Dissection of the Genetic Basis of Resistance to Stem Rot in Cultivated Peanuts (Arachis hypogaea L.) through Genome-Wide Association Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Evaluation of Stem Rot Resistance
2.2. Samples Preparation and Genotyping
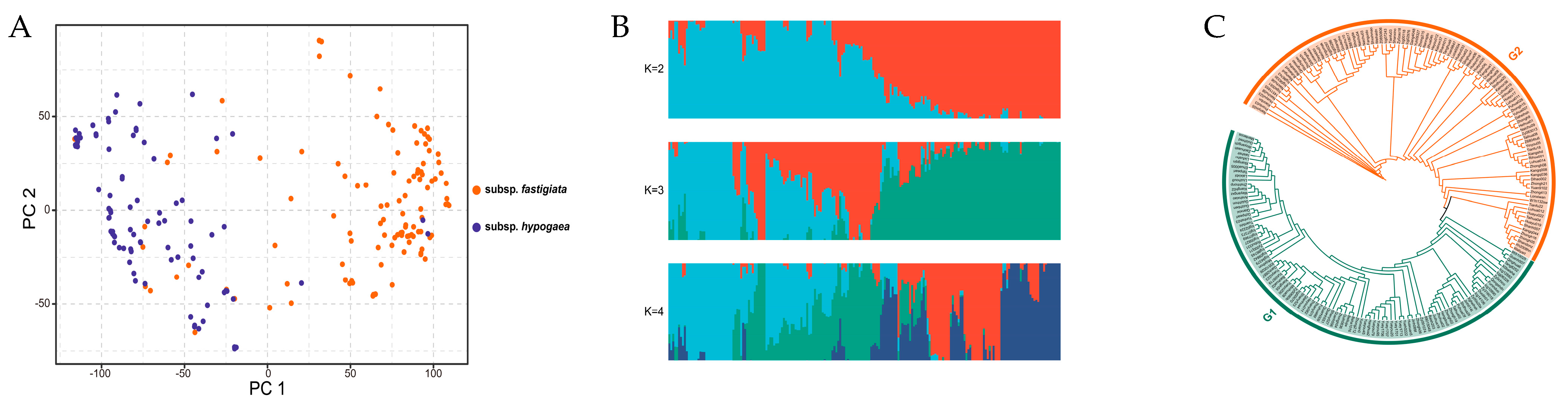
2.3. Population Structure Analysis and Phylogenetic Tree Construction
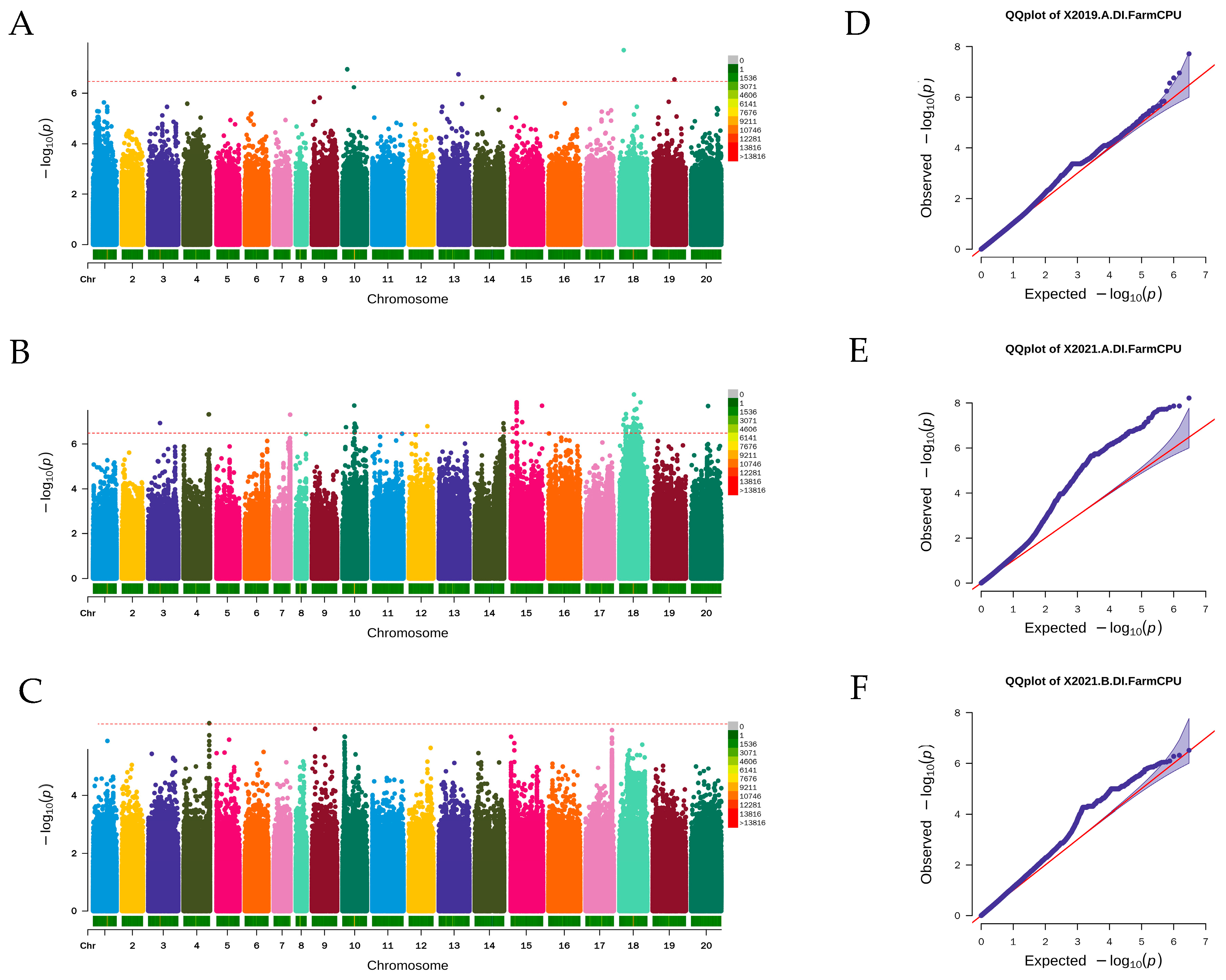
2.4. Genome-Wide Association Analysis and Candidate Genes’ Predication
3. Results
3.1. Phenotypic Variation among Peanut Accessions
3.2. Genome-Wide Distribution of SNPs
3.3. Population Structure and Phylogenetic Tree
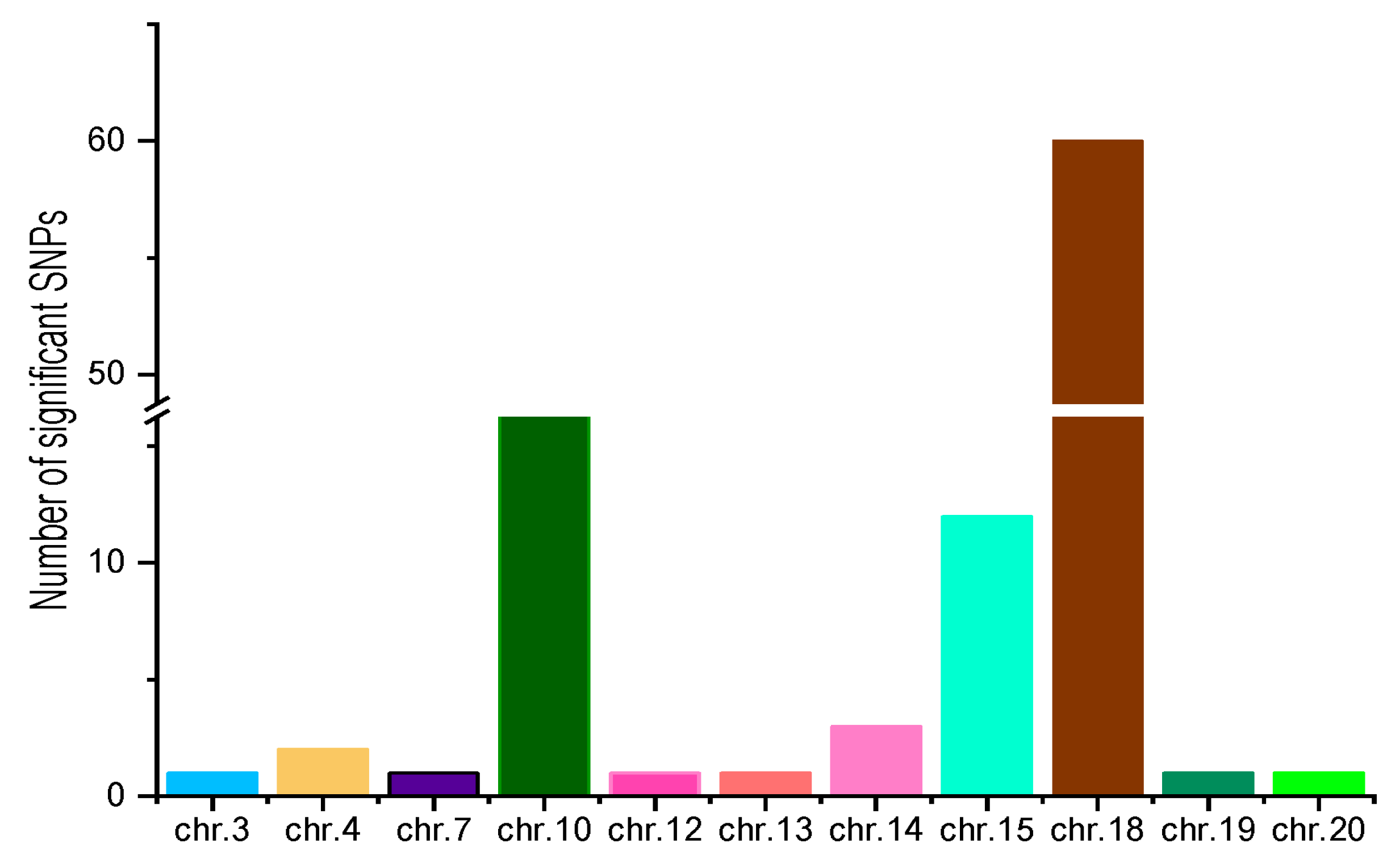
3.4. Identification of Loci Associated with Stem Rot Resistance Using GWAS
3.5. Identification of Putative Candidate Genes for Stem Rot Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, R.H.; Fang, X.L.; Zeng, S.C.; Li, S.Y. Occurrence and control measures of main peanut diseases in Jiangxi. Acta Agric. Jiangxi 2009, 21, 106–109. [Google Scholar] [CrossRef]
- Fu, J.F.; Liu, B.; Zhou, R.J.; Wang, S.W. Identification of biological characteristics of Sclerotium rolfsii causing peanut stem rot in Liaoning Province. Chin. J. Oil Crop Sci. 2014, 36, 635–640. [Google Scholar] [CrossRef]
- Li, M.; Li, S.; Zhang, Z.X.; Du, P.Q.; Lin, Z.; Dong, W.Z. Sensitivity to carboxin and population diversity of Sclerotium rolfsii from peanut in Henan province. J. Henan Agric. Sci. 2021, 50, 64–73. [Google Scholar] [CrossRef]
- Xiao, X.; Chen, X.L.; Deng, M.G.; Xu, D.G.; Pan, R.Q. Preliminary research on distribution and biological characteristics of Sclerotium rolfsii, the pathogen of peanut stem rot in Guangdong province. Guangdong Agric. Sci. 2012, 17, 71–73. [Google Scholar] [CrossRef]
- Chen, K.R.; Ren, L.; Xu, L.; Liu, F.; Fang, X.P. Research progress on peanut southern stem rot caused by Sclerotium rolfsii. Chin. J. Oil Crop Sci. 2018, 40, 302–308. [Google Scholar] [CrossRef]
- Yan, L.Y.; Song, W.D.; Lei, Y.; Wan, L.Y.; Huai, D.X.; Kang, Y.P.; Chen, Y.P.; Liao, B.S. Evaluation of peanut accessions for re-sistance to Sclerotium stem rot. Chin. J. Oil Crop Sci. 2019, 41, 781–787. [Google Scholar] [CrossRef]
- Branch, W.D.; Culbreath, A.K. Registration of ‘Georgia-10T’ Peanut. J. Plant Regist. 2011, 5, 279–281. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-12Y’ Peanut. J. Plant Regist. 2013, 7, 151–153. [Google Scholar] [CrossRef]
- Tillman, B.L. Registration of ‘FloRun ‘331’ peanut. J. Plant Regist. 2021, 15, 294–299. [Google Scholar] [CrossRef]
- Dodia, S.M.; Joshi, B.; Gangurde, S.S.; Thirumalaisamy, P.P.; Mishra, G.P.; Narandrakumar, D.; Soni, P.; Rathnakumar, A.L.; Dobaria, J.R.; Sangh, C.; et al. Genotyping-by-sequencing based genetic mapping reveals large number of epistatic interactions for stem rot resistance in groundnut. Theor. Appl. Genet. 2019, 132, 1001–1016. [Google Scholar] [CrossRef]
- Bera, S.K.; Kamdar, J.H.; Kasundra, S.V.; Ajay, B.C. A novel QTL governing resistance to stem rot disease caused by Sclerotium rolfsii in peanut. Australas. Plant Pathol. 2016, 45, 1–8. [Google Scholar] [CrossRef]
- Luo, Z.; Cui, R.; Chavarro, C.; Tseng, Y.-C.; Zhou, H.; Peng, Z.; Chu, Y.; Yang, X.; Lopez, Y.; Tillman, B.; et al. Mapping quantitative trait loci (QTLs) and estimating the epistasis controlling stem rot resistance in cultivated peanut (Arachis hypogaea). Theor. Appl. Genet. 2020, 133, 1201–1212. [Google Scholar] [CrossRef]
- Cui, R.; Clevenger, J.; Chu, Y.; Brenneman, T.; Isleib, T.G.; Holbrook, C.C.; Ozias-Akins, P. QTL-seq derived molecular markers for selection of stem rot (Sclerotium rolfsii) resistance in peanut (Arachis hypogaea). Crop. Sci. 2020, 60, 2008–2018. [Google Scholar] [CrossRef]
- Zou, K.; Kim, K.S.; Kim, K.; Kang, D.; Park, Y.H.; Sun, H.; Ha, B.K.; Ha, J.; Jun, T.H. Genetic Diversity and Genome-Wide Association Study of Seed Aspect Ratio Using a High-Density SNP Array in Peanut (Arachis hypogaea L.). Genes 2020, 12, 2. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Sha, J.; Liu, S.; Bao, L.; Zhang, J.; Wang, R.; Yao, J.; Li, C.; Feng, J.; Sun, F.; et al. A genome-wide association study in catfish reveals the presence of functional hubs of related genes within QTLs for columnaris disease resistance. BMC Genom. 2015, 16, 196. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Schmutz, J. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.P.; Lu, Q.; Liu, H.; Zhang, J.N.; Hong, Y.B.; Lan, H.F.; Li, H.F.; Wang, J.P.; Liu, H.Y.; Li, S.X.; et al. Sequencing of Cultivated Peanut, Arachis hypogaea, Yields Insights into Genome Evolution and Oil Improvement. Mol. Plant 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Liu, N.; Huang, L.; Chen, W.G.; Wu, B.; Jiang, H.F. Dissection of the genetic basis of oil content in Chinese peanut cultivars through association mapping. BMC Genet. 2020, 21, 60. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; He, X.; Wang, Y.; Ma, X.; Yin, D. Genome-wide association -study of major agronomic traits related to domestication in peanut. Front. Plant Sci. 2017, 8, 1611. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, C.; Shi, D.; Zhao, X.; Yuan, C.; Sun, Q.; Mou, Y.; Chen, H.; Li, Y.; Li, C.; et al. The genetic base for peanut height-related traits revealed by a meta-analysis. Plants 2021, 10, 1058. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Upadhyaya, H.D.; Rathore, A.; Vadez, V.; Sheshshayee, M.S.; Sriswathi, M.; Govil, M.; Kumar, A.; Gowda, M.V.C.; Sharma, S. Genomewide association studies for 50 agronomic traits in peanut using the ‘Reference Set’ comprising 300 genotypes from 48 countries of the Semi-Arid Tropics of the world. PLoS ONE 2014, 9, e105228. [Google Scholar] [CrossRef]
- Wang, J.; Yan, C.; Li, Y.; Li, C.; Zhao, X.; Yuan, C.; Sun, Q.; Shan, S. GWAS discovery of candidate genes for yield-related traits in peanut and support from earlier QTL mapping studies. Genes 2019, 10, 803. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Guo, J.; Pandey, M.K.; Varshney, R.K.; Huang, L.; Luo, H.; Liu, N.; Chen, W.; Lei, Y.; Liao, B.; et al. Dissection of the genetic basis of yield-related traits in the Chinese peanut mini-Core collection through genome-wide association studies. Front. Plant Sci. 2021, 12, 637284. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.B.; Qiu, X.K.; Luo, H.Y.; Huang, L.; Guo, J.B.; Yu, B.L.; Sudini, H.; Pandey, M.; Kang, Y.P.; Liu, N.; et al. Comprehensive evaluation of Chinese peanut mini-mini core collection and QTL mapping for aflatoxin resistance. BMC Plant Biol. 2022, 22, 207. [Google Scholar] [CrossRef]
- Yu, B.; Jiang, H.; Pandey, M.K.; Huang, L.; Huai, D.; Zhou, X.; Kang, Y.; Varshney, R.K.; Sudini, H.K.; Ren, X.; et al. Identification of two novel peanut genotypes resistant to aflatoxin production and their SNP markers associated with resistance. Toxins 2020, 12, 156. [Google Scholar] [CrossRef]
- Zhang, H.; Chu, Y.; Dang, P.; Tang, Y.Y.; Chen, C. Identification of QTLs for resistance to leaf spots in cultivated peanut (Arachis hypogaea L.) through GWAS analysis. Theor. Appl. Genet. 2020, 133, 2051–2061. [Google Scholar] [CrossRef]
- Fan, P.; Song, W.; Kang, Y.; Wan, L.; Lei, Y.; Huai, D.; Chen, Y.; Wang, X.; Jiang, H.; Yan, L. Phenotypic identification of peanut germplasm for resistance to southern stem rot. Oil Crop. Sci. 2020, 5, 174–179. [Google Scholar] [CrossRef]
- Shokes, F.M.; Weber, Z.; Gorbet, D.W.; Pudelko, H.A.; Taczanowski, M. Evaluation of peanut genotypes for resistance to southern stem rot using an agar disk technique1. Peanut Sci. 1998, 25, 12–17. [Google Scholar] [CrossRef]
- Yan, L.; Song, W.; Yu, D.; Sudini, H.K.; Kang, Y.; Lei, Y.; Huai, D.; Wang, Z.; Chen, Y.; Wang, X.; et al. Genetic, phenotypic, and pathogenic variation among Athelia rolfsii, the causal agent of peanut stem rot in China. Plant Dis. 2022, 106, 2722–2729. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wuspler, A.; Fennell, T.M.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Sardos, J.; Rouard, M.; Hueber, Y.; Cenci, A.; Hyma, K.E.; van den Houwe, I.; Hribova, R.; Courtois, B.; Roux, N. A Genome-wide association study on the seedless phenotype in Banana (Musa spp.) reveals the potential of a selected panel to detect candidate genes in a vegetatively propagated crop. PLoS ONE 2016, 11, e0154448. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Mehan, V.K.; Mayee, C.D.; McDonald, D.; Ramakrishna, N.; Jayanthi, S. Resistance in groundnut to Sclerotium rolfsii caused stem and pod rot. Int. J. Pest Manag. 1995, 41, 79–83. [Google Scholar] [CrossRef]
- Bennett, R.; Chamberlin, K. Resistance to Athelia rolfsii and web blotch in the U.S. mini-core collection. Peanut Sci. 2020, 47, 17–24. [Google Scholar] [CrossRef]
- Jiang, H.F.; Wang, S.Y.; Ren, X.P. Reaction of groundnut germplasm to Aspergillus flavus invasion. Chin. J. Oil Crop Sci. 2002, 24, 23–25. [Google Scholar]
- Guo, Z.L.; Liu, X.; Zhang, B.; Yuan, X.J.; Xing, Y.Z.; Liu, H.Y.; Luo, L.J.; Chen, G.X.; Xiong, L.Z. Genetic analyses of lodging resistance and yield provide insights into post-Green-Revolution breeding in rice. Plant Biotechnol. J. 2021, 19, 814–829. [Google Scholar] [CrossRef]
- Pang, Y.L.; Wu, Y.Y.; Liu, C.X.; Li, W.H.; Amand, P.S.; Bernardo, A.; Wang, D.F.; Dong, L.; Yuan, X.F.; Zhang, H.R.; et al. High-resolution genome-wide association study and genomic prediction for disease resistance and cold tolerance in wheat. Theor. Appl. Genet. 2021, 134, 2857–2873. [Google Scholar] [CrossRef]
- Zhao, N.; Wang, W.R.; Grover, C.E.; Jiang, K.Y.; Pan, Z.X.; Guo, B.S.; Zhu, J.H.; Su, Y.; Wang, M.; Nie, H.S.; et al. Genomic and GWAS analyses demonstrate phylogenomic relationships of Gossypium barbadense in China and selection for fibre length, lint percentage and Fusarium wilt resistance. Plant Biotechnol. J. 2022, 20, 691–710. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Chen, W.; Cui, Y.L.; Sang, X.H.; Lu, J.H.; Jing, H.J.; Wang, W.J.; Zhao, P.; Wang, H.M. Detection of candidate genes and development of KASP markers for Verticillium wilt resistance by combining genome-wide association study, QTL-seq and transcriptome sequencing in cotton. Theor. Appl. Genet. 2021, 134, 1063–1081. [Google Scholar] [CrossRef]


| Environment | Minimum | Maximum | Mean | SD | CV | Skew | Kurt | H2 (%) |
|---|---|---|---|---|---|---|---|---|
| 2019-A | 27.03 | 83.75 | 48.52 | 12.33 | 0.25 | 0.72 | 0.23 | 67.1 |
| 2021-A | 28.71 | 97.34 | 52.62 | 13.77 | 0.26 | 0.85 | 0.50 | |
| 2021-B | 34.26 | 94.29 | 60.06 | 12.85 | 0.21 | 0.21 | −0.65 |
| Subspecies | Environment | |||
|---|---|---|---|---|
| 2019-A | 2021-A | 2021-B | Mean | |
| subsp. fastigiata | 47.86 ± 12.23 a | 52.06 ± 13.52 a | 58.47 ± 12.80 a | 52.8 ± 9.59 a |
| subsp. hypogaea | 49.34 ± 12.47 a | 53.31 ± 14.11 a | 61.95 ± 12.71 a | 54.89 ± 9.89 a |
| Originated Place | Environment | |||
|---|---|---|---|---|
| 2019-A | 2021-A | 2021-B | Mean | |
| Southern China | 45.39 ± 10.60 a | 46.20 ± 11.22 a | 54.92 ± 11.54 a | 48.89 ± 8.22 a |
| Yangtze River region | 51.17 ± 14.48 b | 54.59 ± 15.03 b | 63.08 ± 14.75 b | 56.28 ± 11.41 b |
| Northern China | 45.68 ± 9.29 a | 50.09 ± 9.47 ab | 60.04 ± 10.42 b | 51.98 ± 5.65 a |
| Other countries | 52.64 ± 13.06 b | 61.71 ± 15.10 c | 62.09 ± 13.07 b | 58.82 ± 10.40 c |
| Function | Annotation | Number of Genes |
|---|---|---|
| Recognition (12) | disease-resistance protein (TIR-NBS-LRR) | 1 |
| LRR receptor-like kinase | 1 | |
| protein kinase superfamily protein | 2 | |
| receptor kinase | 2 | |
| receptor-like protein | 1 | |
| receptor-like protein kinase | 3 | |
| receptor-like serine/threonine kinase | 2 | |
| Signal transduction (13) | ATPase | 1 |
| ATP-binding/protein serine/threonine kinase | 1 | |
| calcium-binding EF-hand family protein | 1 | |
| glutathione S-transferase | 1 | |
| myb transcription factor | 1 | |
| signal peptide peptidase | 1 | |
| thioredoxin superfamily protein | 1 | |
| transcription factor | 1 | |
| WRKY family transcription factor | 2 | |
| zinc finger family protein | 3 | |
| Defense (2) | peroxidase superfamily protein | 1 |
| terpene synthase | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, L.; Song, W.; Wang, Z.; Yu, D.; Sudini, H.; Kang, Y.; Lei, Y.; Huai, D.; Chen, Y.; Wang, X.; et al. Dissection of the Genetic Basis of Resistance to Stem Rot in Cultivated Peanuts (Arachis hypogaea L.) through Genome-Wide Association Study. Genes 2023, 14, 1447. https://doi.org/10.3390/genes14071447
Yan L, Song W, Wang Z, Yu D, Sudini H, Kang Y, Lei Y, Huai D, Chen Y, Wang X, et al. Dissection of the Genetic Basis of Resistance to Stem Rot in Cultivated Peanuts (Arachis hypogaea L.) through Genome-Wide Association Study. Genes. 2023; 14(7):1447. https://doi.org/10.3390/genes14071447
Chicago/Turabian StyleYan, Liying, Wanduo Song, Zhihui Wang, Dongyang Yu, Hari Sudini, Yanping Kang, Yong Lei, Dongxin Huai, Yuning Chen, Xin Wang, and et al. 2023. "Dissection of the Genetic Basis of Resistance to Stem Rot in Cultivated Peanuts (Arachis hypogaea L.) through Genome-Wide Association Study" Genes 14, no. 7: 1447. https://doi.org/10.3390/genes14071447
APA StyleYan, L., Song, W., Wang, Z., Yu, D., Sudini, H., Kang, Y., Lei, Y., Huai, D., Chen, Y., Wang, X., Wang, Q., & Liao, B. (2023). Dissection of the Genetic Basis of Resistance to Stem Rot in Cultivated Peanuts (Arachis hypogaea L.) through Genome-Wide Association Study. Genes, 14(7), 1447. https://doi.org/10.3390/genes14071447






