.; Nam, H.G.; Hwang, I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 814–819. [Google Scholar] [CrossRef]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef]
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. Cell Mol. Biol. 2006, 47, 907–916. [Google Scholar] [CrossRef]
- Takahashi, N.; Kajihara, T.; Okamura, C.; Kim, Y.; Katagiri, Y.; Okushima, Y.; Matsunaga, S.; Hwang, I.; Umeda, M. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 2013, 23, 1812–1817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Q.; Lian, H.; Zhou, C.M.; Xu, L.; Jiao, Y.; Wang, J.W. A Two-Step Model for de Novo Activation of WUSCHEL during Plant Shoot Regeneration. Plant Cell 2017, 29, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Ramireddy, E.; Brenner, W.G.; Pfeifer, A.; Heyl, A.; Schmulling, T. In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 2013, 54, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- El Deeb, S.; Al-Harrasi, A.; Khan, A.; Al-Broumi, M.; Al-Thani, G.; Alomairi, M.; Elumalai, P.; Sayed, R.A.; Ibrahim, A.E. Microscale thermophoresis as a powerful growing analytical technique for the investigation of biomolecular interaction and the determination of binding parameters. Methods Appl. Fluores 2022, 10, 042001. [Google Scholar] [CrossRef] [PubMed]
- Asmari, M.; Michalcova, L.; Ibrahim, A.E.; Glatz, Z.; Watzig, H.; El Deeb, S. Studying molecular interactions via capillary electrophoresis and microscale thermophoresis: A review. Electrophoresis 2023, 44, 1114–1142. [Google Scholar] [CrossRef]
- Tschammer, N.; Galinec, S.; Weigert, S.; Muller, Y.; You, C.; Piehler, J.; Breitsprecher, D. One-Step, Purification-Free and Site-Specific Labeling of Polyhistidine-Tagged Proteins for MST; NanoTemper Technologies, Inc.: South San Francisco, CA, USA, 2017. [Google Scholar]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Mendez, B.; Uebel, S.; Lundgren, L.P.; Sedivy, A. Microscale Thermophoresis and additional effects measured in NanoTemper Monolith instruments. Eur. Biophys. J. 2021, 50, 653–660. [Google Scholar] [CrossRef]
- Brautigam, C.A. Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data. Methods Enzymol. 2015, 562, 109–133. [Google Scholar] [CrossRef]
- Blanco, A.G.; Sola, M.; Gomis-Ruth, F.X.; Coll, M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 2002, 10, 701–713. [Google Scholar] [CrossRef]
- Gao, R.; Mack, T.R.; Stock, A.M. Bacterial response regulators: Versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007, 32, 225–234. [Google Scholar] [CrossRef]
- Pekarova, B.; Szmitkowska, A.; Dopitova, R.; Degtjarik, O.; Zidek, L.; Hejatko, J. Structural Aspects of Multistep Phosphorelay-Mediated Signaling in Plants. Mol. Plant 2016, 9, 71–85. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, I.B.; Deruere, J.; Kieber, J.J. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000, 124, 1706–1717. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The Antagonistic Action of Abscisic Acid and Cytokinin Signaling Mediates Drought Stress Response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.; Haberer, G.; Ferreira, F.J.; Deruere, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef]
- Sakai, H.; Aoyama, T.; Bono, H.; Oka, A. Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol. 1998, 39, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Bandilla, P.; von Reutern, M.; Schnepf, M.; Rieder, S.; Unnerstall, U.; Gaul, U. True equilibrium measurement of transcription factor-DNA binding affinities using automated polarization microscopy. Nat. Commun. 2018, 9, 1605. [Google Scholar] [CrossRef]
- André, T.; Breitsprecher, D. Thermodynamic Characterization of DNA Hybridization; NanoTemper Technologies GmbH: Munich, Germany, 2013. [Google Scholar]
- Bartoschik, T.; Galinec, S.; Kleusch, C.; Walkiewicz, K.; Breitsprecher, D.; Weigert, S.; Muller, Y.A.; You, C.; Piehler, J.; Vercruysse, T.; et al. Near-native, site-specific and purification-free protein labeling for quantitative protein interaction analysis by MicroScale Thermophoresis. Sci. Rep. 2018, 8, 4977. [Google Scholar] [CrossRef]
- Bartoschik, T.A. Development and Validation of New Approaches for Quantitative Intermolecular Interaction Analysis by MicroScale Thermophoresis under Near-Native Experimental Conditions. Ph.D Thesis, Faculty of Biology and Preclinical Medicine at the University of Regensburg, Regensburg, Germany, 2018. [Google Scholar]
- Gudim, I.; Lofstad, M.; Hammerstad, M.; Hersleth, H.-P. Measurement of FNR-NrdI Interaction by Microscale Thermophoresis (MST). Bio-Protocol 2017, 7, e2223. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- Xie, M.T.; Chen, H.Y.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018, 9, 1604. [Google Scholar] [CrossRef] [PubMed]
- Zubo, Y.O.; Blakley, I.C.; Yamburenko, M.V.; Worthen, J.M.; Street, I.H.; Franco-Zorrilla, J.M.; Zhang, W.J.; Hill, K.; Raines, T.; Solano, R.; et al. Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E5995–E6004. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Johnen, P.; Azevedo, C.; Dynowski, M.; Weiss, M.; Capolicchio, S.; Mao, H.; Iven, T.; Steenbergen, M.; Freyer, M.; et al. VIH2 Regulates the Synthesis of Inositol Pyrophosphate InsP8 and Jasmonate-Dependent Defenses in Arabidopsis. Plant Cell 2015, 27, 1082–1097. [Google Scholar] [CrossRef] [PubMed]
- Bleicken, S.; Hantusch, A.; Das, K.K.; Frickey, T.; Garcia-Saez, A.J. Quantitative interactome of a membrane Bcl-2 network identifies a hierarchy of complexes for apoptosis regulation. Nat. Commun. 2017, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Flores-Romero, H.; Landeta, O.; Ugarte-Uribe, B.; Cosentino, K.; Garcia-Porras, M.; Garcia-Saez, A.J.; Basanez, G. BFL1 modulates apoptosis at the membrane level through a bifunctional and multimodal mechanism showing key differences with BCLXL. Cell Death Differ. 2019, 26, 1880–1894. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef]
- Hildebrandt, A.; Dehof, A.K.; Rurainski, A.; Bertsch, A.; Schumann, M.; Toussaint, N.C.; Moll, A.; Stockel, D.; Nickels, S.; Mueller, S.C.; et al. BALL--biochemical algorithms library 1.3. BMC Bioinform. 2010, 11, 531. [Google Scholar] [CrossRef]
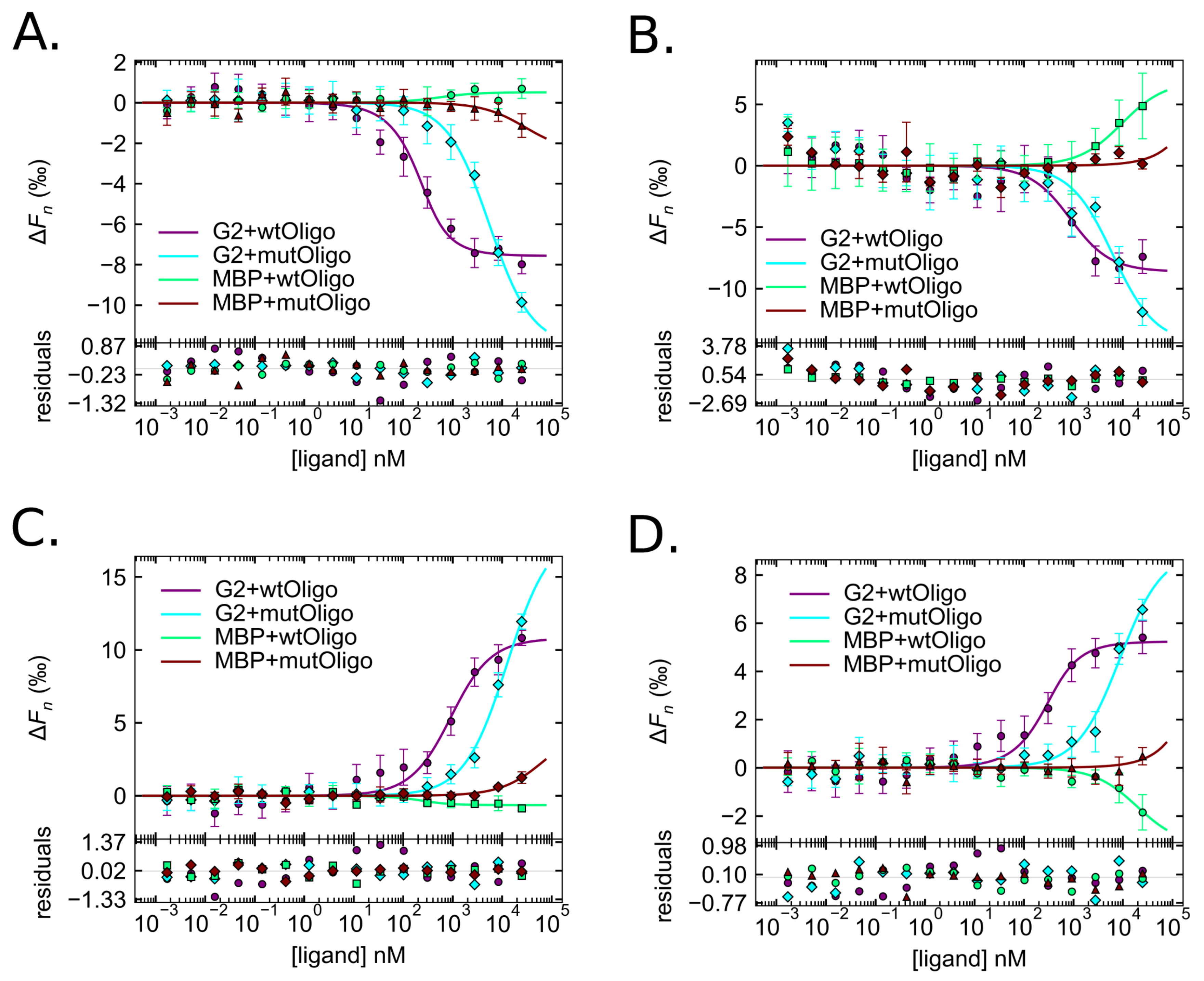
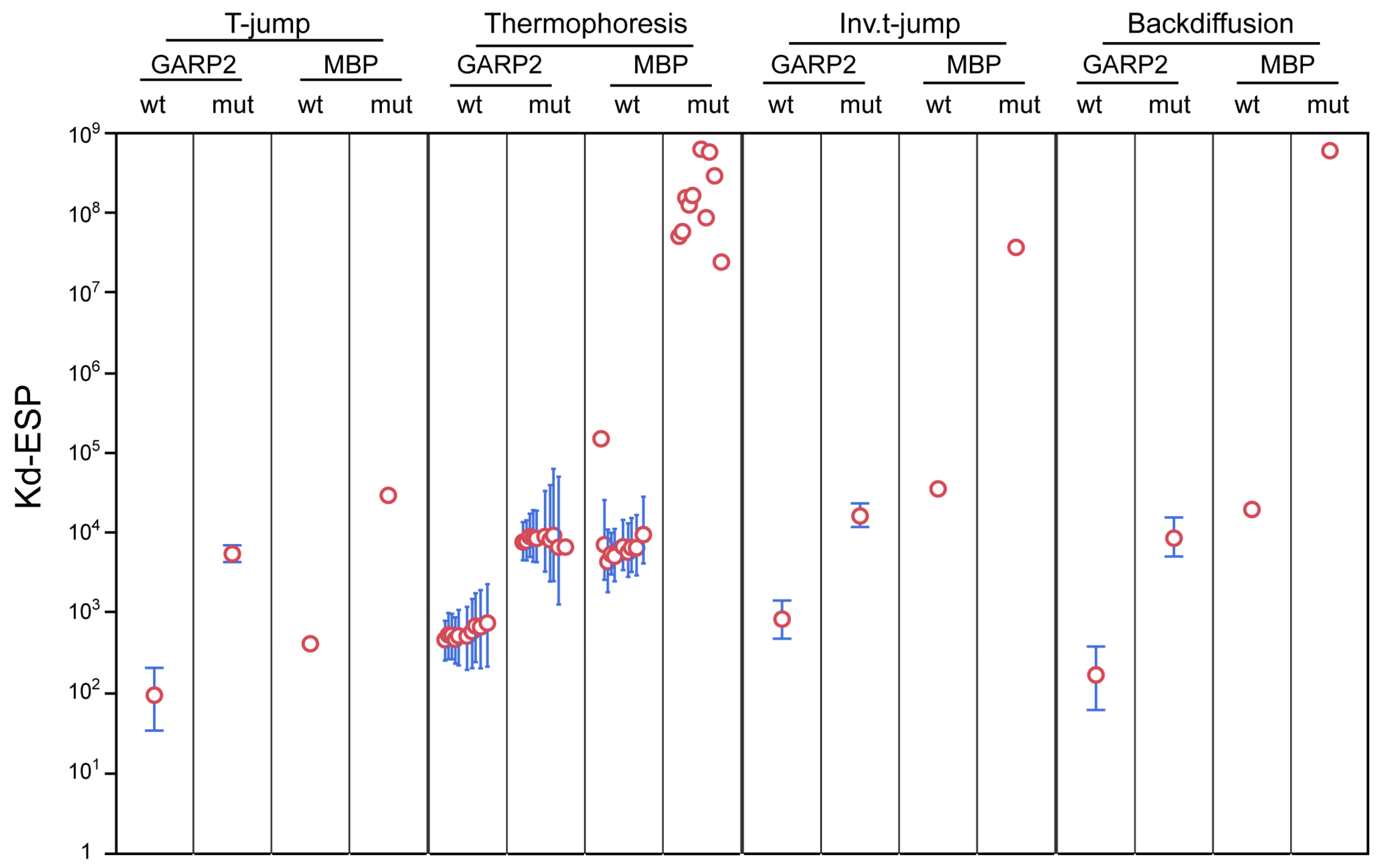
| Type | Fusion | Oligo | Regions | KD [bounds] (nM) | S.D. (nM) |
|---|---|---|---|---|---|
| T-jump | GARP2 | wt | 2/1 | 93.3 [33.9, 203.5] | 40.3 |
| mut | 5422.9 [4271.8, 6928.7] | 661.5 | |||
| MBP | wt | 406.2 [Un, Un] | 764.9 | ||
| mut | 29,278.0 [2846.3, Un] | 73167.8 | |||
| Thermo | GARP2 | wt | 41/3 | 455.8 [250.9, 787.3] | 131.5 |
| wt | 42/3 | 525.6 [259.5, 988.2] | 178.3 | ||
| wt | 43/3 | 519.6 [260.4, 960.9] | 172.4 | ||
| wt | 44/3 | 462.5 [229.7, 868.7] | 163.0 | ||
| wt | 45/3 | 510.1 [218.2, 1077.2] | 211.7 | ||
| wt | 46/3 | 506.0 [192.7, 1173.8] | 245.8 | ||
| wt | 47/3 | 578.5 [202.0, 1474.6] | 316.9 | ||
| wt | 48/3 | 679.3 [240.8, 1742.8] | 375.6 | ||
| wt | 49/3 | 660.3 [200.6, 1912.2] | 419.4 | ||
| wt | 410/3 | 738.2 [211.1, 2267.5] | 504.6 | ||
| GARP2 | mut | 41/3 | 7621.0 [4510.1, 13,538.4] | 2179.6 | |
| mut | 42/3 | 7820.1 [4494.1, 14,274.6] | 2247.5 | ||
| mut | 43/3 | 8941.7 [4960.4, 17,313.3] | 2733.3 | ||
| mut | 44/3 | 8617.7 [4314.9, 19,073.8] | 3035.9 | ||
| mut | 45/3 | 8456.3 [4257.3, 18,824.4] | 3061.7 | ||
| mut | 46/3 | 8938.1 [3245.4, 33,206.7] | 4792.4 | ||
| mut | 47/3 | 8121.0 [2446.3, 39,503.1] | 5003.2 | ||
| mut | 48/3 | 9239.0 [2472.7, 62,858.3] | 6153.9 | ||
| mut | 49/3 | 6562.0 [1260.5, 50,038.4] | 4654.2 | ||
| mut | 410/3 | 6602.7 [1179.3, Un] | 5150.7 | ||
| MBP | wt | 41/3 | 149,761.5 [19,415.2, Un] | 1,116,236.4 | |
| wt | 42/3 | 7064.6 [2568.2, 25,579.5] | 3852.5 | ||
| wt | 43/3 | 4315.4 [1794.4, 10,952.0] | 1826.6 | ||
| wt | 44/3 | 5387.0 [2993.7, 9992.5] | 1583.2 | ||
| wt | 45/3 | 5063.2 [2455.4, 11,136.9] | 1878.1 | ||
| wt | 46/3 | 6648.7 [3395.5, 14,432.7] | 2424.9 | ||
| wt | 47/3 | 5672.9 [2791.3, 13,055.6] | 2399.1 | ||
| wt | 48/3 | 6505.9 [3207.5, 15,237.1] | 2870.7 | ||
| wt | 49/3 | 6462.8 [2914.2, 16,616.1] | 3089.9 | ||
| wt | 410/3 | 9453.1 [4116.0, 28,162.2] | 4808.7 | ||
| MBP | mut | 41/3 | 51,212,374.7 [Un, Un] | 9,394,498.2 | |
| mut | 42/3 | 58,673,041.9 [Un, Un] | 10,990,771.3 | ||
| mut | 43/3 | 155,534,044.7 [Un, Un] | 14,545,170.7 | ||
| mut | 44/3 | 126,324,766.5 [Un, Un] | 26,533,955.0 | ||
| mut | 45/3 | 166,306,517.3 [Un, Un] | 7,086,615.9 | ||
| mut | 46/3 | 630,007,997.6 [Un, Un] | 3,188,549.4 | ||
| mut | 47/3 | 87,588,060.8 [Un, Un] | 17,699,022.8 | ||
| mut | 48/3 | 581,508,613.5 [Un, Un] | 3,533,413.5 | ||
| mut | 49/3 | 293,033,378.5 [Un, Un] | 5,205,908.5 | ||
| mut | 410/3 | 24,422,340.4 [Un, Un] | 51,603,646.7 | ||
| Inv.t-jump | GARP2 | wt | 6/5 | 826.5 [471.3, 1412.8] | 222.3 |
| mut | 16,158.1 [11,753.7, 23,231.6] | 2872.7 | |||
| MBP | wt | 35,248.3 [721.8, Un] | 94,944.9 | ||
| mut | 37,243,141.3 [Un, Un] | 2,581,304.9 | |||
| Backdiff | GARP2 | wt | 8/7 | 166.4 [61.0, 374.4] | 74.3 |
| mut | 8516.4 [5019.5, 15,514.5] | 2569.0 | |||
| MBP | wt | 19,483.5 [3759.8, Un] | 17,709.6 | ||
| mut | 604,733,167.6 [Un, Un] | 1,193,501.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rieger, J.; Fitz, M.; Fischer, S.M.; Wallmeroth, N.; Flores-Romero, H.; Fischer, N.M.; Brand, L.H.; García-Sáez, A.J.; Berendzen, K.W.; Mira-Rodado, V. Exploring the Binding Affinity of the ARR2 GARP DNA Binding Domain via Comparative Methods. Genes 2023, 14, 1638. https://doi.org/10.3390/genes14081638
Rieger J, Fitz M, Fischer SM, Wallmeroth N, Flores-Romero H, Fischer NM, Brand LH, García-Sáez AJ, Berendzen KW, Mira-Rodado V. Exploring the Binding Affinity of the ARR2 GARP DNA Binding Domain via Comparative Methods. Genes. 2023; 14(8):1638. https://doi.org/10.3390/genes14081638
Chicago/Turabian StyleRieger, Janine, Michael Fitz, Stefan Markus Fischer, Niklas Wallmeroth, Hector Flores-Romero, Nina Monika Fischer, Luise Helene Brand, Ana J. García-Sáez, Kenneth Wayne Berendzen, and Virtudes Mira-Rodado. 2023. "Exploring the Binding Affinity of the ARR2 GARP DNA Binding Domain via Comparative Methods" Genes 14, no. 8: 1638. https://doi.org/10.3390/genes14081638
APA StyleRieger, J., Fitz, M., Fischer, S. M., Wallmeroth, N., Flores-Romero, H., Fischer, N. M., Brand, L. H., García-Sáez, A. J., Berendzen, K. W., & Mira-Rodado, V. (2023). Exploring the Binding Affinity of the ARR2 GARP DNA Binding Domain via Comparative Methods. Genes, 14(8), 1638. https://doi.org/10.3390/genes14081638





