Aicardi Syndrome Is a Genetically Heterogeneous Disorder
Abstract
1. Introduction
2. Materials and Methods
2.1. Human and Animal Ethics
2.2. Exome and Genome Sequencing
2.3. Candidate Variant Prioritization and Filtering
2.4. Structural Variants
2.5. Molecular Modelling of Missense Variants
2.6. Plasmids and Cloning
2.7. Cell Culture and Transfection
2.8. TOPflash Dual Luciferase Assay
2.9. Protein Extraction
2.10. Western Blot
2.11. Zebrafish Morphant Analyses
2.12. Fetal Mouse Histology
3. Results
3.1. Phenotypic Descriptions of Individuals Diagnosed with AIC
3.2. Exome and Genome Sequencing Excludes X-Linked Candidate Genes
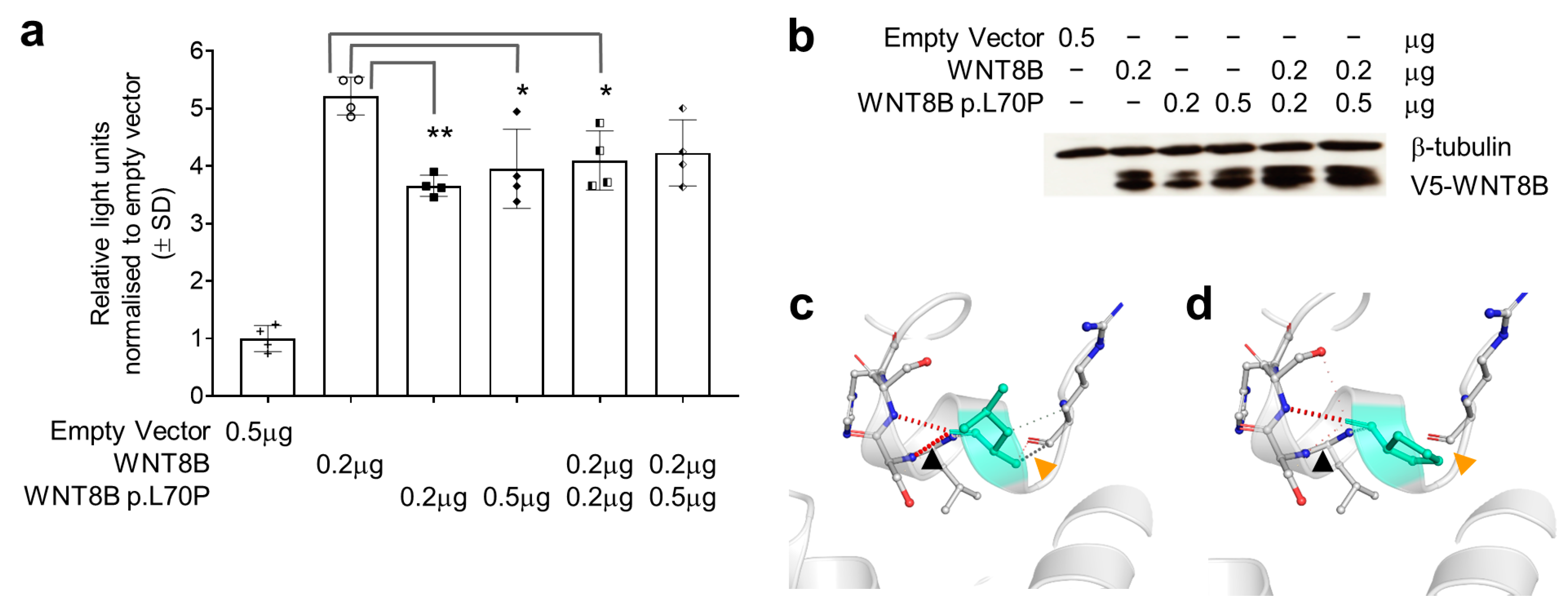
3.3. The WNT8B p.Leu70Pro Variant Has a Dominant Negative Effect on Wnt Signalling
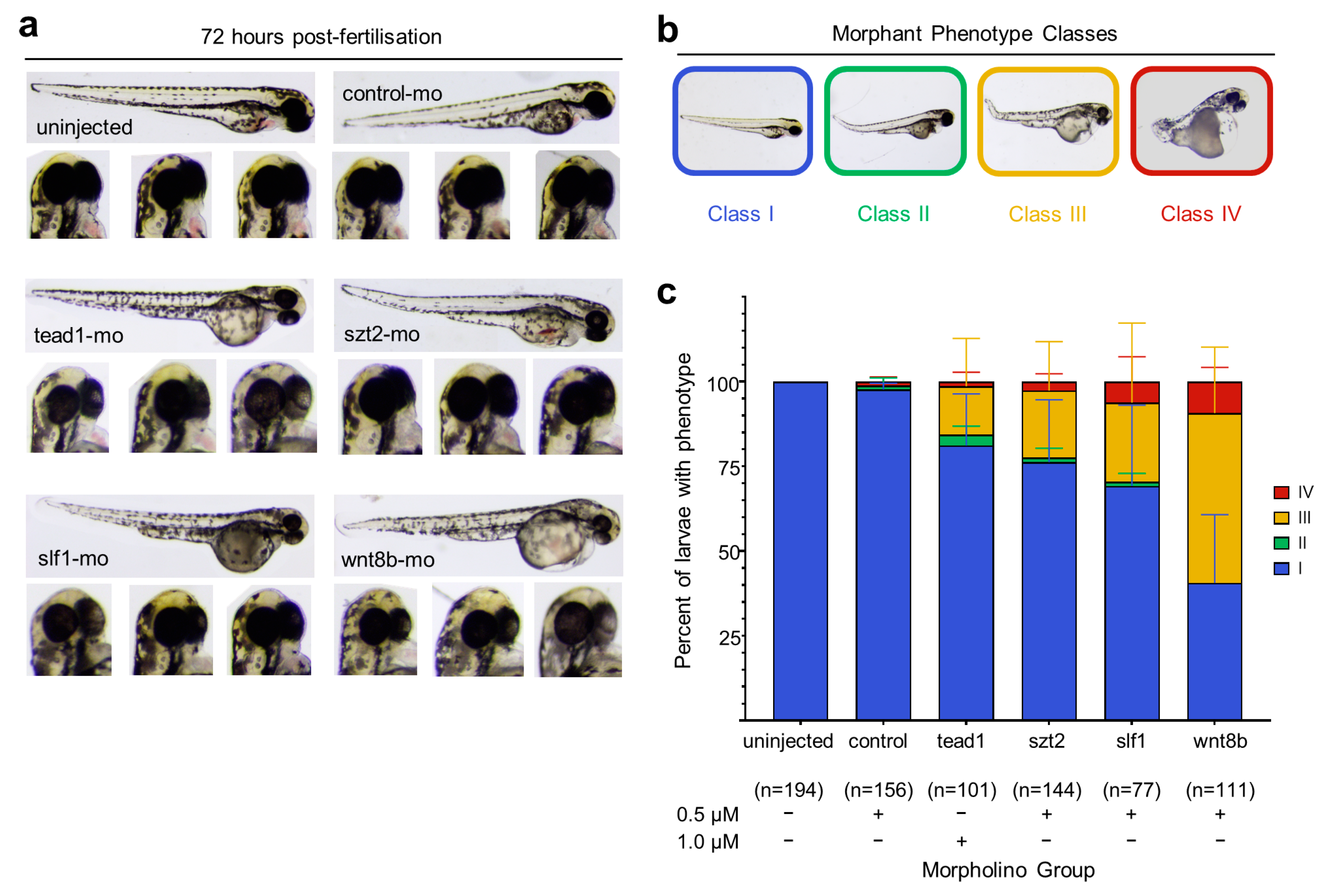
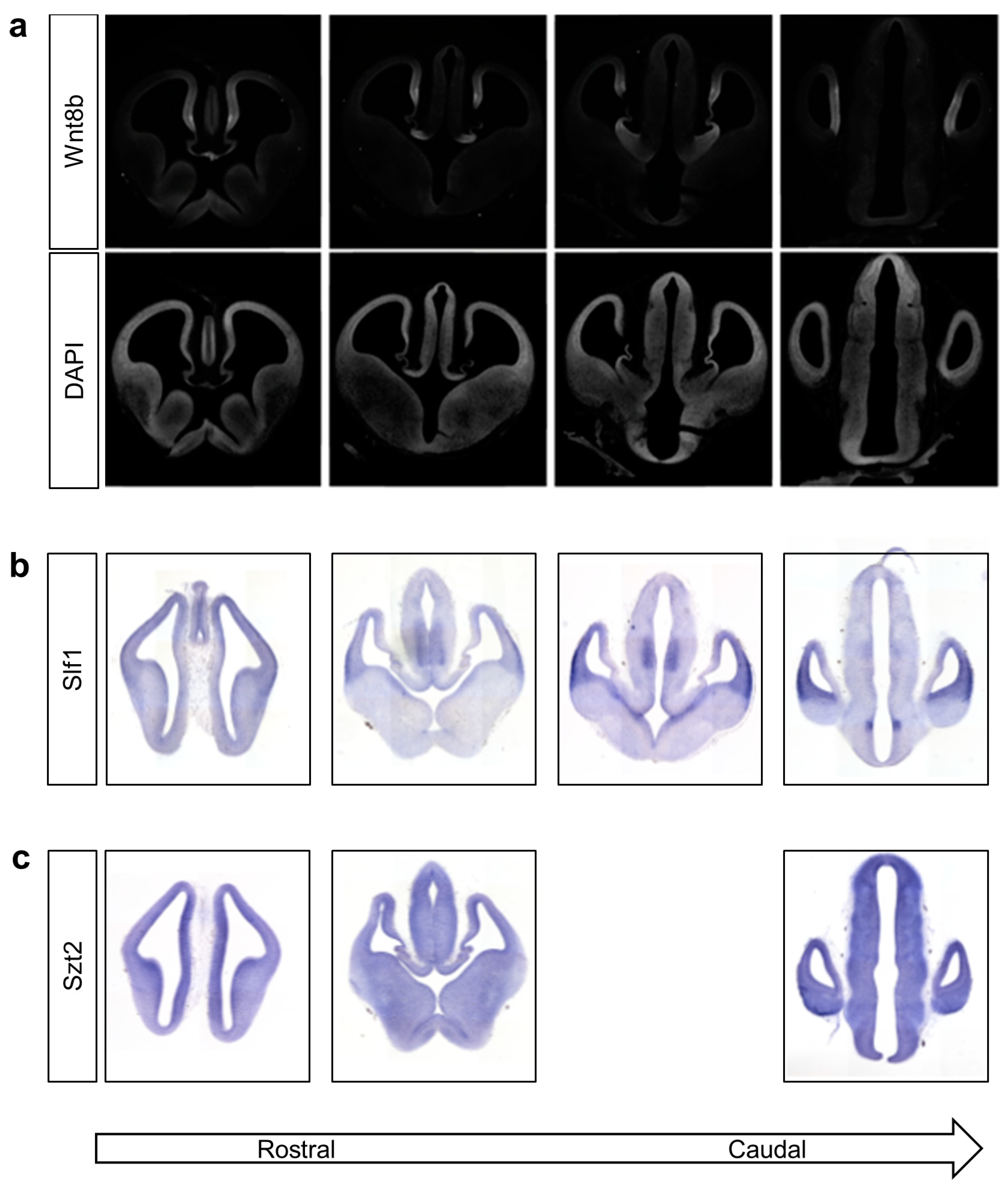
3.4. Knockdown of wnt8b and slf1 in Zebrafish Leads to Altered Eye and Body Development
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kroner, B.L.; Preiss, L.R.; Ardini, M.-A.; Gaillard, W.D. New Incidence, Prevalence, and Survival of Aicardi Syndrome from 408 Cases. J. Child. Neurol. 2008, 23, 531–535. [Google Scholar] [CrossRef]
- Aicardi, J. Aicardi Syndrome. Brain Dev. 2005, 27, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Fruhman, G.; Eble, T.N.; Gambhir, N.; Sutton, V.R.; Van den Veyver, I.B.; Lewis, R.A. Ophthalmologic Findings in Aicardi Syndrome. J. AAPOS 2012, 16, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Masnada, S.; Pichiecchio, A.; Formica, M.; Arrigoni, F.; Borrelli, P.; Accorsi, P.; Bonanni, P.; Borgatti, R.; Bernardina, B.D.; Danieli, A.; et al. Basal Ganglia Dysmorphism in Patients with Aicardi Syndrome. Neurology 2021, 96, e1319–e1333. [Google Scholar] [CrossRef]
- Neidich, J.A.; Nussbaum, R.L.; Packer, R.J.; Emanuel, B.S.; Puck, J.M. Heterogeneity of Clinical Severity and Molecular Lesions in Aicardi Syndrome. J. Pediatr. 1990, 116, 911–917. [Google Scholar] [CrossRef]
- Nielsen, K.B.; Anvret, M.; Flodmark, O.; Furuskog, P.; Bohman-Valis, K. Aicardi Syndrome: Early Neuroradiological Manifestations and Results of DNA Studies in One Patient. Am. J. Med. Genet. 1991, 38, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Hoag, H.M.; Taylor, S.A.; Duncan, A.M.; Khalifa, M.M. Evidence That Skewed X Inactivation Is Not Needed for the Phenotypic Expression of Aicardi Syndrome. Hum. Genet. 1997, 100, 459–464. [Google Scholar] [CrossRef]
- Yilmaz, S.; Fontaine, H.; Brochet, K.; Grégoire, M.-J.; Devignes, M.-D.; Schaff, J.-L.; Philippe, C.; Nemos, C.; McGregor, J.L.; Jonveaux, P. Screening of Subtle Copy Number Changes in Aicardi Syndrome Patients with a High Resolution X Chromosome Array-CGH. Eur. J. Med. Genet. 2007, 50, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Eble, T.N.; Sutton, V.R.; Sangi-Haghpeykar, H.; Wang, X.; Jin, W.; Lewis, R.A.; Fang, P.; Van den Veyver, I.B. Non-Random X Chromosome Inactivation in Aicardi Syndrome. Hum. Genet. 2009, 125, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Pai, C.-P.; Wang, C.-Y.; Kuo, Y.-T.; Liang, J.-S. Aicardi Syndrome without CDKL5 Gene Mutation. J. Exp. Clin. Med. 2013, 5, 81–82. [Google Scholar] [CrossRef]
- Hopkins, I.J.; Humphrey, I.; Keith, C.G.; Susman, M.; Webb, G.C.; Turner, E.K. The Aicardi Syndrome in a 47, XXY Male. Aust. Paediatr. J. 1979, 15, 278–280. [Google Scholar] [CrossRef]
- Chen, T.-H.; Chao, M.-C.; Lin, L.-C.; Jong, Y.-J.; Yang, S.N.; Lai, Y.-H.; Chen, H.-L. Aicardi Syndrome in a 47, XXY Male Neonate with Lissencephaly and Holoprosencephaly. J. Neurol. Sci. 2009, 278, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Zubairi, M.S.; Carter, R.F.; Ronen, G.M. A Male Phenotype with Aicardi Syndrome. J. Child. Neurol. 2009, 24, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Shetty, J.; Fraser, J.; Goudie, D.; Kirkpatrick, M. Aicardi Syndrome in a 47 XXY Male—A Variable Developmental Phenotype? Eur. J. Paediatr. Neurol. 2014, 18, 529–531. [Google Scholar] [CrossRef]
- Ropers, H.H.; Zuffardi, O.; Bianchi, E.; Tiepolo, L. Agenesis of Corpus Callosum, Ocular, and Skeletal Anomalies (X-Linked Dominant Aicardi’s Syndrome) in a Girl with Balanced X/3 Translocation. Hum. Genet. 1982, 61, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, T.; Momoi, M.; Miyamoto, S.; Kobayashi, S.; Kamoshita, S. Multi-Institutional Survey of the Aicardi Syndrome in Japan. Brain Dev. 1990, 12, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Donnenfeld, A.E.; Packer, R.J.; Zackai, E.H.; Chee, C.M.; Sellinger, B.; Emanuel, B.S. Clinical, Cytogenetic, and Pedigree Findings in 18 Cases of Aicardi Syndrome. Am. J. Med. Genet. 1989, 32, 461–467. [Google Scholar] [CrossRef]
- Kalscheuer, V.M.; Tao, J.; Donnelly, A.; Hollway, G.; Schwinger, E.; Kubart, S.; Menzel, C.; Hoeltzenbein, M.; Tommerup, N.; Eyre, H.; et al. Disruption of the Serine/Threonine Kinase 9 Gene Causes Severe X-Linked Infantile Spasms and Mental Retardation. Am. J. Hum. Genet. 2003, 72, 1401–1411. [Google Scholar] [CrossRef]
- Bursztejn, A.-C.; Bronner, M.; Peudenier, S.; Grégoire, M.-J.; Jonveaux, P.; Nemos, C. Molecular Characterization of a Monosomy 1p36 Presenting as an Aicardi Syndrome Phenocopy. Am. J. Med. Genet. A 2009, 149A, 2493–2500. [Google Scholar] [CrossRef]
- Prontera, P.; Bartocci, A.; Ottaviani, V.; Isidori, I.; Rogaia, D.; Ardisia, C.; Guercini, G.; Mencarelli, A.; Donti, E. Aicardi Syndrome Associated with Autosomal Genomic Imbalance: Coincidence or Evidence for Autosomal Inheritance with Sex-Limited Expression? Mol. Syndromol. 2013, 4, 197–202. [Google Scholar] [CrossRef]
- Broomall, E.; Renaud, D.; Ghadban, R.; Gavrilova, R.; Brodsky, M.C. Peripapillary Chorioretinal Lacunae in a Girl with 3q21.3 to 3q22.1 Microdeletion with Features of Aicardi Syndrome. JAMA Ophthalmol. 2013, 131, 1485–1487. [Google Scholar] [CrossRef]
- Spennato, P.; La Porta, A.; Varone, A.; Ruggiero, C.; Buono, S.; Cinalli, G. Aicardi and Turner Syndrome in a 45,X0/46,XX Female. Clin. Neurol. Neurosurg. 2013, 115, 820–822. [Google Scholar] [CrossRef] [PubMed]
- Schrauwen, I.; Szelinger, S.; Siniard, A.L.; Corneveaux, J.J.; Kurdoglu, A.; Richholt, R.; De Both, M.; Malenica, I.; Swaminathan, S.; Rangasamy, S.; et al. A De Novo Mutation in TEAD1 Causes Non-X-Linked Aicardi Syndrome. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3896–3904. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lund, C.; Striano, P.; Sorte, H.S.; Parisi, P.; Iacomino, M.; Sheng, Y.; Vigeland, M.D.; Øye, A.-M.; Møller, R.S.; Selmer, K.K.; et al. Exome Sequencing Fails to Identify the Genetic Cause of Aicardi Syndrome. Mol. Syndromol. 2016, 7, 234–238. [Google Scholar] [CrossRef]
- Paine, I.; Posey, J.E.; Grochowski, C.M.; Jhangiani, S.N.; Rosenheck, S.; Kleyner, R.; Marmorale, T.; Yoon, M.; Wang, K.; Robison, R.; et al. Paralog Studies Augment Gene Discovery: DDX and DHX Genes. Am. J. Hum. Genet. 2019, 105, 302–316. [Google Scholar] [CrossRef]
- Yavuz Saricay, L.; Hoyek, S.; Ashit Parikh, A.; Baldwin, G.; Bodamer, O.A.; Gonzalez, E.; Patel, N.A. A Case of Aicardi Syndrome Associated with Duplication Event of Xp22 Including SHOX. Ophthalmic Genet. 2023, 1–4. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 11, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Petrovski, S.; Wang, Q.; Heinzen, E.L.; Allen, A.S.; Goldstein, D.B. Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes. PLoS Genet. 2013, 9, e1003709. [Google Scholar] [CrossRef]
- Ha, T.T.; Sadleir, L.G.; Mandelstam, S.A.; Paterson, S.J.; Scheffer, I.E.; Gecz, J.; Corbett, M.A. A Mutation in COL4A2 Causes Autosomal Dominant Porencephaly with Cataracts. Am. J. Med. Genet. A 2016, 170A, 1059–1063. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef] [PubMed]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Keane, T.M.; Wong, K.; Adams, D.J. RetroSeq: Transposable Element Discovery from next-Generation Sequencing Data. Bioinformatics 2013, 29, 389–390. [Google Scholar] [CrossRef]
- Coe, B.P.; Witherspoon, K.; Rosenfeld, J.A.; van Bon, B.W.M.; Vulto-van Silfhout, A.T.; Bosco, P.; Friend, K.L.; Baker, C.; Buono, S.; Vissers, L.E.L.M.; et al. Refining Analyses of Copy Number Variation Identifies Specific Genes Associated with Developmental Delay. Nat. Genet. 2014, 46, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, D.I.; Wijmenga, C.; Slagboom, E.P.; Swertz, M.A.; Karssen, L.C.; Abdellaoui, A.; Ye, K.; Guryev, V.; Vermaat, M.; van Dijk, F.; et al. The Genome of The Netherlands: Design, and Project Goals. Eur. J. Hum. Genet. 2014, 22, 221–227. [Google Scholar] [CrossRef]
- Riggs, E.R.; Jackson, L.; Miller, D.T.; Van Vooren, S. Phenotypic Information in Genomic Variant Databases Enhances Clinical Care and Research: The International Standards for Cytogenomic Arrays Consortium Experience. Hum. Mutat. 2012, 33, 787–796. [Google Scholar] [CrossRef][Green Version]
- Deciphering Developmental Disorders Study Large-Scale Discovery of Novel Genetic Causes of Developmental Disorders. Nature 2015, 519, 223–228. [CrossRef]
- Huang, N.; Lee, I.; Marcotte, E.M.; Hurles, M.E. Characterising and Predicting Haploinsufficiency in the Human Genome. PLoS Genet. 2010, 6, e1001154. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the Impact of Mutations on Protein Conformation, Flexibility and Stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Hien, A.; Zhang, X.; Iranloye, O.; Virshup, D.M.; Waterman, M.L.; He, X. Disulfide Bond Requirements for Active Wnt Ligands. J. Biol. Chem. 2014, 289, 18122–18136. [Google Scholar] [CrossRef]
- Veeman, M.T.; Slusarski, D.C.; Kaykas, A.; Louie, S.H.; Moon, R.T. Zebrafish Prickle, a Modulator of Noncanonical Wnt/Fz Signaling, Regulates Gastrulation Movements. Curr. Biol. 2003, 13, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon: Eugene, OR, USA, 2000. [Google Scholar]
- Moldrich, R.X.; Gobius, I.; Pollak, T.; Zhang, J.; Ren, T.; Brown, L.; Mori, S.; De Juan Romero, C.; Britanova, O.; Tarabykin, V.; et al. Molecular Regulation of the Developing Commissural Plate. J. Comp. Neurol. 2010, 518, 3645–3661. [Google Scholar] [CrossRef]
- Zech, M.; Lam, D.D.; Winkelmann, J. Update on KMT2B-Related Dystonia. Curr. Neurol. Neurosci. Rep. 2019, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Abela, L.; Kurian, M.A. KMT2B-Related Dystonia. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Tsurusaki, Y.; Okamoto, N.; Ohashi, H.; Kosho, T.; Imai, Y.; Hibi-Ko, Y.; Kaname, T.; Naritomi, K.; Kawame, H.; Wakui, K.; et al. Mutations Affecting Components of the SWI/SNF Complex Cause Coffin-Siris Syndrome. Nat. Genet. 2012, 44, 376–378. [Google Scholar] [CrossRef] [PubMed]
- Dolzhenko, E.; van Vugt, J.J.F.A.; Shaw, R.J.; Bekritsky, M.A.; van Blitterswijk, M.; Narzisi, G.; Ajay, S.S.; Rajan, V.; Lajoie, B.R.; Johnson, N.H.; et al. Detection of Long Repeat Expansions from PCR-Free Whole-Genome Sequence Data. Genome Res. 2017, 27, 1895–1903. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Monjo, T.; Hoang, P.H.; Yoshimura, J.; Yurino, H.; Mitsui, J.; Ishiura, H.; Takahashi, Y.; Ichikawa, Y.; Goto, J.; et al. Rapid Detection of Expanded Short Tandem Repeats in Personal Genomics Using Hybrid Sequencing. Bioinformatics 2014, 30, 815–822. [Google Scholar] [CrossRef]
- Zook, J.M.; Catoe, D.; McDaniel, J.; Vang, L.; Spies, N.; Sidow, A.; Weng, Z.; Liu, Y.; Mason, C.E.; Alexander, N.; et al. Extensive Sequencing of Seven Human Genomes to Characterize Benchmark Reference Materials. Sci. Data 2016, 3, 160025. [Google Scholar] [CrossRef]
- Kim, S.-H.; Shin, J.; Park, H.-C.; Yeo, S.-Y.; Hong, S.-K.; Han, S.; Rhee, M.; Kim, C.-H.; Chitnis, A.B.; Huh, T.-L. Specification of an Anterior Neuroectoderm Patterning by Frizzled8a-Mediated Wnt8b Signalling during Late Gastrulation in Zebrafish. Development 2002, 129, 4443–4455. [Google Scholar] [CrossRef] [PubMed]
- Cavodeassi, F.; Carreira-Barbosa, F.; Young, R.M.; Concha, M.L.; Allende, M.L.; Houart, C.; Tada, M.; Wilson, S.W. Early Stages of Zebrafish Eye Formation Require the Coordinated Activity of Wnt11, Fz5, and the Wnt/β-Catenin Pathway. Neuron 2005, 47, 43–56. [Google Scholar] [CrossRef]
- Hofmeister, W.; Key, B. Frizzled-3a and Wnt-8b Genetically Interact during Forebrain Commissural Formation in Embryonic Zebrafish. Brain Res. 2013, 1506, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, W. Focus on Molecules: Wnt8b: A Suppressor of Early Eye and Retinal Progenitor Formation. Exp. Eye Res. 2012, 101, 113–114. [Google Scholar] [CrossRef]
- Kok, F.O.; Shin, M.; Ni, C.-W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef]
- Miesfeld, J.B.; Gestri, G.; Clark, B.S.; Flinn, M.A.; Poole, R.J.; Bader, J.R.; Besharse, J.C.; Wilson, S.W.; Link, B.A. Yap and Taz Regulate Retinal Pigment Epithelial Cell Fate. Development 2015, 142, 3021–3032. [Google Scholar] [CrossRef] [PubMed]
- Fotaki, V.; Price, D.J.; Mason, J.O. Wnt/β-Catenin Signaling Is Disrupted in the Extra-Toes (Gli3(Xt/Xt) ) Mutant from Early Stages of Forebrain Development, Concomitant with Anterior Neural Plate Patterning Defects. J. Comp. Neurol. 2011, 519, 1640–1657. [Google Scholar] [CrossRef]
- Snijders Blok, L.; Madsen, E.; Juusola, J.; Gilissen, C.; Baralle, D.; Reijnders, M.R.F.; Venselaar, H.; Helsmoortel, C.; Cho, M.T.; Hoischen, A.; et al. Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am. J. Hum. Genet. 2015, 97, 343–352. [Google Scholar] [CrossRef]
- Murtaza, M.; Jolly, L.A.; Gecz, J.; Wood, S.A. La FAM Fatale: USP9X in Development and Disease. Cell. Mol. Life Sci. 2015, 72, 2075–2089. [Google Scholar] [CrossRef]
- Grzeschik, K.-H.; Bornholdt, D.; Oeffner, F.; König, A.; del Carmen Boente, M.; Enders, H.; Fritz, B.; Hertl, M.; Grasshoff, U.; Höfling, K.; et al. Deficiency of PORCN, a Regulator of Wnt Signaling, Is Associated with Focal Dermal Hypoplasia. Nat. Genet. 2007, 39, 833–835. [Google Scholar] [CrossRef]
- Koth, M.L.; Garcia-Moreno, S.A.; Novak, A.; Holthusen, K.A.; Kothandapani, A.; Jiang, K.; Taketo, M.M.; Nicol, B.; Yao, H.H.-C.; Futtner, C.R.; et al. Canonical Wnt/β-Catenin Activity and Differential Epigenetic Marks Direct Sexually Dimorphic Regulation of Irx3 and Irx5 in Developing Mouse Gonads. Development 2020, 147, dev183814. [Google Scholar] [CrossRef]
- Lyraki, R.; Grabek, A.; Tison, A.; Weerasinghe Arachchige, L.C.; Peitzsch, M.; Bechmann, N.; Youssef, S.A.; de Bruin, A.; Bakker, E.R.M.; Claessens, F.; et al. Crosstalk between Androgen Receptor and WNT/β-Catenin Signaling Causes Sex-Specific Adrenocortical Hyperplasia in Mice. Dis. Models Mech. 2023, 16, dmm050053. [Google Scholar] [CrossRef]
- Kim, M.; Jho, E.-H. Cross-Talk between Wnt/β-Catenin and Hippo Signaling Pathways: A Brief Review. BMB Rep. 2014, 47, 540–545. [Google Scholar] [CrossRef]
- Gupta, P.D.; Johar, K.; Nagpal, K.; Vasavada, A.R. Sex Hormone Receptors in the Human Eye. Surv. Ophthalmol. 2005, 50, 274–284. [Google Scholar] [CrossRef]
- Wagner, H.; Fink, B.A.; Zadnik, K. Sex- and Gender-Based Differences in Healthy and Diseased Eyes. Optometry 2008, 79, 636–652. [Google Scholar] [CrossRef]
- Sobreira, N.; Schiettecatte, F.; Valle, D.; Hamosh, A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015, 36, 928–930. [Google Scholar] [CrossRef]
- Wang, X.; Sutton, V.R.; Eble, T.N.; Lewis, R.A.; Gunaratne, P.; Patel, A.; Van den Veyver, I.B. A Genome-Wide Screen for Copy Number Alterations in Aicardi Syndrome. Am. J. Med. Genet. A 2009, 149A, 2113–2121. [Google Scholar] [CrossRef][Green Version]
- Masnada, S.; Chiara, D.; Giana, I.; Manuela, F.; Marco, S.; Andrea, A.; Patrizia, A.; Nadia, B.-B.; Valeria, C.; Mara, C.; et al. Aicardi Syndrome: Key Fetal MRI Features and Prenatal Differential Diagnosis. Neuropediatrics 2020, 51, 276–285. [Google Scholar] [CrossRef]
- Pomar, L.; Ochoa, J.; Cabet, S.; Huisman, T.A.G.M.; Paladini, D.; Klaritsch, P.; Galmiche, A.; Prayer, F.; Gacio, S.; Haratz, K.; et al. Prenatal Diagnosis of Aicardi Syndrome Based on a Suggestive Imaging Pattern: A Multicenter Case-Series. Prenat. Diagn. 2022, 42, 484–494. [Google Scholar] [CrossRef]
| Genetic Variant [Reference] | Typical AIC Traits | Atypical AIC Traits | Diagnosis |
|---|---|---|---|
| 46,X,t(X;3)(p22;q12) [15] | Scoliosis, Rib anomalies, Reduced muscle tone, Bilateral ptosis, Microopthalmia, Severe DD | Symblepharon, Corneal Ulceration, Lagopthalmus, “Clods” of retinal pigment | Suspected |
| 46,XX,t(12;21)(q13.3;q11.2) [16] | CRL, Coloboma, ACC, Infantile spasm, Hypsarrhythmia | Definite | |
| Xp22.2pter Deletion and Partial 3p Trisomy [17] | Micropthalmia, Ventriculomegaly | Sclerocornea, Posterior hair whorl, Teratoma/lipoma in optic chiasm, Sharply demarcated chorioretinal defects, No ACC, Costovertebral anomalies, Infantile spasms or Intracranial heterotopias | Suspected |
| 46,X,t(X;7)(p22.3;p15) disrupting CDKL5 [18] | CC hypoplasia, Infantile spasm | Pale fundi, Microcephaly, Choreoform and Myoclonic dyskinesia | Suspected |
| 1p36pter Deletion [19] | Infantile spasm, bilateral papillary coloboma, ACC Ventricular dilatation, Delayed psychomotor milestone | Brachydactyly, Hypertrichosis, Deep set eyes, Posterior rotated ears | Phenocopy |
| 6q27qter Deletion (includes DLL1) and 12q24.32q24.33 Duplication [20] | Neurodevelopmental delay, Infantile spasm, Partial ACC, Colpocephaly, Ventriculomegaly, Gross cerebral asymmetry, Other cortical malformations, Coloboma, CRL, Scoliosis, Craniofacial abnormalities | Definite | |
| 3q21.3q22.1 Deletion [21] | Preaxial polydactyly, Muscular hypotonia, Abnormal corpus callosum, Chorioretinal lacunae, Partial ACC, Ventricular dilatation, Mild cortical thickening, Cavum septum pellucidum | Retromicrognathia, Atrial-septal defect, Mild pulmonary valvular stenosis | Suspected |
| 45,X0/46,XX [22] | Ventricular dilatation, Intraventricular cysts, Muscular hypotonia, Optic nerve coloboma, ACC, Intracranial cysts, Multiform epileptic seizures, Chorioretinal atrophy | Short neck, Pterigium colli, Teletelia, Barrel-shaped thorax, Lymphedema, Hemispondylia, Patent foramen ovale, Minimal persistent arterial duct, Hydrocephalus, Intracranial hypertension, Deafness | Aicardi and Turner Mosaic |
| NM_021961.5 (TEAD1):c.618G > A; p.Trp206Ter [23] | CRL, Infantile spasm, Cerebellar cysts, Periventricular heterotopias | Definite | |
| NM_024578.1 (OCEL1):c.499G > A; p.Ala167Thr [23] | Partial ACC, CRL, Infantile spasm, Posterior fossa arachnoid cyst | Definite | |
| NM_002518.4 (NPAS2):c.2465C > T; p.Pro822Leu [24] | CRL, Infantile spasm, ACC | Definite | |
| NM_032656.3 (DHX37):c.1145A > G; p.Asp382Gly [25] | CRL, Infantile spasm, ACC, Coloboma, Colpocephaly, Ventriculomegaly, Intracranial cysts, Polymicrogyria, Heterotopia, Severe DD and ID, Dysmorphic features | Definite | |
| Xp22.33 duplication (SHOX) [26] | CRL, Infantile spasm, Thin corpus callosum, Frontal polymicrogyria, Ventricular dysmorphism, Seizures | Definite |
| Proband ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Candidate variant | NM_003393.3 (WNT8B):c.209T > C:p.(Leu70Pro) | NM_032290.3 (SLF1): c.1409T > C: p.(Leu470Ser) | NM_015284.3 (SZT2): c.9103C > T: p.(His3035Tyr) | NM_014727.1 (KMT2B): c.6418C > G: p.(Pro2140Ala) | NM_003073.3(SMARCB1):c.1091_1093del | |||||
| Aicardi diagnosis | Classical | Classical | Classical | Classical | Classical | Likely | Likely | Suspected | Suspected | Suspected |
| Evidence for AIC diagnosis | ACC, CRL, Spasms | ACC, CRL, Spasms | pACC, CRL, Spasms | ACC, CRL, Spasms | ACC, CRL, Spasms | CRL, Spasms, Cortical Malformation (Mj), Intracranial cyst (Mj). Two classical, two major. | Spasms. Cortical Malformation (Mj) CPP (Mj), Optic disc coloboma (Mj), microphthalmia (min). One classical, three major, one minor. | CRL, Spasms. Cortical Malformation (Mj). Two classical, one major. | CRL, Spasms, CPP (Mj), Vertebral and costal abnormalities (Min). Two classical, one major, one minor | ACC, CRL, Scoliosis (min). Two classical, one minor. |
| Sex | F | F | F | F | F | F | F | F | F | F |
| Age at latest examination | 9 years | 17 years | 23 years | 11.5 years | 4 years | 8 years | 19 months (deceased) | 7 years | 23 years | 7.5 years |
| MRI | ACC and interhemispheric cyst; PNH; subcortical heterotopia; PMG | ACC; choroid plexus cysts; PNH | pACC | ACC | ACC; PMG; subcortical heterotopias; septated cyst beneath L cerebellar hemisphere | Thin CC; PMG; posterior fossa arachnoid cyst | Thin CC; cortical dysplasia; bilateral CPP | Thin CC; extensive cortical dysplasia | Bilateral CPP | Corpus callosum dysgenesis, |
| Ocular findings | Bilateral CRL, strabismus, nystagmus, astigmatism, hypermetropia | Bilateral CRL, left optic nerve coloboma | Bilateral CRL, exotropia, hypermetropia | CRL, peri-papillary chorioretinal atrophy | Bilateral CRL, left optic disc coloboma | CRL | Bilateral choroid-retinal abnormalities, bilateral optic disc coloboma, focal nodular retinal thickening on the left, bilateral microphthalmia, nystagmus and downward gaze | Unilateral CRL (left eye only) | Bilateral CRL, pigmented dystrophic right disc and mild tilt of left disc | Unilateral CRL (right eye only), high myopia |
| Intellectual disability | Profound (non-verbal) | Profound (non-verbal) | Profound: At 12 years; 4 words. Single words; 2 years | Profound (non-verbal) | Mild/moderate: 3 months fix and follow, 5 months—grabbing, babbling, 15 months no words, 17 months “dad” but not specific. | Severe: At 8 years; non-verbal | Profound. | Moderate; At 21 months, understood 5 words, said “ba” “ma”. 27 months, babbling but no words. 7 years: talking in sentences of 6–8 words, can read and write. | Mild | Severe 3 years: non-verbal, 7.5 years: Special school, one word only. |
| Motor | Non-ambulatory, spastic quadriplegia | Non-ambulatory, hypotonia | Walks with supervision. Sat 17 months, walked 4 years | Hypotonia. Minimal independent mobilisation, waddling gait, decreased tone and reflexes in upper limbs, paucity of movements | At 5 months rolling, 6 months delayed head control, rolling front to back but not back to front, 8 months pulling up, 17 months taking steps with walker. | At 2 years 8 months, not crawling, able to sit. At 8 years, able to walk assisted | At 10 months trying but unable to roll, can only lift head for a brief period | At 21 months, still learning to pass objects between hands, can pick up small objects but not hold a pencil correctly, she could throw a ball and take steps forward but not backwards nor kick a ball. Able to run at 7 years | Sat at 9 months. Walked at 15 months | At 3 years, sitting, not walking, rides a tricycle. At 7.5 years: commando crawling, dependent for all activities of daily living, not toilet trained. |
| Development prior to seizure onset | Abnormal | Abnormal | Abnormal | Abnormal | Normal | Normal | Delayed | Normal | Normal | N/A |
| Development after seizure onset | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed; | Delayed | Delayed | Mild delay | N/A |
| Autism Spectrum Disorder | No | No | No | No | No | No | No | Yes | No | No |
| Developmental Regression | Yes. Regressed with introduction of clonazepam at 5 months | Yes. Stopped rolling 18 months, lost single word at 2 years, further loss of motor milestones and interaction | Yes. Gait deteriorated at 19 years | Yes. Loss of smiling at seizure onset (2–3 months) | No | Yes at seizure onset | No | Yes. Anecdotally began to progress when she came off medication. | No | No |
| Seizure onset age | 2 weeks | 3.5 months | 4 years | 2 months | 5 months | 10 weeks | 5 months | 3 months | 6 months | N/A |
| Initial seizure type | Right facial clonic | Infantile spasms | Focal Status | Infantile spasms | Infantile spasms | Hemiclonic | Infantile spasms | Asymmetric infantile spasms | Predominantly right sided Flexion Spasms | N/A |
| Seizure types | Infantile spasms, FBTC, tonic seizure, focal motor seizures, >3 per week. | Infantile spasms, TCS, tonic seizures, myoclonic seizure, absence, atonic, hemiclonic | Infantile spasms (ongoing at 23 years), focal seizures, febrile seizure at 1 year | Infantile spasms, FBTC, TCS, focal seizures, myoclonic seizures, gelastic, SE, atonic | Infantile spasms, focal seizures | Infantile spasm, SE, hemiclonic, head drops | Infantile spasms, focal tonic. | Infantile spasms, tonic seizures | Infantile spasm, TCS, possible aura before the infantile spasms, SE, PNES | N/A |
| EEG | Multifocal discharges. Electroclinical focal seizure captured at left central region. | Multifocal discharges. Tonic and atonic seizures recorded | Bilateral fronto-temporal and fronto-central independent sharp and sharp-slow waves, predominantly from the left. Spasms captured. | Multifocal discharges, modified hypsarrhythmia. Tonic spasms recorded. | Epileptiform activity over left posterior-temporal region, modified hypsarrhythmia. Spasms recorded. | Predominant epileptiform activity from left frontal region, with generalised spread, hypsarrhythmia | Posterior spike/slow discharges, biposterior sharp activity and bursts of posterior fast activity. High amplitude delta slowing over bilateral posterior quadrants and infrequent sharp slow waves over the left occipital regions. Focal tonic seizure captured. | Left frontal and anterior temporal activity. Tonic, focal and spasms captured on EEG, modified Hypsarrhythmia | Focal discharges, most prominent in right posterior quadrant and left frontal regions. Bitemporal discharges. Generalised bursts of polyspike wave and irregular spike and slow wave. Spasms captured with associated with slow wave discharges. Hypsarrhythmia | Normal |
| Family history of seizures | No | Yes (maternal second cousin with well controlled TCS) | No, brother (who does not carry the SLF1 variant) with FCD and ID | No | No | No | No | No | No | No |
| Other | PEG, scoliosis, dental caries, drooling, hip dysplasia, pelvic deformity, microcephaly | PEG, scoliosis, failure to thrive, irritability, constipation, drooling, chronic pain, hip dislocation | Scoliosis, Becker-Nevus Syndrome, Poland Syndrome, clinodactyly, syndactyly, short forearm, hypopigmentation, skin tags, hirsute patches, facial dysmorphism, short stature, sleep apnea, torticollis, vertebral fusion | Asthma, dental caries, constipation, drooling, finger sucking, excoriation of her fingers and palms, facial features: short philtrum, upturned nasal tip and somewhat large pinnae. | Vertebrate abnormalities, torticollis, physio from 2 months. | Hemiparesis, hypotonia, nephrocalcinosis, hypermobile joints | Subdural shunt, naso-gastric tube | Hypotonia, mild facial dysmorphisms | Scoliosis, depression | Bilateral hip dysplasia, scoliosis, pectus excavatum, growth retardation, previous PEG, conductive hearing issues, delayed dentition, sparse scalp hair, hirsutism, thin upper lips, small ears, absent/hypoplastic nail 5th toes, 4th toe clinodactyly, 2nd toes override 3rd toes, 2× café au lait macules |
| Individual | Symbol | hg38 | RefSeq | cDNA Variant | Protein Variant | PolyPhen2 | CADD | GERP+ | gnomAD v2.1.1 | DynaMut ΔΔG (kJmol−1) | Zygosity | Inheritance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | SZT2 | chr1:43447156 | NM_015284.3 | c.9103C > T | p.His3035Tyr | 0.997 (D) | 18.18 | 5.43 | 0 | No available structure | Heterozygous | De novo |
| 3 | SLF1 | chr5:94665901 | NM_032290.3 | c.1409T > C | p.Leu470Ser | 0.946 (D) | 21.10 | 5.41 | 0 | −13.832 | Heterozygous | De novo |
| 1 | WNT8B | chr10:100479980 | NM_003393.3 | c.209 T > C | p.Leu70Pro | 0.962 (D) | 21.20 | 5.70 | 0 | −1.251 | Heterozygous | De novo |
| 9 | KMT2B | chr19:35732967 | NM_014727.1 | c.6418C > G | p.Pro2140Ala | 0.677(P) | 15.08 | 3.99 | 0 | No available structure | Heterozygous | De novo |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, T.T.; Burgess, R.; Newman, M.; Moey, C.; Mandelstam, S.A.; Gardner, A.E.; Ivancevic, A.M.; Pham, D.; Kumar, R.; Smith, N.; et al. Aicardi Syndrome Is a Genetically Heterogeneous Disorder. Genes 2023, 14, 1565. https://doi.org/10.3390/genes14081565
Ha TT, Burgess R, Newman M, Moey C, Mandelstam SA, Gardner AE, Ivancevic AM, Pham D, Kumar R, Smith N, et al. Aicardi Syndrome Is a Genetically Heterogeneous Disorder. Genes. 2023; 14(8):1565. https://doi.org/10.3390/genes14081565
Chicago/Turabian StyleHa, Thuong T., Rosemary Burgess, Morgan Newman, Ching Moey, Simone A. Mandelstam, Alison E. Gardner, Atma M. Ivancevic, Duyen Pham, Raman Kumar, Nicholas Smith, and et al. 2023. "Aicardi Syndrome Is a Genetically Heterogeneous Disorder" Genes 14, no. 8: 1565. https://doi.org/10.3390/genes14081565
APA StyleHa, T. T., Burgess, R., Newman, M., Moey, C., Mandelstam, S. A., Gardner, A. E., Ivancevic, A. M., Pham, D., Kumar, R., Smith, N., Patel, C., Malone, S., Ryan, M. M., Calvert, S., van Eyk, C. L., Lardelli, M., Berkovic, S. F., Leventer, R. J., Richards, L. J., ... Corbett, M. A. (2023). Aicardi Syndrome Is a Genetically Heterogeneous Disorder. Genes, 14(8), 1565. https://doi.org/10.3390/genes14081565







