Bi-Directional Interactions between Glucose-Lowering Medications and Gut Microbiome in Patients with Type 2 Diabetes Mellitus: A Systematic Review
Abstract
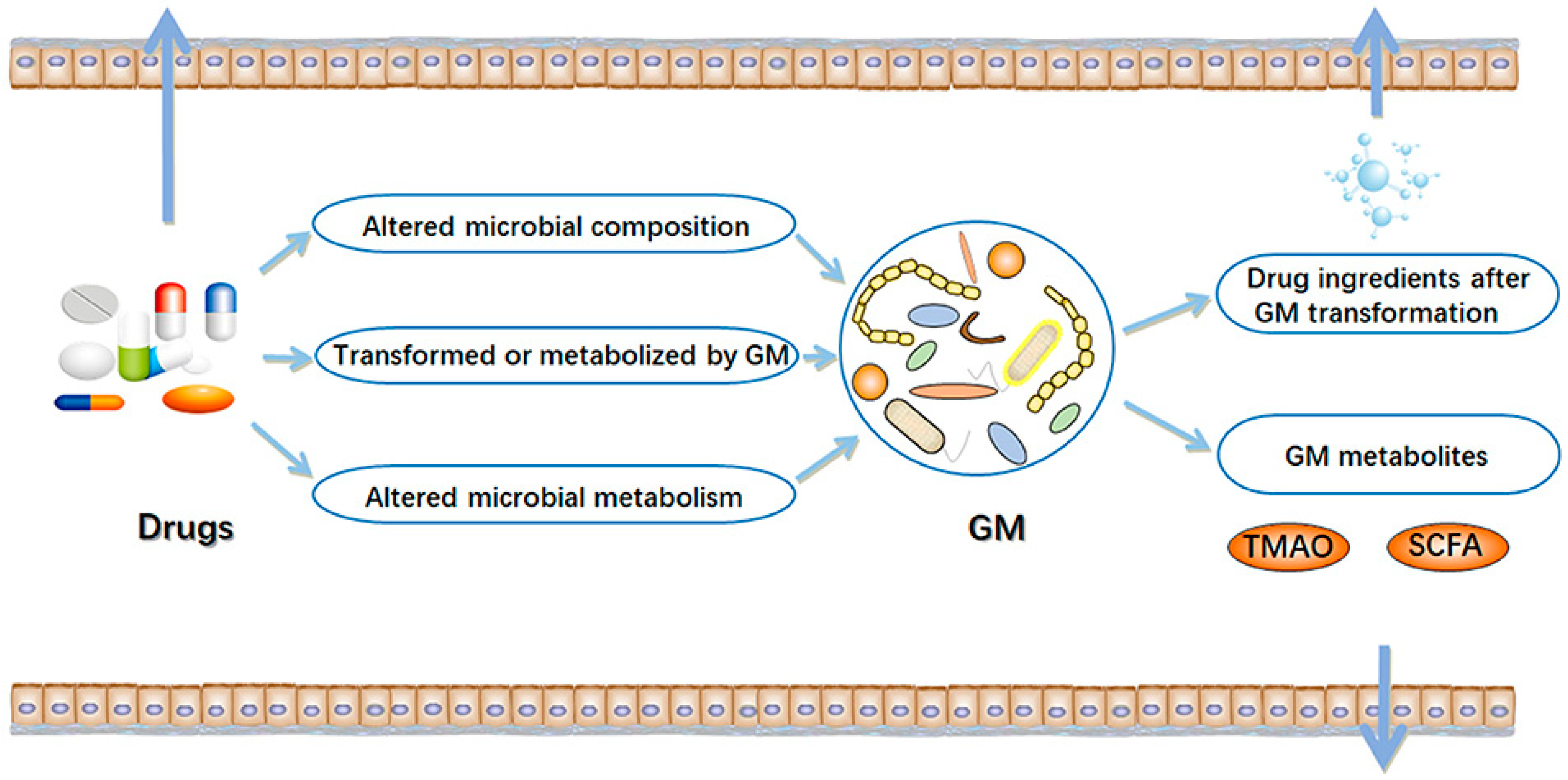
1. Introduction
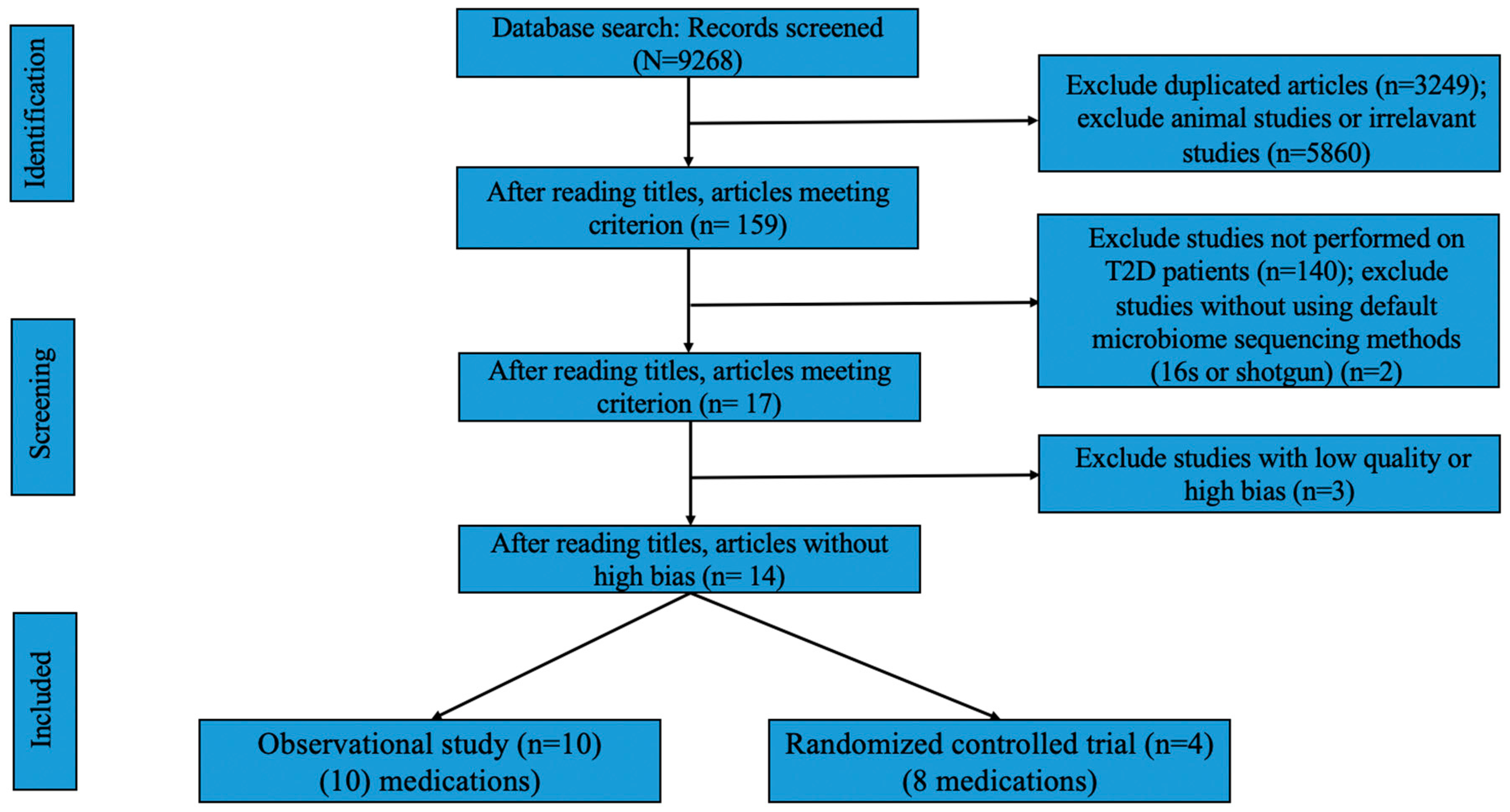
2. Methods
2.1. Search Strategy for the Systematic Review
2.2. Inclusion and Exclusion Criterion
2.3. Data Extraction
2.4. Quality and Risk of Bias Assessment
3. Results
3.1. Effects of Glucose-Lowering Medication on Gut Microbiome Composition
3.1.1. Metformin
Results from Observational Studies
Results from RCTs
3.1.2. α-Glucosidase Inhibitors (α-GI)
Results from Observational Studies
Results from RCTs
3.1.3. Glucagon-Like Peptide-1 Receptor Agonist (GLP-1RA)
Results from Observational Studies
3.1.4. Sulfonylureas
Results from RCTs
3.1.5. SGLT2 Inhibitor
Results from RCTs
3.1.6. Dipeptidyl Peptidase 4 Inhibitor
Results from RCTs
3.2. Effects of the Gut Microbiome on Glycemic Control
3.2.1. Metformin
Results from Observational Studies
Results from RCTs
3.2.2. Other Drugs
α-Glucosidase Inhibitor and Dipeptidyl Peptidase 4 Inhibitor
SGLT2 Inhibitor and Sulfonylureas
4. Discussion
4.1. Effects of Glucose-Lowering Drugs on Gut Microbiome Composition
4.1.1. Metformin
4.1.2. Other Drugs
4.2. Effects of the Gut Microbiome on Glycemic Control
4.3. Summary and Conclusions
5. Limitations, Clinical Implications, and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Maskarinec, G.; Raquinio, P.; Kristal, B.S.; Setiawan, V.W.; Wilkens, L.R.; Franke, A.A.; Lim, U.; Le Marchand, L.; Randolph, T.W.; Lampe, J.W.; et al. The gut microbiome and type 2 diabetes status in the Multiethnic Cohort. PLoS ONE 2021, 16, e0250855. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Radjabzadeh, D.; Chen, L.; Kurilshikov, A.; Kavousi, M.; Ahmadizar, F.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Association of Insulin Resistance and Type 2 Diabetes With Gut Microbial Diversity: A Microbiome-Wide Analysis from Population Studies. JAMA Netw. Open 2021, 4, e2118811. [Google Scholar] [CrossRef]
- Huda, M.N.; Kim, M.; Bennett, B.J. Modulating the Microbiota as a Therapeutic Intervention for Type 2 Diabetes. Front. Endocrinol. 2021, 12, 632335. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, R.; Kurilshikov, A.; Vila, A.V.; Sinha, T.; Klaassen, M.A.Y.; Bolte, L.A.; Andreu-Sánchez, S.; Chen, L.; Collij, V.; Hu, S.; et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 2022, 604, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Kim, H.G.; Kim, J.S.; Oh, D.G.; Um, Y.J.; Seo, C.S.; Han, J.W.; Cho, H.J.; Kim, G.H.; Jeong, T.C.; et al. The effect of gut microbiota on drug metabolism. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1295–1308. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Jia, W. The influence of gut microbiota on drug metabolism and toxicity. Expert Opin. Drug Metab. Toxicol. 2016, 12, 31–40. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, J.; Wang, R. Gut microbiota modulates drug pharmacokinetics. Drug Metab. Rev. 2018, 50, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Whang, A.; Nagpal, R.; Yadav, H. Bi-directional drug-microbiome interactions of anti-diabetics. EBioMedicine 2019, 39, 591–602. [Google Scholar] [CrossRef]
- Ahmad, T.; Li, G.; Wang, J.; Li, M.; Xiao, Y.; Yu, X.; Zheng, Y.; Moosa, A.; Nie, C.; Liu, Y. First Report of Bacterial Leaf Spot of Ficus benghalensis Caused by Pseudomonas cichorii in Pakistan. Plant Dis. 2022, 107, 552. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; the PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, M.; Yang, J.; Xu, Q.; Liang, C.; Chen, B.; Zhang, J.; Yang, Y.; Wang, H.; Shang, Y.; et al. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 2019, 66, 485–493. [Google Scholar] [CrossRef]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Krogh Pedersen, H.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velásquez-Mejía, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin Is Associated with Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef]
- Napolitano, A.; Miller, S.; Nicholls, A.W.; Baker, D.; Van Horn, S.; Thomas, E.; Rajpal, D.; Spivak, A.; Brown, J.R.; Nunez, D.J. Novel Gut-Based Pharmacology of Metformin in Patients with Type 2 Diabetes Mellitus. PLoS ONE 2014, 9, e100778. [Google Scholar] [CrossRef]
- Elbere, I.; Silamikelis, I.; Dindune, I.I.; Kalnina, I.; Ustinova, M.; Zaharenko, L.; Silamikele, L.; Rovite, V.; Gudra, D.; Konrade, I.; et al. Baseline gut microbiome composition predicts metformin therapy short-term efficacy in newly diagnosed type 2 diabetes patients. PLoS ONE 2020, 15, e0241338. [Google Scholar] [CrossRef]
- Nakajima, H.; Takewaki, F.; Hashimoto, Y.; Kajiyama, S.; Majima, S.; Okada, H.; Senmaru, T.; Ushigome, E.; Nakanishi, N.; Hamaguchi, M.; et al. The Effects of Metformin on the Gut Microbiota of Patients with Type 2 Diabetes: A Two-Center, Quasi-Experimental Study. Life 2020, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Xu, J.; Lian, F.; Yu, X.; Zhao, Y.; Xu, L.; Zhang, M.; Zhao, X.; Shen, J.; Wu, S.; et al. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: A Multicenter, Randomized, Open Label Clinical Trial. mBio 2018, 9, e02392-17. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.M.; Serino, M.; Planas-Fèlix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wang, X.; Li, J.; Zhang, Y.; Zhong, H.; Liu, R.; Zhang, D.; Feng, Q.; Xie, X.; Hong, J.; et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat. Commun. 2017, 8, 1785. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Liu, F.; Zhang, B.; Dong, K.; Lu, M.; Jiang, R.; Xu, Y.; Diao, L.; Zhao, J.; Tang, H. Liraglutide-induced structural modulation of the gut microbiota in patients with type 2 diabetes mellitus. PeerJ 2021, 9, e11128. [Google Scholar] [CrossRef]
- van Bommel, E.J.M.; Herrema, H.; Davids, M.; Kramer, M.H.H.; Nieuwdorp, M.; van Raalte, D.H. Effects of 12-week treatment with dapagliflozin and gliclazide on faecal microbiome: Results of a double-blind randomized trial in patients with type 2 diabetes. Diabetes Metab. 2020, 46, 164–168. [Google Scholar] [CrossRef]
- Wang, Z.; Saha, S.; Van Horn, S.; Thomas, E.; Traini, C.; Sathe, G.; Rajpal, D.K.; Brown, J.R. Gut microbiome differences between metformin- and liraglutide-treated T2DM subjects. Endocrinol. Diabetes Metab. 2018, 1, e00009. [Google Scholar] [CrossRef]
- Almugadam, B.S.; Liu, Y.; Chen, S.-M.; Wang, C.-H.; Shao, C.-Y.; Ren, B.-W.; Tang, L. Alterations of Gut Microbiota in Type 2 Diabetes Individuals and the Confounding Effect of Antidiabetic Agents. J. Diabetes Res. 2020, 2020, 7253978. [Google Scholar] [CrossRef]
- Takewaki, F.; Nakajima, H.; Takewaki, D.; Hashimoto, Y.; Majima, S.; Okada, H.; Senmaru, T.; Ushigome, E.; Hamaguchi, M.; Yamazaki, M.; et al. Habitual Dietary Intake Affects the Altered Pattern of Gut Microbiome by Acarbose in Patients with Type 2 Diabetes. Nutrients 2021, 13, 2107. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, H.; Zhao, C.; Shi, Z.; Qiu, L.; Yang, F.; Zhou, X.; Han, X.; Wu, K.; Zhong, H.; et al. Metagenomic analysis reveals crosstalk between gut microbiota and glucose-lowering drugs targeting the gastrointestinal tract in Chinese patients with type 2 diabetes: A 6 month, two-arm randomised trial. Diabetologia 2022, 65, 1613–1626. [Google Scholar] [CrossRef]
- He, D.; Han, H.; Fu, X.; Liu, A.; Zhan, Y.; Qiu, H.; Ma, L.; Zhang, X.; Wang, X. Metformin Reduces Blood Glucose in Treatment-Naive Type 2 Diabetes by Altering the Gut Microbiome. Can. J. Diabetes 2022, 46, 150–156. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.-Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.M.; Buse, J.B.; Davidson, M.B.; Ferrannini, E.; Holman, R.R.; Sherwin, R.; Zinman, B. Medical management of hyperglycemia in type 2 diabetes: A consensus algorithm for the initiation and adjustment of therapy: A consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care 2009, 32, 193–203. [Google Scholar] [CrossRef]
- Pernicova, I.; Korbonits, M. Metformin—Mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014, 10, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Foretz, M.; Hébrard, S.; Leclerc, J.; Zarrinpashneh, E.; Soty, M.; Mithieux, G.; Sakamoto, K.; Andreelli, F.; Viollet, B. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Investig. 2010, 120, 2355–2369. [Google Scholar] [CrossRef] [PubMed]
- Madiraju, A.K.; Erion, D.M.; Rahimi, Y.; Zhang, X.-M.; Braddock, D.T.; Albright, R.A.; Prigaro, B.J.; Wood, J.L.; Bhanot, S.; MacDonald, M.J.; et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 2014, 510, 542–546. [Google Scholar] [CrossRef]
- Miller, R.A.; Chu, Q.; Xie, J.; Foretz, M.; Viollet, B.; Birnbaum, M.J. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 2013, 494, 256–260. [Google Scholar] [CrossRef]
- Guo, G.L.; Xie, W. Metformin action through the microbiome and bile acids. Nat. Med. 2018, 24, 1789–1790. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhang, H.; Lu, Y. Molecular Mechanisms of Metformin for Diabetes and Cancer Treatment. Front. Physiol. 2018, 9, 1039. [Google Scholar] [CrossRef]
- Li, R.; Medina-Gomez, C.; Rivadeneira, F. Down-to-Earth Studies of the Gut Microbiome in Bone Health and Disease. J. Bone Miner. Res. 2022, 37, 595–596. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Chen, C.Y.; Knox, N.C.; Marrie, R.A.; El-Gabalawy, H.; de Kievit, T.; Alfa, M.; Bernstein, C.N.; Van Domselaar, G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 2018, 6, 221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yun, Y.; An, H.; Zhao, W.; Ma, T.; Wang, Z.; Yang, F. Gut Microbiome Composition Associated With Major Depressive Disorder and Sleep Quality. Front. Psychiatry 2021, 12, 645045. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Xiao, Y.; Wu, R.T.; Xie, D.; Zhao, H.H.; Shen, G.Y.; Wu, E.Q. Comparative analysis of type 2 diabetes-associated gut microbiota between Han and Mongolian people. J. Microbiol. 2021, 59, 693–701. [Google Scholar] [CrossRef]
- Nah, G.; Park, S.-C.; Kim, K.; Kim, S.; Park, J.; Lee, S.; Won, S. Type-2 Diabetics Reduces Spatial Variation of Microbiome Based on Extracellular Vesicles from Gut Microbes across Human Body. Sci. Rep. 2019, 9, 20136. [Google Scholar] [CrossRef]
- Lam, Y.Y.; Ha, C.W.Y.; Campbell, C.R.; Mitchell, A.J.; Dinudom, A.; Oscarsson, J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Increased Gut Permeability and Microbiota Change Associate with Mesenteric Fat Inflammation and Metabolic Dysfunction in Diet-Induced Obese Mice. PLoS ONE 2012, 7, e34233. [Google Scholar] [CrossRef]
- O’Neill, I.; Schofield, Z.; Hall, L.J. Exploring the role of the microbiota member Bifidobacterium in modulating immune-linked diseases. Emerg. Top. Life Sci. 2017, 1, 333–349. [Google Scholar] [CrossRef]
- Pinzone, M.R.; Celesia, B.M.; Di Rosa, M.; Cacopardo, B.; Nunnari, G. Microbial translocation in chronic liver diseases. Int. J. Microbiol. 2012, 2012, 694629. [Google Scholar] [CrossRef] [PubMed]
- Le, T.K.; Hosaka, T.; Nguyen, T.T.; Kassu, A.; Dang, T.O.; Tran, H.B.; Pham, T.P.; Tran, Q.B.; Le, T.H.; Pham, X.D. Bifidobacterium species lower serum glucose, increase expressions of insulin signaling proteins, and improve adipokine profile in diabetic mice. Biomed. Res. 2015, 36, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, S.; Araoka, R.; Kajihara, Y.; Ito, T.; Miyamoto, H.; Kodama, H. Valerate production by Megasphaera elsdenii isolated from pig feces. J. Biosci. Bioeng. 2018, 125, 519–524. [Google Scholar] [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Firouzi, S.; Majid, H.A.; Ismail, A.; Kamaruddin, N.A.; Barakatun-Nisak, M.-Y. Effect of multi-strain probiotics (multi-strain microbial cell preparation) on glycemic control and other diabetes-related outcomes in people with type 2 diabetes: A randomized controlled trial. Eur. J. Nutr. 2017, 56, 1535–1550. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Xiao, X.; Yu, S.; Huang, W.; Rao, B.; Chen, F. A Single Strain of Lactobacillus (CGMCC 21661) Exhibits Stable Glucose-and Lipid-Lowering Effects by Regulating Gut Microbiota. Nutrients 2023, 15, 670. [Google Scholar] [CrossRef]
- Allin, K.H.; Nielsen, T.; Pedersen, O. Mechanisms in endocrinology: Gut microbiota in patients with type 2 diabetes mellitus. Eur. J. Endocrinol. 2015, 172, R167–R177. [Google Scholar] [CrossRef]
- Bornet, E.; Westermann, A.J. The ambivalent role of Bacteroides in enteric infections. Trends Microbiol. 2022, 30, 104–108. [Google Scholar] [CrossRef]
- Clostridium-Wikipedia. Available online: https://en.wikipedia.org/wiki/Clostridium (accessed on 4 March 2023).
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef] [PubMed]
- Utzschneider, K.M.; Kratz, M.; Damman, C.J.; Hullar, M. Mechanisms Linking the Gut Microbiome and Glucose Metabolism. J. Clin. Endocrinol. Metab. 2016, 101, 1445–1454. [Google Scholar] [CrossRef]
- Zhou, Y.-D.; Liang, F.-X.; Tian, H.-R.; Luo, D.; Wang, Y.-Y.; Yang, S.-R. Mechanisms of gut microbiota-immune-host interaction on glucose regulation in type 2 diabetes. Front. Microbiol. 2023, 14, 1121695. [Google Scholar] [CrossRef]
- Zhang, F.; Qiu, L.; Xu, X.; Liu, Z.; Zhan, H.; Tao, X.; Shah, N.P.; Wei, H. Beneficial effects of probiotic cholesterol-lowering strain of Enterococcus faecium WEFA23 from infants on diet-induced metabolic syndrome in rats. J. Dairy Sci. 2017, 100, 1618–1628. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, J.; Hou, J.; Dong, Y.; Lu, Y.; Jin, L.; Cao, R.; Li, T.; Wu, J. Oral administration of recombinant Lactococcus lactis expressing HSP65 and tandemly repeated P277 reduces the incidence of type I diabetes in non-obese diabetic mice. PLoS ONE 2014, 9, e105701. [Google Scholar] [CrossRef]
- Zeng, Z.; Yu, R.; Zuo, F.; Zhang, B.; Ma, H.; Chen, S. Recombinant Lactococcus lactis expressing bioactive exendin-4 to promote insulin secretion and beta-cell proliferation in vitro. Appl. Microbiol. Biotechnol. 2017, 101, 7177–7186. [Google Scholar] [CrossRef]
- Yamashita, M.; Okubo, H.; Kobuke, K.; Ohno, H.; Oki, K.; Yoneda, M.; Tanaka, J.; Hattori, N. Alteration of gut microbiota by a Westernized lifestyle and its correlation with insulin resistance in non-diabetic Japanese men. J. Diabetes Investig. 2019, 10, 1463–1470. [Google Scholar] [CrossRef]
- Walter, J.; Martínez, I.; Rose, D.J. Holobiont nutrition: Considering the role of the gastrointestinal microbiota in the health benefits of whole grains. Gut Microbes 2013, 4, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Hosomi, K.; Saito, M.; Park, J.; Murakami, H.; Shibata, N.; Ando, M.; Nagatake, T.; Konishi, K.; Ohno, H.; Tanisawa, K.; et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun. 2022, 13, 4477. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Doumatey, A.P.; Adeyemo, A.; Zhou, J.; Lei, L.; Adebamowo, S.N.; Adebamowo, C.; Rotimi, C.N. Gut Microbiome Profiles Are Associated with Type 2 Diabetes in Urban Africans. Front. Cell. Infect. Microbiol. 2020, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Toejing, P.; Khampithum, N.; Sirilun, S.; Chaiyasut, C.; Lailerd, N. Influence of Lactobacillus paracasei HII01 Supplementation on Glycemia and Inflammatory Biomarkers in Type 2 Diabetes: A Randomized Clinical Trial. Foods 2021, 10, 1455. [Google Scholar] [CrossRef]
- Bock, P.M.; Telo, G.H.; Ramalho, R.; Sbaraini, M.; Leivas, G.; Martins, A.F.; Schaan, B.D. The effect of probiotics, prebiotics or synbiotics on metabolic outcomes in individuals with diabetes: A systematic review and meta-analysis. Diabetologia 2021, 64, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Pham, N.H.T.; Joglekar, M.V.; Wong, W.K.M.; Nassif, N.T.; Simpson, A.M.; Hardikar, A.A. The role of short-chain fatty acids on insulin sensitivity: A systematic review and meta-analysis. Nat. Rev. 2023, nuad042. [Google Scholar] [CrossRef]
| Source [Reference] | Study Type | Intervention Group (Sample Size) | Control Group (Sample Size) | Follow-Up | R | D | Mi | Me | S | Co | O |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang et al. [17] | Observational study | T2D patients with metformin treatment (n = 51) | T2D patients without metformin treatment (n = 26) | >3 months | - | - | |||||
| Forslund et al. [18] | Observational study | T2D patients with metformin treatment (n = 93) | T2D patients without metformin treatment (n = 106) | NA | - | - | |||||
| Cuesta-Zuluaga et al. [19] | Observational study | T2D patients with metformin treatment (n = 14) | T2D patients without metformin treatment (n = 14) | NA | - | - | |||||
| Sun et al. [20] | Observational study | T2D patients with metformin treatment (n = 30) | The same patients before treatment | 3 days | - | - | |||||
| Napolitano et al. [21] | Observational study | T2D patients with metformin treatment (n = 14) | The same patients before treatment | 3 months | - | - | |||||
| Elbere et al. [14] | Observational study | T2D patients with metformin treatment (n = 50) | The same patients before treatment | 7 days | - | - | |||||
| Nakajima et al. [22] | Observational study | T2D patients with metformin treatment (n = 21) | The same patients before treatment | 4 weeks | - | - | |||||
| Zhang et al. [17] | Observational study | T2D patients with α-GI treatment (n = 17) | T2D patients without α-GI treatment (n = 26) | >3 months | - | - | |||||
| Shang et al. [25] | Observational study | T2D patients with GLP-1RA (Liraglutide) treatment (n = 40) | The same patients before treatment | 4 months | - | - | |||||
| Takewaki et al. [29] | Observational study | T2D patients with α-GI treatment (n = 18) | The same patients before treatment | 4 weeks | - | - | |||||
| Tong et al. [15] | RCT | T2D patients with metformin treatment(n = 100) | The same patients before treatment | 12 weeks | |||||||
| Wu et al. [23] | RCT | T2D patients with metformin treatment (n = 22) | T2D patients with placebo treatment (n = 18) | 4 months | |||||||
| Gu et al. [24] | RCT | T2D patients with α-GI treatment (n = 51) | The same patients before treatment | 3 months | |||||||
| Gu et al. [24] | RCT | T2D patients with Sulfonylureas treatment (n = 43) | The same patients before treatment | 3 months | |||||||
| Bommel EJM et al. [26] | RCT | T2D patients with Sulfonylureas treatment (n = 20) | The same patients before treatment | 12 weeks | |||||||
| Bommel EJM et al. [26] | RCT | T2D patients with SGLT2 Inhibitor treatment (n = 24) | The same patients before treatment | 12 weeks | |||||||
| Zhang et al. [30] | RCT | T2D patients with α-GI treatment (n = 44) | The same patients before treatment | 6 months | |||||||
| Zhang et al. [30] | RCT | T2D patients with DPP-4 inhibitor treatment (n = 48) | The same patients before treatment | 6 months |
| Glucose Lowering Drug | Increased Taxa | Decreased Taxa | α Diversity | Study Design [Reference] |
|---|---|---|---|---|
| Metformin | Spirochaete and Turicibacter | Erysipelotrichi and Ruminococcus | No effect | Observational study [17] |
| Escherichia spp. | Intestinibacter spp. | NA | Observational study [18] | |
| Prevotella, Megasphaera | Oscillospira, Barnesiellaceae, Clostridiaceae 02d06 | NA | Observational study [19] | |
| NA | Bacteroides fragilis (B. fragilis) | ↓ | Observational study [20] | |
| o__Bifidobacteriales, f__Bifidobacteriaceae, g__Bifidobacterium, s__Bifidobacterium_adolescentis, t__Bifidobacterium_adolescentis_unclassified, g__Barnesiella, s__Barnesiella_intestinihominis, s__Clostridium_bartlettii | f__Clostridiaceae, g__Lactococcus, g__Clostridium, s__Parabacteroides_distasonis, t__Parabacteroides_distasonis_unclassified, s__Lactococcus_lactis, t__Lactococcus_lactis_unclassified, f__Oscillospiraceae, g__Oscillibacter, s__Oscillibacter_unclassified, s__Enterococcus_faecium, t__Enterococcus_faecium_unclassified, s__Bacteroides_vulgatus, t__Bacteroides_vulgatus_unclassified, f__Enterococcaceae, and g__Enterococcusbacteriales | No effect | Observational study [14] | |
| NA | Firmicutes, Firmicutes/Bacteroidetes ratio | No effect | Observational study [22] | |
| Clostridium XIVa, Erysipelotrichaceae incertae sedis, Escherichia-Shigella, Fusobacterium, Flavonifractor, Clostridium XVIII and IV, Blautia spp. and Anaerostipes | Bacteroides, Parabacteroides, Alistipes, Oscillibacter, and un-Ruminococcaceae | ↑ | RCT [15] | |
| γ-Proteobacteria, Escherichia coli, and Firmicutes | Intestinibacter | NA | RCT [23] | |
| α-glucosidase inhibitor | Bifidobacterium and Lactobacillus | NA | No effect | Observational study [15] |
| Actinobacteria, Bifidobacterium, Eubacterium, Megasphaera, and Lactobacillus | Bacteroidetes, Bacteroides, Blautia, Prevotella, Clostridium, Phascolarctobacterium, and Lachnoclostridium | No effect | Observational study [29] | |
| Lactobacillus and Bifidobacterium | Bacteroides, Alistipes and Clostridium | ↓ | RCT [24] | |
| Bifidobacterium, Lactobacillus, Solobacterium, Streptococcus, Actinomyces, Acidaminococcus, Megasphaera, Veillonella, Haemophilus, Granulicatella, Collinsella, Gemella, Anaerostipes, Rothia, Enterococcus, and 30 species | Bacteroides, Roseburia, Alistipes, Bilophila, Oscillibacter, Parabacteroides, Clostridium, Odoribacter, Holdemania, Adlercreutzia, Barnesiella, Flavonifractor, Subdoligranulum, Ruminococcus, Oxalobacter, Parasutterella, Anaerotruncus, Akkermansia, and 46 species | ↓ | RCT [30] | |
| Liraglutide/ GLP-1RA | Streptococcaceae, Bacilli, Verrucomicrobia, Coriobacteriia, Coriobacteriaceae, Collinsella, Akkermansia, Verrucomicrobiaceae, Coriobacteriales, Lactobacillales, Verrucomicrobiae, Clostridium, Clostridiaceae, Verrucomicrobiales, Actinobacteria | Acinetobacter, Oscillospira, Desulfovibrionales, S24_7, Fusobacteriaceae, Rikenellaceae, Pseudomonadaceae, Pseudomonadales, Desulfovibrionaceae, Acidaminococcus, Fusobacteriales, Succinatimonas, Deltaproteobacteria, Fusobacteriia, Moraxellaceae, Megamonas, Alistipes, Fusobacteria, Fusobacterium, Megasphaera | ↓ | Observational study [25] |
| Sulfonylu-reas | No significant changes | No effect | RCT [24,26] | |
| SGLT2 Inhibitor | No significant changes | No effect | RCT [26] | |
| Dipeptidyl peptidase 4 inhibitor | Bifidobacterium, and 2 species (Clostridium bartlettii, and Bifidobacterium adolescentis) | Paraprevotella, Fusobacterium, Parabacteroides, Bacteroides, and 8 species | No effect | RCT [30] |
| Glucose Lowering Drug | Taxa Positively Correlate with Glycemic Control | Taxa Negatively Correlate with Glycemic Control | If α Diversity Influences Glycemic Control | Study Design [Reference] |
|---|---|---|---|---|
| Metformin | E. faecium, Lactococcus lactis, Odoribacter, and Dialister | Prevotella copri | No | Observational study [14] |
| Blautia, Anaerostipes, Clostridium XIVa, Erysipelotrichaceae incertae sedis, Escherichia-Shigella, Fusobacterium, Flavonifractor, Lachnospiraceae, Lachnospiracea incertae sedis, Clostridium XVIII and IV | Bacteroides, Parabacteroides, Alistipes, Oscillibacter, and un-Ruminococcaceae | NA | RCT [15] | |
| SGLT2 Inhibitor | Not associated | Not associated | NA | RCT [26] |
| Sulfonylureas | Not associated | Not associated | NA | RCT [26] |
| α-glucosidase inhibitor | Not significant | Not significant | NA | RCT [30] |
| Dipeptidyl peptidase 4 inhibitor | Not significant | Not significant | NA | RCT [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Shokri, F.; Rincon, A.L.; Rivadeneira, F.; Medina-Gomez, C.; Ahmadizar, F. Bi-Directional Interactions between Glucose-Lowering Medications and Gut Microbiome in Patients with Type 2 Diabetes Mellitus: A Systematic Review. Genes 2023, 14, 1572. https://doi.org/10.3390/genes14081572
Li R, Shokri F, Rincon AL, Rivadeneira F, Medina-Gomez C, Ahmadizar F. Bi-Directional Interactions between Glucose-Lowering Medications and Gut Microbiome in Patients with Type 2 Diabetes Mellitus: A Systematic Review. Genes. 2023; 14(8):1572. https://doi.org/10.3390/genes14081572
Chicago/Turabian StyleLi, Ruolin, Fereshteh Shokri, Alejandro Lopez Rincon, Fernando Rivadeneira, Carolina Medina-Gomez, and Fariba Ahmadizar. 2023. "Bi-Directional Interactions between Glucose-Lowering Medications and Gut Microbiome in Patients with Type 2 Diabetes Mellitus: A Systematic Review" Genes 14, no. 8: 1572. https://doi.org/10.3390/genes14081572
APA StyleLi, R., Shokri, F., Rincon, A. L., Rivadeneira, F., Medina-Gomez, C., & Ahmadizar, F. (2023). Bi-Directional Interactions between Glucose-Lowering Medications and Gut Microbiome in Patients with Type 2 Diabetes Mellitus: A Systematic Review. Genes, 14(8), 1572. https://doi.org/10.3390/genes14081572







