Genome-Wide Identification and Characterization of the Phytochrome Gene Family in Peanut
Abstract
1. Introduction
2. Materials and Methods
2.1. Prediction of PHY Genes in Peanut
2.2. Sequence Structure Analysis
2.3. Chromosomal Localization and Syntenic Analysis
2.4. Cis-Elements Prediction and Expression Pattern Analysis
3. Results
3.1. Identification of the PHY Gene Family in Peanut
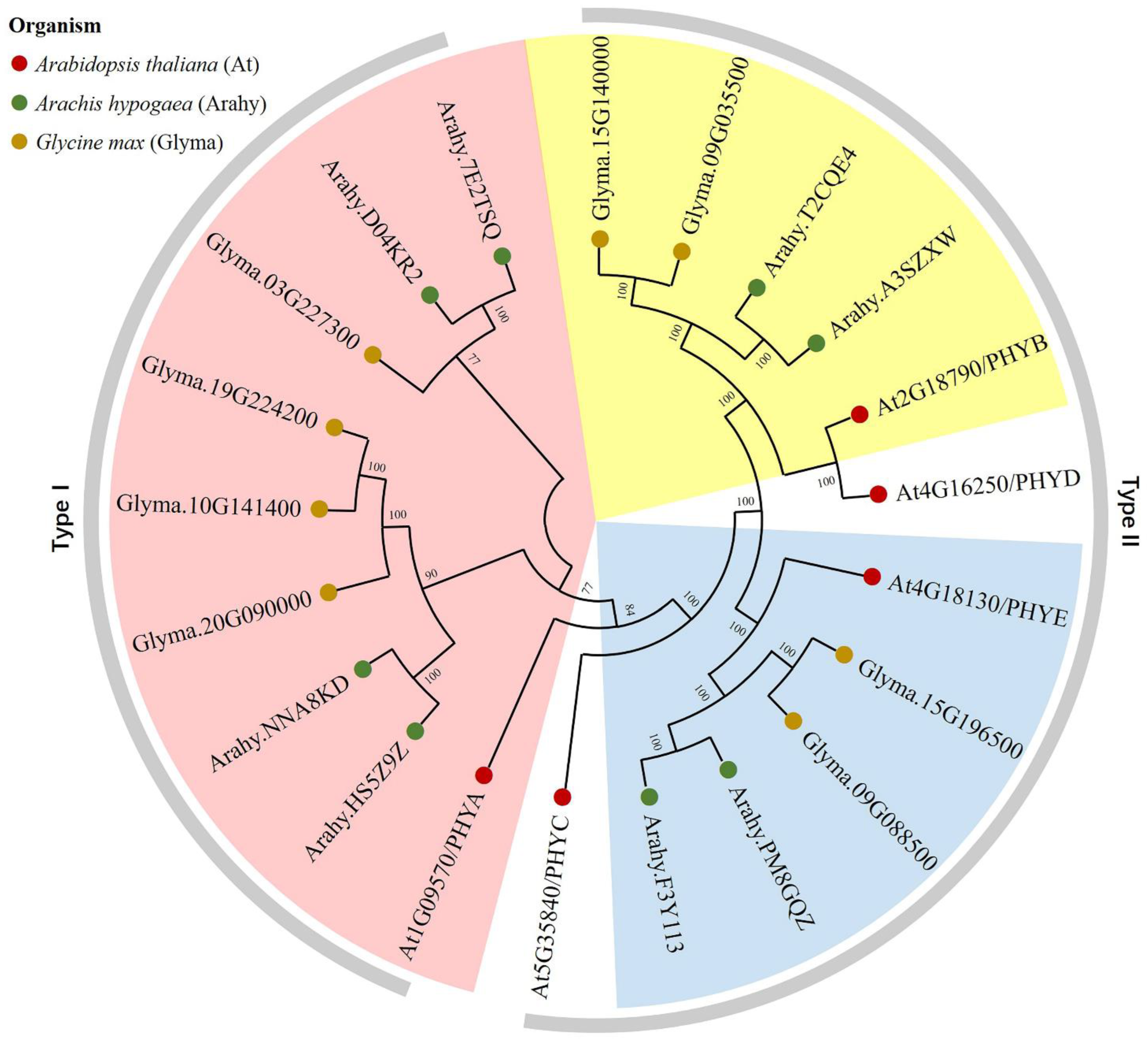
3.2. Phylogenetic Analysis and Classification of AhPHY Genes
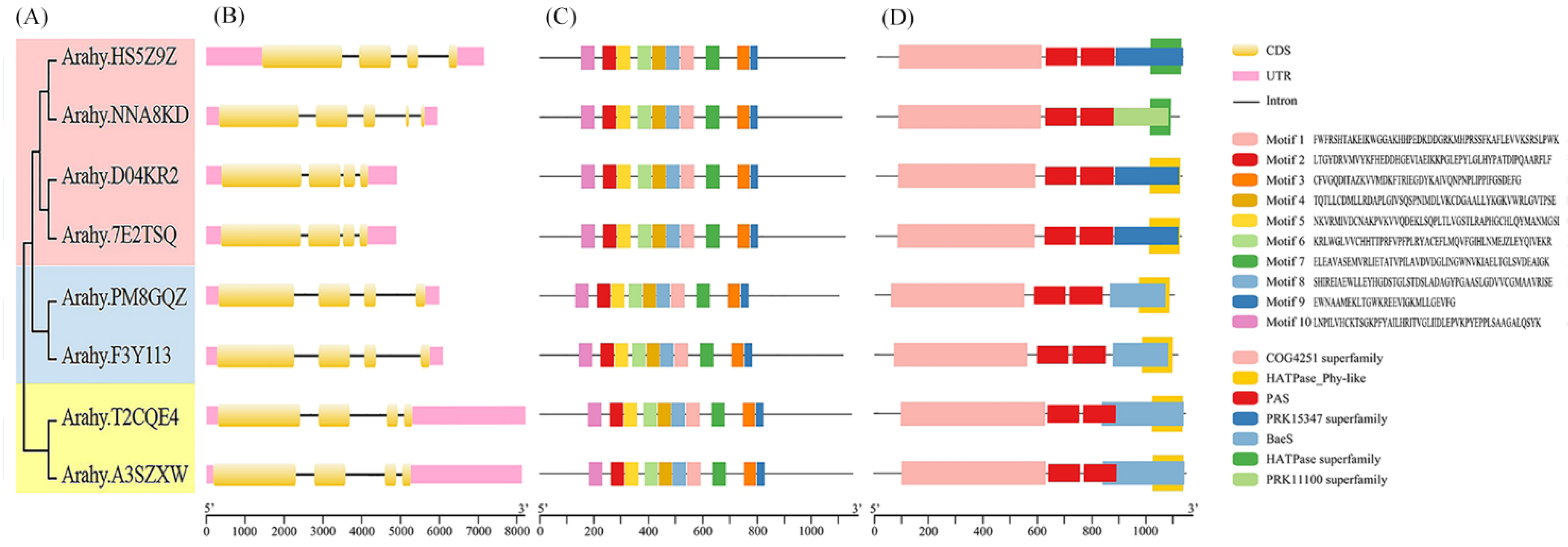
3.3. Conserved Motif, Domain, and Structure Analysis
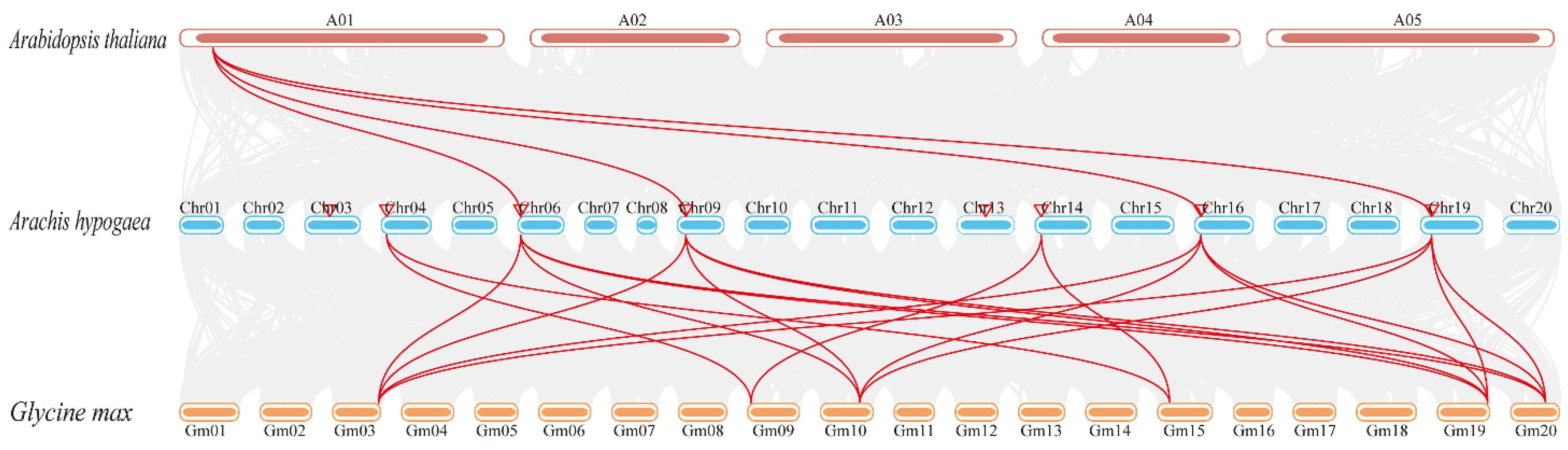
3.4. Genome Distribution and Syntenic Analysis of AhPHY Genes
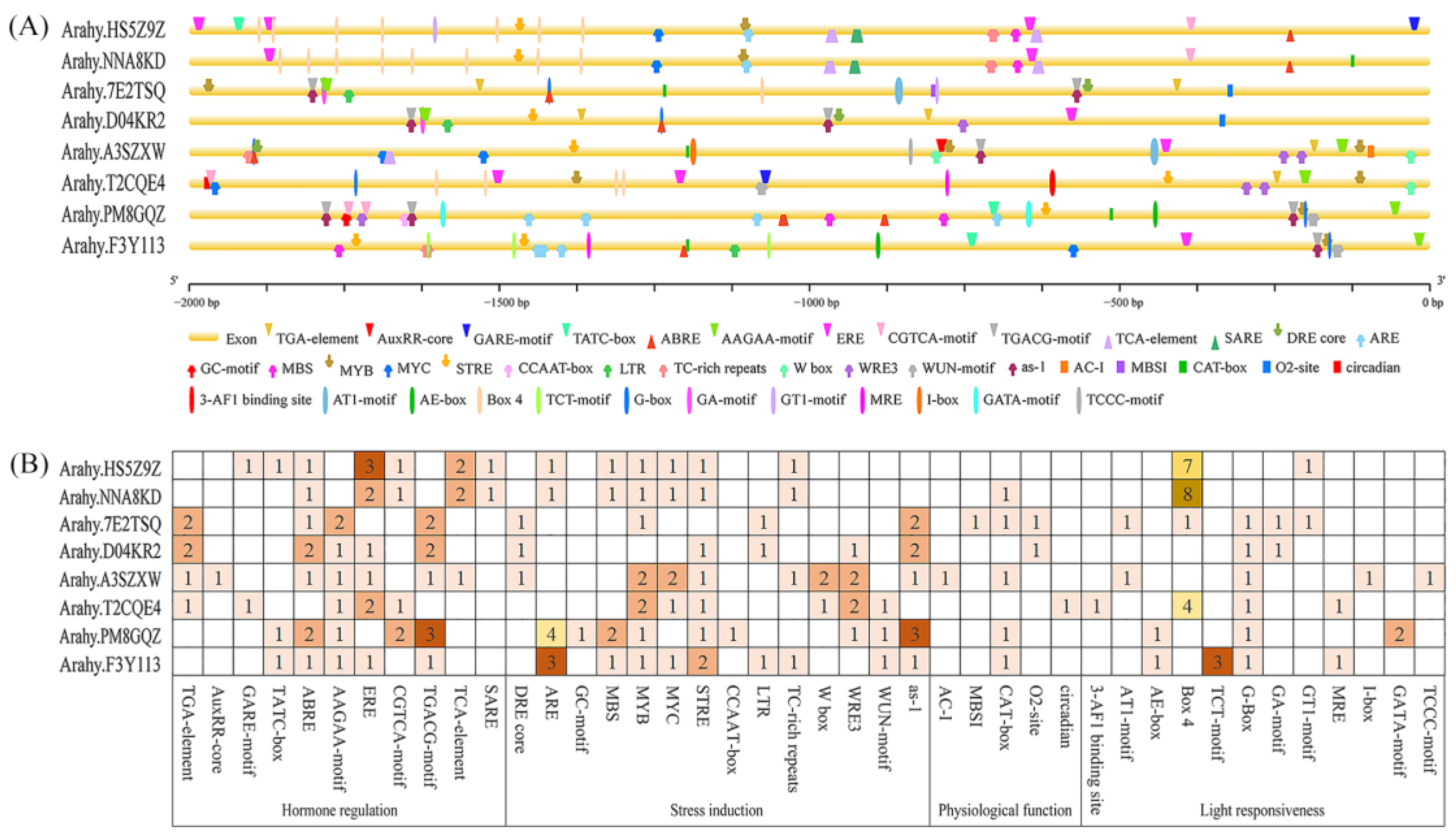
3.5. Promoter Cis-Element Analysis of AhPHY Genes
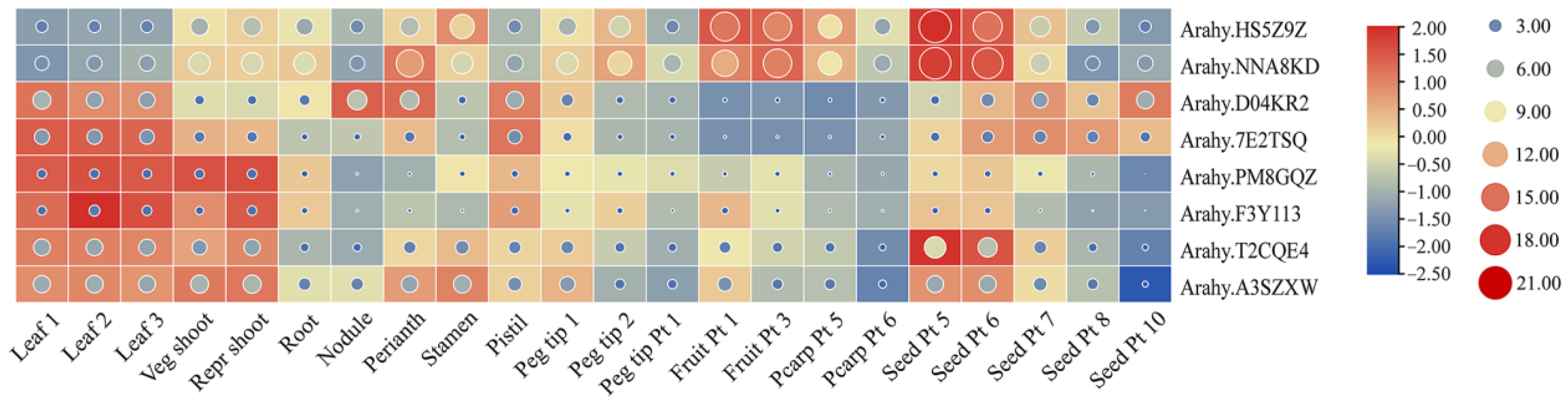
3.6. Expression Pattern Analysis of AhPHY Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schäfer, E.; Nagy, F. (Eds.) Photomorphogenesis in Plants and Bacteria: Fumction and Signal Transduction Mechanisms, 3rd ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 41–247. [Google Scholar]
- Arana, M.V.; Sánchez-Lamas, M.; Strasser, B.; Ibarra, S.E.; Cerdán, P.D.; Botto, J.F.; Sánchez, R.A. Functional diversity of phytochrome family in the control of light and gibberellin-mediated germination in Arabidopsis. Plant Cell Environ. 2014, 37, 2014–2023. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.Y.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef]
- Lamparter, T.; Carrascal, M.; Michael, N.; Martinez, E.; Rottwinkel, G.; Abian, J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry 2004, 43, 3659–3669. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J.; Luccioni, L.G.; Oliverio, K.A.; Boccalandro, H.E. Light, phytochrome signalling and photomorphogenesis in Arabidopsis. Photochem. Photobiol. Sci. 2003, 2, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Kircher, S.; Bauer, D.; Schäfer, E.; Nagy, F. Intramolecular uncoupling of chromophore photoconversion from structural signaling determinants drive mutant phytochrome B photoreceptor to far-red light perception. Plant Signal. Behav. 2012, 7, 904–906. [Google Scholar] [CrossRef]
- Casal, J.J.; Candia, A.N.; Sellaro, R. Light perception and signalling by phytochrome A. J. Exp. Bot. 2014, 65, 2835–2845. [Google Scholar] [CrossRef]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef]
- Piao, W.; Kim, E.Y.; Han, S.H.; Sakuraba, Y.; Paek, N.C. Rice phytochrome B (OsPhyB) negatively regulates dark- and starvation-induced leaf senescence. Plants 2015, 4, 644–663. [Google Scholar] [CrossRef]
- Wang, H.; Jia, G.; Zhang, N.; Zhi, H.; Xing, L.; Zhang, H.; Sui, Y.; Tang, S.; Li, M.; Zhang, H.; et al. Domestication-associated PHYTOCHROME C is a flowering time repressor and a key factor determining Setaria as a short-day plant. New Phytol. 2022, 236, 1809–1823. [Google Scholar] [CrossRef]
- Germán, W.; Ida, M.A.; José, C.J.; Ángel, M.G. Phytochrome B enhances plant growth, biomass and grain yield in field-grown maize. Ann. Bot. 2019, 123, 1079–1088. [Google Scholar]
- Li, Q.; Wu, G.; Zhao, Y.; Wang, B.; Zhao, B.; Kong, D.; Wei, H.; Chen, C.; Wang, H. CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol. J. 2020, 18, 2520–2532. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.J.; Ha, J.; Lee, T.; Jeong, H.; Kim, M.Y.; Kim, S.K.; Lee, Y.H.; Jung, J.W.; Lee, S.H. A candidate flowering gene in mungbean is homologous to a soybean Phytochrome A gene. Euphytica 2017, 213, 79. [Google Scholar] [CrossRef]
- Wu, F.Q.; Fan, C.M.; Zhang, X.M.; Fu, Y.F. The phytochrome gene family in soybean and a dominant negative effect of a soybean PHYA transgene on endogenous Arabidopsis PHYA. Plant Cell Rep. 2013, 32, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Dong, L.; Tang, Y.; Li, H.; Cheng, Q.; Li, H.; Zhang, T.; Ma, L.; Xiang, H.; Chen, L.; et al. Novel and multifaceted regulations of photoperiodic flowering by phytochrome A in soybean. Proc. Natl. Acad. Sci. USA 2022, 119, e2208708119. [Google Scholar] [CrossRef]
- Zhou, T.; Song, B.; Liu, T.; Shen, Y.; Dong, L.; Jing, S.; Xie, C.; Liu, J. Phytochrome F plays critical roles in potato photoperiodic tuberization. Plant J. 2019, 98, 42–54. [Google Scholar] [CrossRef]
- Carlson, K.D.; Bhogale, S.; Anderson, D.; Zaragoza-Mendoza, A.; Madlung, A. Subfunctionalization of phytochrome B1/B2 leads to differential auxin and photosynthetic responses. Plant Direct 2020, 4, e00205. [Google Scholar] [CrossRef]
- Shahzad, R.; Ahmed, F.; Wang, Z.; Harlina, P.W.; Nishawy, E.; Ayaad, M.; Manan, A.; Maher, M.; Ewas, M. Comparative analysis of two phytochrome mutants of tomato (Micro-Tom cv.) reveals specific physiological, biochemical, and molecular responses under chilling stress. J. Genet. Eng. Biotechnol. 2020, 18, 77. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Brasileiro, A.C.M.; Morgante, C.V.; Araujo, A.C.G.; Leal-Bertioli, S.C.M.; Silva, A.K.; Martins, A.C.Q.; Vinson, C.C.; Santos, C.M.R.; Bonfim, O.; Togawa, R.C.; et al. Transcriptome profiling of wild Arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Mol. Biol. Rep. 2015, 33, 1876–1892. [Google Scholar] [CrossRef]
- Guimaraes, P.M.; Guimaraes, L.A.; Morgante, C.V.; Silva, O.B.; Araujo, A.C.G.; Martins, A.C.Q.; Saraiva, M.A.P.; Oliveira, T.N.; Togawa, R.C.; Leal-Bertioli, S.C.M.; et al. Root transcriptome analysis of wild peanut reveals candidate genes for nematode resistance. PLoS ONE 2015, 10, e0140937. [Google Scholar] [CrossRef]
- Shen, Y.; Zhiguo, E.; Liu, Y.; Chen, Z. Screening and transcriptome analysis of water deficiency tolerant germplasms in peanut (Arachis hypogaea). Acta Physiol. Plant. 2015, 37, 103. [Google Scholar] [CrossRef]
- Whitelam, G.C.; Devlin, P.F. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 1997, 20, 752–758. [Google Scholar] [CrossRef]
- Olsen, J.E. Mechanisms of dormancy regulation. Acta Hortic. 2006, 727, 157–166. [Google Scholar] [CrossRef]
- Vitaly, S.; Larissa, K.; Michael, R.; Peter, N. Phytochrome A and its functional manifestations in etiolated and far-red light-grown seedlings of the wild-type rice and its hebiba and cpm2 mutants deficient in the defense-related phytohormone jasmonic acid. Photochem. Photobiol. 2020, 97, 335–342. [Google Scholar]
- Ruberti, I.; Sessa, G.; Ciolfi, A.; Possenti, M.; Carabelli, M.; Morelli, G. Plant adaptation to dynamically changing environment: The shade avoidance response. Biotechnol. Adv. 2012, 30, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, F.; Zhou, J.; Fan, Z.; Chen, F.; Ma, H.; Xie, X. Overexpression of a phytochrome-regulated tandem zinc finger protein gene, OsTZF1, confers hypersensitivity to ABA and hyposensitivity to red light and far-red light in rice seedlings. Plant Cell Rep. 2012, 31, 1333–1343. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Quecini, V.; Peres, L.E.P. Hormonal modulation of photomorphogenesis-controlled anthocyanin accumulation in tomato (Solanum lycopersicum L. cv Micro-Tom) hypocotyls: Physiological and genetic studies. Plant Sci. 2010, 178, 258–264. [Google Scholar] [CrossRef]
- Mitsunori, S.; Eiji, N.; Giltsu, C.; Shinjiro, Y. Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 2009, 69, 463–472. [Google Scholar]
- Kircher, S.; Kozma-Bognár, L.; Kim, L.; Adám, E.; Harter, K.; Schäfer, E.; Nagy, F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 1999, 11, 1445–1456. [Google Scholar]
- Yamaguchi, R.; Nakamura, M.; Mochizuki, N.; Kay, S.A.; Nagatani, A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 1999, 145, 437–445. [Google Scholar] [CrossRef]
- Kim, L.; Kircher, S.; Toth, R.; Adam, E.; Schäfer, E.; Nagy, F. Light-induced nuclear import of phytochrome-A, GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 2000, 22, 125–133. [Google Scholar] [CrossRef]
- Kircher, S.; Gil, P.; Kozma-Bognár, L.; Fejes, E.; Speth, V.; Husselstein-Muller, T.; Bauer, D.; Adám, E.; Schäfer, E.; Nagy, F. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 2002, 14, 1541–1555. [Google Scholar] [CrossRef]
- Furuya, M.; Schäfer, E. Photoperception and signalling of induction reactions by different phytochromes. Trends Plant Sci. 1996, 1, 301–307. [Google Scholar] [CrossRef]
- Cheng, Y.; Ahammed, G.J.; Yao, Z.; Ye, Q.; Ruan, M.; Wang, R.; Li, Z.; Zhou, G.; Wan, H. Comparative Genomic Analysis Reveals Extensive Genetic Variations of WRKYs in Solanaceae and Functional Variations of CaWRKYs in Pepper. Front. Genet. 2019, 10, 492. [Google Scholar] [CrossRef]
- Bigeard, J.; Rayapuram, N.; Pflieger, D.; Hirt, H. Phosphorylation-dependent regulation of plant chromatin and chromatin-associated proteins. Proteomics 2014, 14, 2127–2140. [Google Scholar] [CrossRef] [PubMed]
- Adijat, A.A.; Amara, C.; Shakeel, A.; Wang, Y.; Shu, Y.; Li, S.; Liu, X.; Kazeem, B.B.; Muhammad, T.S.; Tong, X.H.; et al. Protein phosphorylation and phosphoproteome: An overview of rice. Rice Sci. 2020, 27, 184–200. [Google Scholar]
- Balcerowicz, M.; Mahjoub, M.; Nguyen, D.; Lan, H.; Stoeckle, D.; Conde, S.; Jaeger, K.E.; Wigge, P.A.; Ezer, D. An early-morning gene network controlled by phytochromes and cryptochromes regulates photomorphogenesis pathways in Arabidopsis. Mol. Plant 2021, 14, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Falleiros, R.C.; Lattarulo, M.C.; Antunes, R.A. The role of phytochrome in stress tolerance. J. Integr. Plant Biol. 2011, 53, 920–929. [Google Scholar]
- Abdellatif, I.M.Y.; Yuan, S.; Yoshihara, S.; Suzaki, T.; Ezura, H.; Miura, K. Stimulation of tomato drought tolerance by PHYTOCHROME A and B1B2 mutations. Int. J. Mol. Sci. 2023, 24, 1560. [Google Scholar] [CrossRef]
- Fichman, Y.; Xiong, H.; Sengupta, S.; Morrow, J.; Loog, H.; Azad, R.K.; Hibberd, J.M.; Liscum, E.; Mittler, R. Phytochrome B regulates reactive oxygen signaling during abiotic and biotic stress in plants. New Phytol. 2022, 237, 1711–1727. [Google Scholar] [CrossRef]
- Halliday, K.J.; Salter, M.G.; Thingnaes, E.; Whitelam, G.C. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 2003, 33, 875–885. [Google Scholar] [CrossRef]
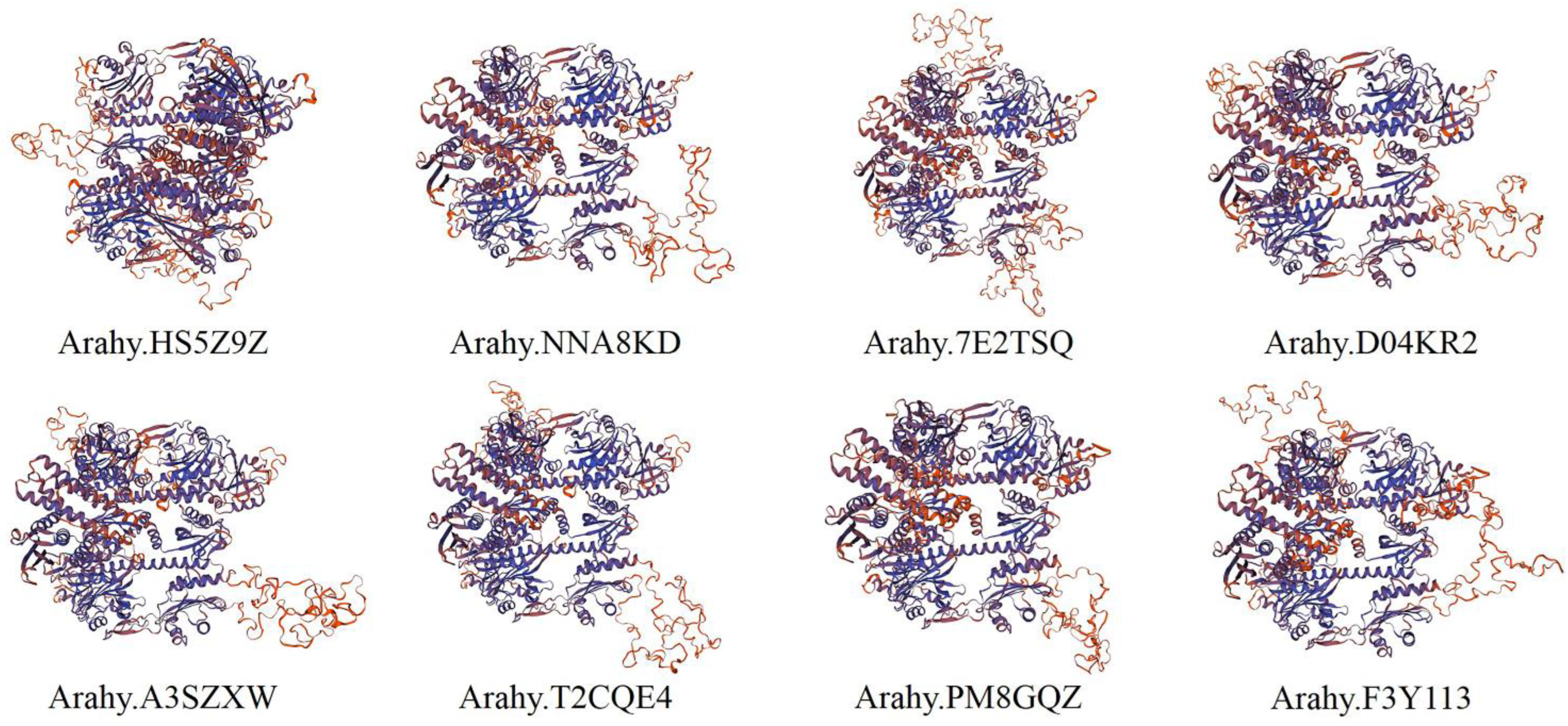

| Gene ID | Amino Acid (aa) | Molecular Weight (Da) | Isoelectric Point (pI) | Instability Index (II) | Aliphatic Index | GRAVY | Subcellular Localization | Phosphorylation Site | |
|---|---|---|---|---|---|---|---|---|---|
| Ser/Thr/Tyr | Total | ||||||||
| Arahy.HS5Z9Z | 1125 | 124,470.30 | 6.01 | 43.54 | 93.95 | −0.081 | Cyt, N | 57/37/10 | 104 |
| Arahy.NNA8KD | 1113 | 123,208.83 | 5.80 | 45.13 | 95.22 | −0.056 | Cyt, N | 58/36/9 | 103 |
| Arahy.7E2TSQ | 1125 | 124,620.66 | 6.05 | 43.27 | 95.07 | −0.055 | Cyt, N | 60/36/8 | 104 |
| Arahy.D04KR2 | 1125 | 124,612.76 | 6.14 | 43.40 | 95.50 | −0.055 | Cyt, N | 61/35/8 | 104 |
| Arahy.A3SZXW | 1151 | 128,157.41 | 5.76 | 44.12 | 91.82 | −0.171 | Cyt, N | 63/30/9 | 102 |
| Arahy.T2CQE4 | 1147 | 127,672.95 | 5.80 | 43.90 | 91.72 | −0.168 | Cyt, N | 64/28/9 | 101 |
| Arahy.PM8GQZ | 1101 | 122,424.19 | 5.80 | 45.79 | 91.04 | −0.132 | Cyt, N | 62/24/10 | 96 |
| Arahy.F3Y113 | 1116 | 123,960.97 | 5.72 | 46.13 | 91.32 | −0.122 | Cyt, N | 61/26/11 | 98 |
| Gene ID | α-Helix (%) | Extended Strand (%) | β-Turn (%) | Random Coil (%) |
|---|---|---|---|---|
| Arahy.HS5Z9Z | 48.00 | 14.31 | 5.42 | 32.27 |
| Arahy.NNA8KD | 48.61 | 14.56 | 5.30 | 31.54 |
| Arahy.7E2TSQ | 47.56 | 14.76 | 4.89 | 32.80 |
| Arahy.D04KR2 | 48.36 | 14.76 | 5.69 | 31.20 |
| Arahy.A3SZXW | 47.35 | 14.68 | 5.30 | 32.67 |
| Arahy.T2CQE4 | 47.69 | 14.73 | 5.58 | 32.00 |
| Arahy.PM8GQZ | 49.05 | 14.80 | 5.81 | 30.34 |
| Arahy.F3Y113 | 49.28 | 14.70 | 5.29 | 30.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Y.; Liu, Y.; Liang, M.; Zhang, X.; Chen, Z.; Shen, Y. Genome-Wide Identification and Characterization of the Phytochrome Gene Family in Peanut. Genes 2023, 14, 1478. https://doi.org/10.3390/genes14071478
Shen Y, Liu Y, Liang M, Zhang X, Chen Z, Shen Y. Genome-Wide Identification and Characterization of the Phytochrome Gene Family in Peanut. Genes. 2023; 14(7):1478. https://doi.org/10.3390/genes14071478
Chicago/Turabian StyleShen, Yue, Yonghui Liu, Man Liang, Xuyao Zhang, Zhide Chen, and Yi Shen. 2023. "Genome-Wide Identification and Characterization of the Phytochrome Gene Family in Peanut" Genes 14, no. 7: 1478. https://doi.org/10.3390/genes14071478
APA StyleShen, Y., Liu, Y., Liang, M., Zhang, X., Chen, Z., & Shen, Y. (2023). Genome-Wide Identification and Characterization of the Phytochrome Gene Family in Peanut. Genes, 14(7), 1478. https://doi.org/10.3390/genes14071478






