Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing
Abstract
1. Introduction
2. RNA Modifications
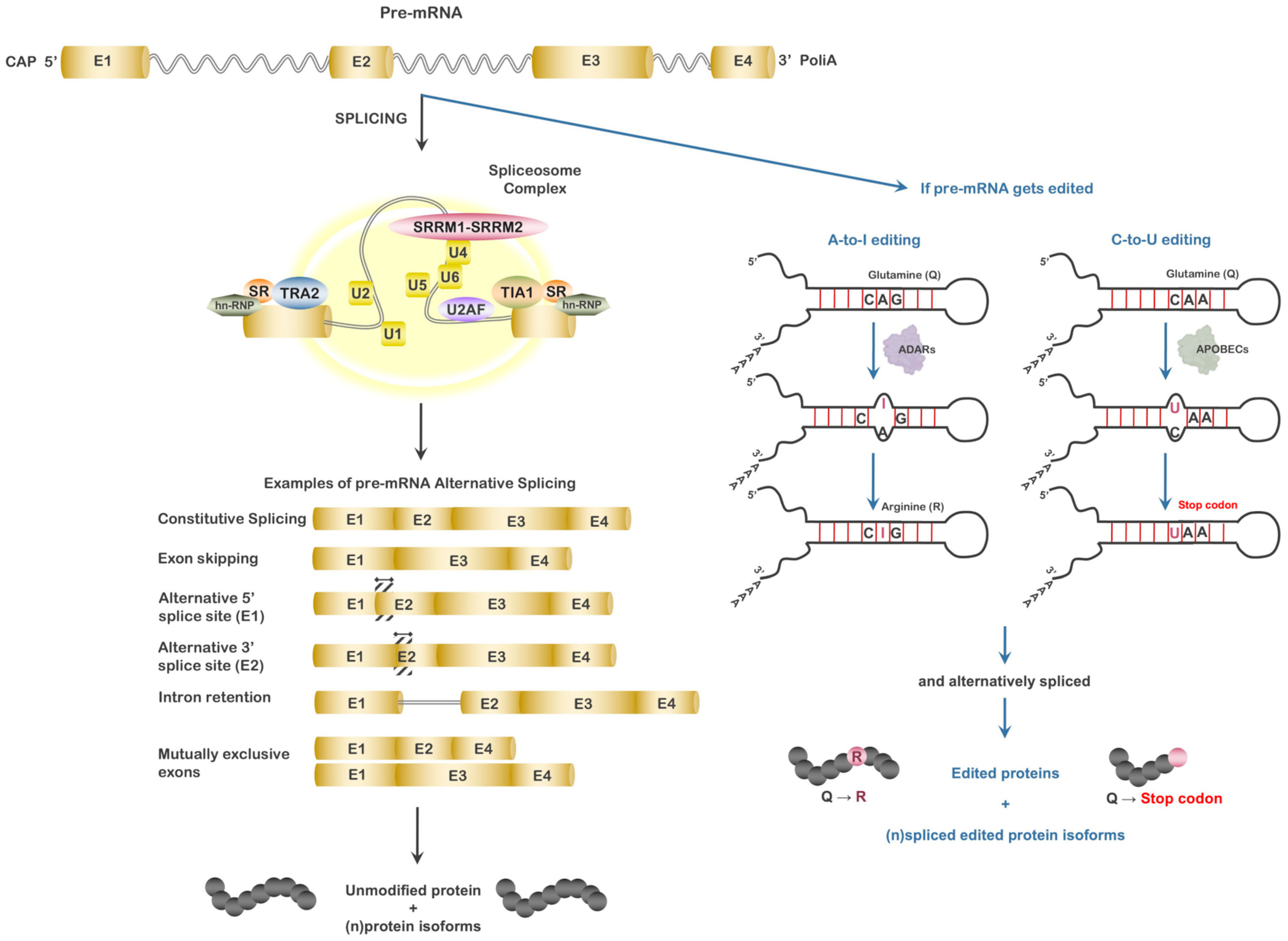
3. mRNA Splicing and Alternative Splicing
3.1. Serine/Arginine-Rich Splicing Factors (SRSFs)
3.2. Heterogeneous Nuclear Ribonucleoproteins (hnRNPs)
3.3. Consequences of Alternative Splicing and Measures to Address the Issue
4. RNA Editing and RNA Editing Enzymes
4.1. Adenosine Deaminases
4.2. Cytidine Deaminases
4.3. Consequences of RNA Editing and Measures to Address the Issue
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lipshutz, R.J.; Morris, D.; Chee, M.; Hubbell, E.; Kozal, M.J.; Shah, N.; Shen, N.; Yang, R.; Fodor, S.P. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques 1995, 19, 442–447. [Google Scholar]
- Emili, A.Q.; Cagney, G. Large-scale functional analysis using peptide or protein arrays. Nat. Biotechnol. 2000, 18, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Masuda, M.; Yamada, T. Signaling pathway profiling by reverse-phase protein array for personalized cancer medicine. Biochim. Biophys. Acta 2015, 1854, 651–657. [Google Scholar] [CrossRef]
- Haga, Y.; Minegishi, Y.; Ueda, K. Frontiers in mass spectrometry-based clinical proteomics for cancer diagnosis and treatment. Cancer Sci. 2023, 114, 1783–1791. [Google Scholar] [CrossRef]
- Kempa, E.E.; Hollywood, K.A.; Smith, C.A.; Barran, P.E. High throughput screening of complex biological samples with mass spectrometry—From bulk measurements to single cell analysis. Analyst 2019, 144, 872–891. [Google Scholar] [CrossRef]
- Liu, J.; Hu, W.; Han, Y.; Nie, H. Recent advances in mass spectrometry imaging of single cells. Anal. Bioanal. Chem. 2023. [Google Scholar] [CrossRef] [PubMed]
- Anaparthy, N.; Ho, Y.J.; Martelotto, L.; Hammell, M.; Hicks, J. Single-Cell Applications of Next-Generation Sequencing. Cold Spring Harb. Perspect. Med. 2019, 9, a026898. [Google Scholar] [CrossRef] [PubMed]
- Lohani, V.; A R, A.; Kundu, S.; Akhter, M.Q.; Bag, S. Single-Cell Proteomics with Spatial Attributes: Tools and Techniques. ACS Omega 2023, 8, 17499–17510. [Google Scholar] [CrossRef]
- Cui, M.; Cheng, C.; Zhang, L. High-throughput proteomics: A methodological mini-review. Lab. Invest. 2022, 102, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Gracia, K.; Gramalla-Schmitz, A.; Weischedel, J.; Chahwan, R. APOBECs orchestrate genomic and epigenomic editing across health and disease. Trends Genet. 2021, 37, 1028–1043. [Google Scholar] [CrossRef]
- Pecori, R.; Di Giorgio, S.; Paulo Lorenzo, J.; Nina Papavasiliou, F. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat. Rev. Genet. 2022, 23, 505–518. [Google Scholar] [CrossRef]
- Peake, J.D.; Noguchi, E. Fanconi anemia: Current insights regarding epidemiology, cancer, and DNA repair. Hum. Genet. 2022, 141, 1811–1836. [Google Scholar] [CrossRef]
- Faenza, I.; Blalock, W.L. Innate Immunity: A Balance between Disease and Adaption to Stress. Biomolecules 2022, 12, 737. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef]
- Devarkar, S.C.; Wang, C.; Miller, M.T.; Ramanathan, A.; Jiang, F.; Khan, A.G.; Patel, S.S.; Marcotrigiano, J. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. USA 2016, 113, 596–601. [Google Scholar] [CrossRef]
- Ren, P.; Lu, L.; Cai, S.; Chen, J.; Lin, W.; Han, F. Alternative Splicing: A New Cause and Potential Therapeutic Target in Autoimmune Disease. Front. Immunol. 2021, 12, 713540. [Google Scholar] [CrossRef]
- Liu, Q.; Fang, L.; Wu, C. Alternative Splicing and Isoforms: From Mechanisms to Diseases. Genes 2022, 13, 401. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Caracausi, M.; Antonaros, F.; Pelleri, M.C.; Vitale, L. GeneBase 1.1: A tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database 2016, 2016, baw153. [Google Scholar] [CrossRef]
- Ergun, A.; Doran, G.; Costello, J.C.; Paik, H.H.; Collins, J.J.; Mathis, D.; Benoist, C.; ImmGen, C. Differential splicing across immune system lineages. Proc. Natl. Acad. Sci. USA 2013, 110, 14324–14329. [Google Scholar] [CrossRef]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Blalock, W.L.; Piazzi, M.; Gallo, A.; Bavelloni, A.; Focaccia, E.; Faenza, I. Ribosome processing and ribosome biogenesis in bone marrow falilure disorders. RNA Dis. 2019, 6, e1531. [Google Scholar]
- Zheng, X.; Peng, Q.; Wang, L.; Zhang, X.; Huang, L.; Wang, J.; Qin, Z. Serine/arginine-rich splicing factors: The bridge linking alternative splicing and cancer. Int. J. Biol. Sci. 2020, 16, 2442–2453. [Google Scholar] [CrossRef]
- Das, S.; Krainer, A.R. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014, 12, 1195–1204. [Google Scholar] [CrossRef]
- Ghosh, G.; Adams, J.A. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011, 278, 587–597. [Google Scholar] [CrossRef]
- Corkery, D.P.; Holly, A.C.; Lahsaee, S.; Dellaire, G. Connecting the speckles: Splicing kinases and their role in tumorigenesis and treatment response. Nucleus 2015, 6, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Bavelloni, A.; Piazzi, M.; Faenza, I.; Raffini, M.; D’Angelo, A.; Cattini, L.; Cocco, L.; Blalock, W.L. Prohibitin 2 represents a novel nuclear AKT substrate during all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. FASEB J. 2014, 28, 2009–2019. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Barnard, D.C.; Li, J.; Peng, R.; Patton, J.G. Regulation of alternative splicing by SRrp86 through coactivation and repression of specific SR proteins. RNA 2002, 8, 526–533. [Google Scholar] [CrossRef]
- Li, J.; Hawkins, I.C.; Harvey, C.D.; Jennings, J.L.; Link, A.J.; Patton, J.G. Regulation of alternative splicing by SRrp86 and its interacting proteins. Mol. Cell. Biol. 2003, 23, 7437–7447. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, B.J.; Issner, R.; Nickerson, J.A.; Sharp, P.A. A coactivator of pre-mRNA splicing. Genes Dev. 1998, 12, 996–1009. [Google Scholar] [CrossRef]
- Aznarez, I.; Barash, Y.; Shai, O.; He, D.; Zielenski, J.; Tsui, L.C.; Parkinson, J.; Frey, B.J.; Rommens, J.M.; Blencowe, B.J. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008, 18, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- Best, A.; Dalgliesh, C.; Kheirollahi-Kouhestani, M.; Danilenko, M.; Ehrmann, I.; Tyson-Capper, A.; Elliott, D.J. Tra2 protein biology and mechanisms of splicing control. Biochem. Soc. Trans. 2014, 42, 1152–1158. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Dragos, V.S.; Stegel, V.; Blatnik, A.; Klancar, G.; Krajc, M.; Novakovic, S. New Approach for Detection of Normal Alternative Splicing Events and Aberrant Spliceogenic Transcripts with Long-Range PCR and Deep RNA Sequencing. Biology 2021, 10, 706. [Google Scholar] [CrossRef] [PubMed]
- Cmero, M.; Schmidt, B.; Majewski, I.J.; Ekert, P.G.; Oshlack, A.; Davidson, N.M. MINTIE: Identifying novel structural and splice variants in transcriptomes using RNA-seq data. Genome Biol. 2021, 22, 296. [Google Scholar] [CrossRef]
- Mertes, C.; Scheller, I.F.; Yepez, V.A.; Celik, M.H.; Liang, Y.; Kremer, L.S.; Gusic, M.; Prokisch, H.; Gagneur, J. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat. Commun. 2021, 12, 529. [Google Scholar] [CrossRef]
- Gerber, A.P.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Piazzi, M.; Bavelloni, A.; Gallo, A.; Blalock, W.L. AKT-Dependent Phosphorylation of ADAR1p110 and ADAR2 Represents a New and Important Link Between Cell Signaling and RNA Editing. DNA Cell Biol. 2020, 39, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Teoh, P.J.; An, O.; Chung, T.H.; Chooi, J.Y.; Toh, S.H.M.; Fan, S.; Wang, W.; Koh, B.T.H.; Fullwood, M.J.; Ooi, M.G.; et al. Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood 2018, 132, 1304–1317. [Google Scholar] [CrossRef]
- Shiromoto, Y.; Sakurai, M.; Minakuchi, M.; Ariyoshi, K.; Nishikura, K. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nat. Commun. 2021, 12, 1654. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Yi, Q.; Tian, M.; Yang, H.T.; Liang, Y.; Huang, J.; Zeng, Q.; Sun, W.; Han, J.; Guo, J.; et al. ADAR1 Prevents R-loop Accumulation-Driven ATR Pathway Activation in Ovarian Cancer. J. Cancer 2022, 13, 2397–2412. [Google Scholar] [CrossRef]
- Lotsof, E.R.; Krajewski, A.E.; Anderson-Steele, B.; Rogers, J.; Zhang, L.; Yeo, J.; Conlon, S.G.; Manlove, A.H.; Lee, J.K.; David, S.S. NEIL1 Recoding due to RNA Editing Impacts Lesion-Specific Recognition and Excision. J. Am. Chem. Soc. 2022, 144, 14578–14589. [Google Scholar] [CrossRef]
- Jimeno, S.; Prados-Carvajal, R.; Fernandez-Avila, M.J.; Silva, S.; Silvestris, D.A.; Endara-Coll, M.; Rodriguez-Real, G.; Domingo-Prim, J.; Mejias-Navarro, F.; Romero-Franco, A.; et al. ADAR-mediated RNA editing of DNA:RNA hybrids is required for DNA double strand break repair. Nat. Commun. 2021, 12, 5512. [Google Scholar] [CrossRef]
- Chiang, C.; Li, Y.; Ng, S.K. The Role of the Z-DNA Binding Domain in Innate Immunity and Stress Granules. Front. Immunol. 2020, 11, 625504. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Pinol-Roma, S.; Kjems, J. Alternative splicing of the ADAR1 transcript in a region that functions either as a 5′-UTR or an ORF. RNA 2007, 13, 1732–1744. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Samuel, C.E. Mechanism of interferon action: Functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 1996, 70, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Ito, K.; Ito, M.; Tsuji, S.; Kwak, S. Novel splice variants of human ADAR2 mRNA: Skipping of the exon encoding the dsRNA-binding domains, and multiple C-terminal splice sites. Gene 2005, 363, 193–201. [Google Scholar] [CrossRef]
- Filippini, A.; Bonini, D.; Giacopuzzi, E.; La Via, L.; Gangemi, F.; Colombi, M.; Barbon, A. Differential Enzymatic Activity of Rat ADAR2 Splicing Variants Is Due to Altered Capability to Interact with RNA in the Deaminase Domain. Genes 2018, 9, 79. [Google Scholar] [CrossRef]
- Gerber, A.; O’Connell, M.A.; Keller, W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA 1997, 3, 453–463. [Google Scholar] [PubMed]
- Chen, C.X.; Cho, D.S.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar] [CrossRef]
- Desterro, J.M.; Keegan, L.P.; Jaffray, E.; Hay, R.T.; O’Connell, M.A.; Carmo-Fonseca, M. SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell 2005, 16, 5115–5126. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qian, G.; Zuo, Y.; Yuan, Y.; Cheng, Q.; Guo, T.; Liu, J.; Liu, C.; Zhang, L.; Zheng, H. Ubiquitin-dependent Turnover of Adenosine Deaminase Acting on RNA 1 (ADAR1) Is Required for Efficient Antiviral Activity of Type I Interferon. J. Biol. Chem. 2016, 291, 24974–24985. [Google Scholar] [CrossRef]
- Sakurai, M.; Shiromoto, Y.; Ota, H.; Song, C.; Kossenkov, A.V.; Wickramasinghe, J.; Showe, L.C.; Skordalakes, E.; Tang, H.Y.; Speicher, D.W.; et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017, 24, 534–543. [Google Scholar] [CrossRef]
- Bavelloni, A.; Focaccia, E.; Piazzi, M.; Raffini, M.; Cesarini, V.; Tomaselli, S.; Orsini, A.; Ratti, S.; Faenza, I.; Cocco, L.; et al. AKT-dependent phosphorylation of the adenosine deaminases ADAR-1 and -2 inhibits deaminase activity. FASEB J. 2019, 33, 9044–9061. [Google Scholar] [CrossRef]
- Shelton, P.M.; Duran, A.; Nakanishi, Y.; Reina-Campos, M.; Kasashima, H.; Llado, V.; Ma, L.; Campos, A.; Garcia-Olmo, D.; Garcia-Arranz, M.; et al. The Secretion of miR-200s by a PKCzeta/ADAR2 Signaling Axis Promotes Liver Metastasis in Colorectal Cancer. Cell Rep. 2018, 23, 1178–1191. [Google Scholar] [CrossRef]
- Torres, A.G.; Pineyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef] [PubMed]
- Dessie, E.Y.; Tu, S.J.; Chiang, H.S.; Tsai, J.J.P.; Chang, Y.S.; Chang, J.G.; Ng, K.L. Construction and Validation of a Prognostic Gene-Based Model for Overall Survival Prediction in Hepatocellular Carcinoma Using an Integrated Statistical and Bioinformatic Approach. Int. J. Mol. Sci. 2021, 22, 1632. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.; Smolka, M.B.; Calabrese, P.; Landolph, A.; Zhang, K.; Zhou, H.; Goodman, M.F. Impact of phosphorylation and phosphorylation-null mutants on the activity and deamination specificity of activation-induced cytidine deaminase. J. Biol. Chem. 2008, 283, 17428–17439. [Google Scholar] [CrossRef]
- Demorest, Z.L.; Li, M.; Harris, R.S. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J. Biol. Chem. 2011, 286, 26568–26575. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Shirakawa, K.; Yokoyama, M.; Fukuda, H.; Sarca, A.D.; Koyabu, S.; Yamazaki, H.; Kazuma, Y.; Matsui, H.; Maruyama, W.; et al. Protein kinase A inhibits tumor mutator APOBEC3B through phosphorylation. Sci. Rep. 2019, 9, 8307. [Google Scholar] [CrossRef]
- Wan, L.; Nagata, T.; Katahira, M. Influence of the DNA sequence/length and pH on deaminase activity, as well as the roles of the amino acid residues around the catalytic center of APOBEC3F. Phys. Chem. Chem. Phys. 2018, 20, 3109–3117. [Google Scholar] [CrossRef]
- Teng, B.; Burant, C.F.; Davidson, N.O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 1993, 260, 1816–1819. [Google Scholar] [CrossRef]
- Chiang, Y.Y.; Nagarajan, D.; Lo, Y.C.; Chen, C.Y.; Ng, I.S.; Chang, C.H.; Lee, D.J.; Chang, J.S. Succinic acid fermentation with immobilized Actinobacillus succinogenes using hydrolysate of carbohydrate-rich microalgal biomass. Bioresour. Technol. 2021, 342, 126014. [Google Scholar] [CrossRef]
- Liu, S.M.; Chang, F.C.; Chen, C.Y.; Shih, S.F.; Meng, B.; Ng, E.; Hsu, C.H.; Chiang, Y.T.; Mao, X.J.; Yi, M.Y.; et al. Effects of Parental Involvement in a Preschool-Based Eye Health Intervention Regarding Children′s Screen Use in China. Int. J. Environ. Res. Public. Health 2021, 18, 11330. [Google Scholar] [CrossRef]
- Ng, D.Z.; Lee, C.Y.; Lam, W.W.; Tong, A.K.; Tan, S.H.; Khoo, L.P.; Tan, Y.H.; Chiang, J.; Chang, E.W.; Chan, J.Y.; et al. Prognostication of diffuse large B-cell lymphoma patients with Deauville score of 3 or 4 at end-of-treatment PET evaluation: A comparison of the Deauville 5-point scale and the DeltaSUVmax method. Leuk. Lymphoma 2022, 63, 256–259. [Google Scholar] [CrossRef]
- Wang, L.; Mai, Z.M.; Ngan, R.K.; Ng, W.T.; Lin, J.H.; Kwong, D.L.; Chiang, S.C.; Yuen, K.T.; Ng, A.W.; Ip, D.K.; et al. Dose-Response Reduction in Risk of Nasopharyngeal Carcinoma From Smoking Cessation: A Multicenter Case-Control Study in Hong Kong, China. Front. Oncol. 2021, 11, 699241. [Google Scholar] [CrossRef] [PubMed]
- Hapuarachchi, H.C.; Wong, W.Y.; Koo, C.; Tien, W.P.; Yeo, G.; Rajarethinam, J.; Tan, E.; Chiang, S.; Chong, C.S.; Tan, C.H.; et al. Transient transmission of Chikungunya virus in Singapore exemplifies successful mitigation of severe epidemics in a vulnerable population. Int. J. Infect. Dis. 2021, 110, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.H.; Lin, Y.J.; Chiu, Y.C.; Chung, W.H.; Ku, C.L.; Ng, C.Y. Feasibility of oral tranexamic acid for vitiligo patients with melasma. Dermatol. Ther. 2021, 34, e15047. [Google Scholar] [CrossRef] [PubMed]
- Malkiel, S.; Sayed, B.A.; Ng, V.; Wall, D.A.; Rozmus, J.; Schreiber, R.A.; Faytrouni, F.; Siddiqui, I.; Chiang, K.Y.; Avitzur, Y. Sequential paternal haploidentical donor liver and HSCT in EPP allow discontinuation of immunosuppression post-organ transplant. Pediatr. Transplant. 2021, 25, e14040. [Google Scholar] [CrossRef] [PubMed]
- Graff, M.; Justice, A.E.; Young, K.L.; Marouli, E.; Zhang, X.; Fine, R.S.; Lim, E.; Buchanan, V.; Rand, K.; Feitosa, M.F.; et al. Discovery and fine-mapping of height loci via high-density imputation of GWASs in individuals of African ancestry. Am. J. Hum. Genet. 2021, 108, 564–582. [Google Scholar] [CrossRef]
- Fang, W.T.; Ng, E.; Liu, S.M.; Chiang, Y.T.; Chang, M.C. Determinants of pro-environmental behavior among excessive smartphone usage children and moderate smartphone usage children in Taiwan. PeerJ 2021, 9, e11635. [Google Scholar] [CrossRef]
- Hong, D.Z.; Goh, J.L.; Ong, Z.Y.; Ting, J.J.Q.; Wong, M.K.; Wu, J.; Tan, X.H.; Toh, R.Q.E.; Chiang, C.L.L.; Ng, C.W.H.; et al. Postgraduate ethics training programs: A systematic scoping review. BMC Med. Educ. 2021, 21, 338. [Google Scholar] [CrossRef]
- Swanton, C.; McGranahan, N.; Starrett, G.J.; Harris, R.S. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. 2015, 5, 704–712. [Google Scholar] [CrossRef]
- Talluri, S.; Samur, M.K.; Buon, L.; Kumar, S.; Potluri, L.B.; Shi, J.; Prabhala, R.H.; Shammas, M.A.; Munshi, N.C. Dysregulated APOBEC3G causes DNA damage and promotes genomic instability in multiple myeloma. Blood Cancer J. 2021, 11, 166. [Google Scholar] [CrossRef]
- Alqassim, E.Y.; Sharma, S.; Khan, A.; Emmons, T.R.; Cortes Gomez, E.; Alahmari, A.; Singel, K.L.; Mark, J.; Davidson, B.A.; Robert McGray, A.J.; et al. RNA editing enzyme APOBEC3A promotes pro-inflammatory M1 macrophage polarization. Commun. Biol. 2021, 4, 102. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Baysal, B.E. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci. Rep. 2016, 6, 39100. [Google Scholar] [CrossRef] [PubMed]
- Buisson, R.; Lawrence, M.S.; Benes, C.H.; Zou, L. APOBEC3A and APOBEC3B Activities Render Cancer Cells Susceptible to ATR Inhibition. Cancer Res. 2017, 77, 4567–4578. [Google Scholar] [CrossRef]
- Sato, Y.; Ohtsubo, H.; Nihei, N.; Kaneko, T.; Sato, Y.; Adachi, S.I.; Kondo, S.; Nakamura, M.; Mizunoya, W.; Iida, H.; et al. Apobec2 deficiency causes mitochondrial defects and mitophagy in skeletal muscle. FASEB J. 2018, 32, 1428–1439. [Google Scholar] [CrossRef]
- Ohtsubo, H.; Sato, Y.; Suzuki, T.; Mizunoya, W.; Nakamura, M.; Tatsumi, R.; Ikeuchi, Y. Data supporting possible implication of APOBEC2 in self-renewal functions of myogenic stem satellite cells: Toward understanding the negative regulation of myoblast differentiation. Data Brief. 2017, 12, 269–273. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Basu, M.K.; Jordan, I.K.; Pavlov, Y.I.; Koonin, E.V. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 2005, 4, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Perkovic, M.; Hain, A.; Jaguva Vasudevan, A.A.; Hofmann, H.; Hanschmann, K.M.; Muhlebach, M.D.; Schumann, G.G.; Konig, R.; Cichutek, K.; et al. APOBEC4 Enhances the Replication of HIV-1. PLoS ONE 2016, 11, e0155422. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; D’Erchia, A.M.; Lo Giudice, C.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017, 45, D750–D757. [Google Scholar] [CrossRef]
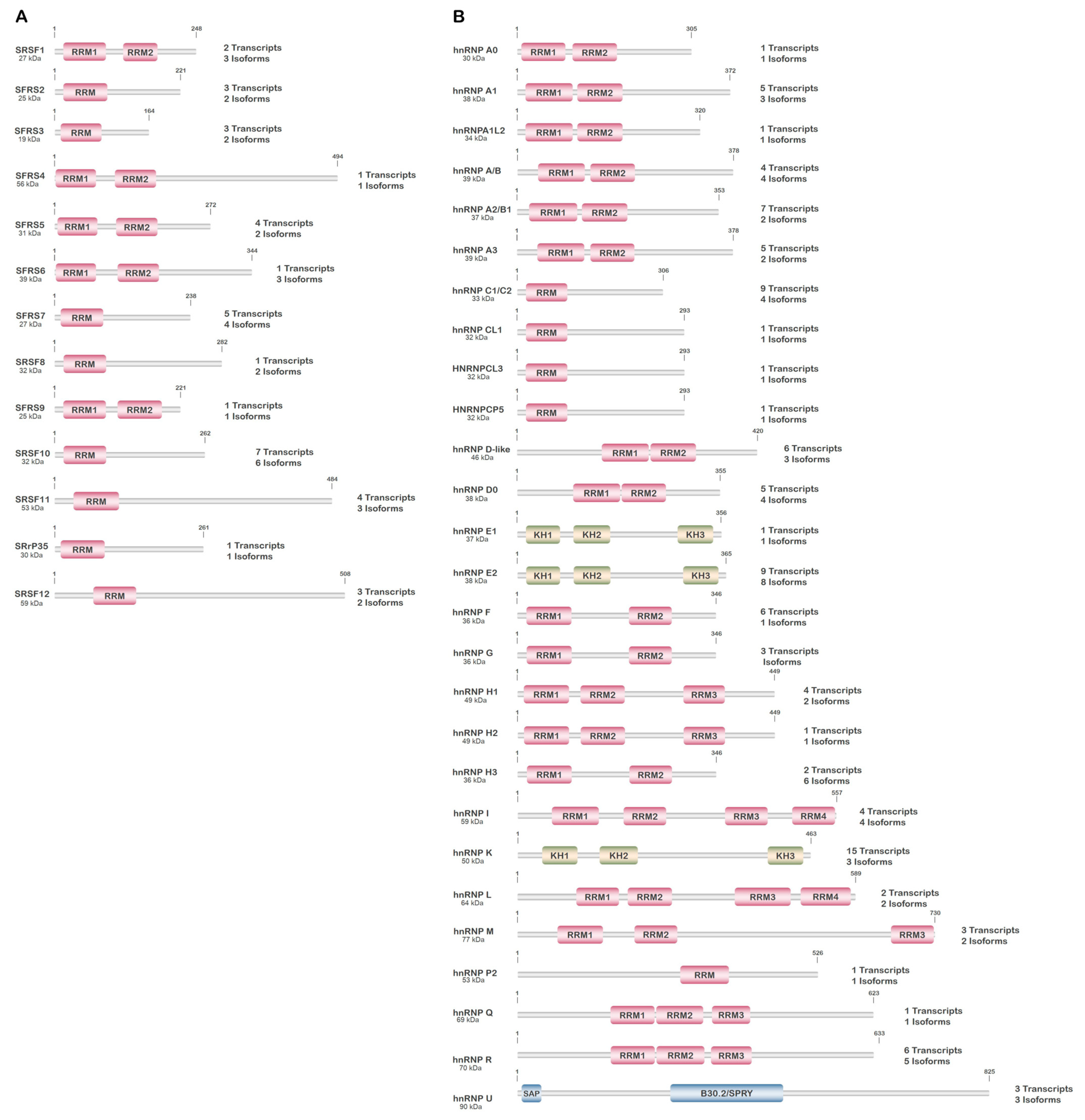
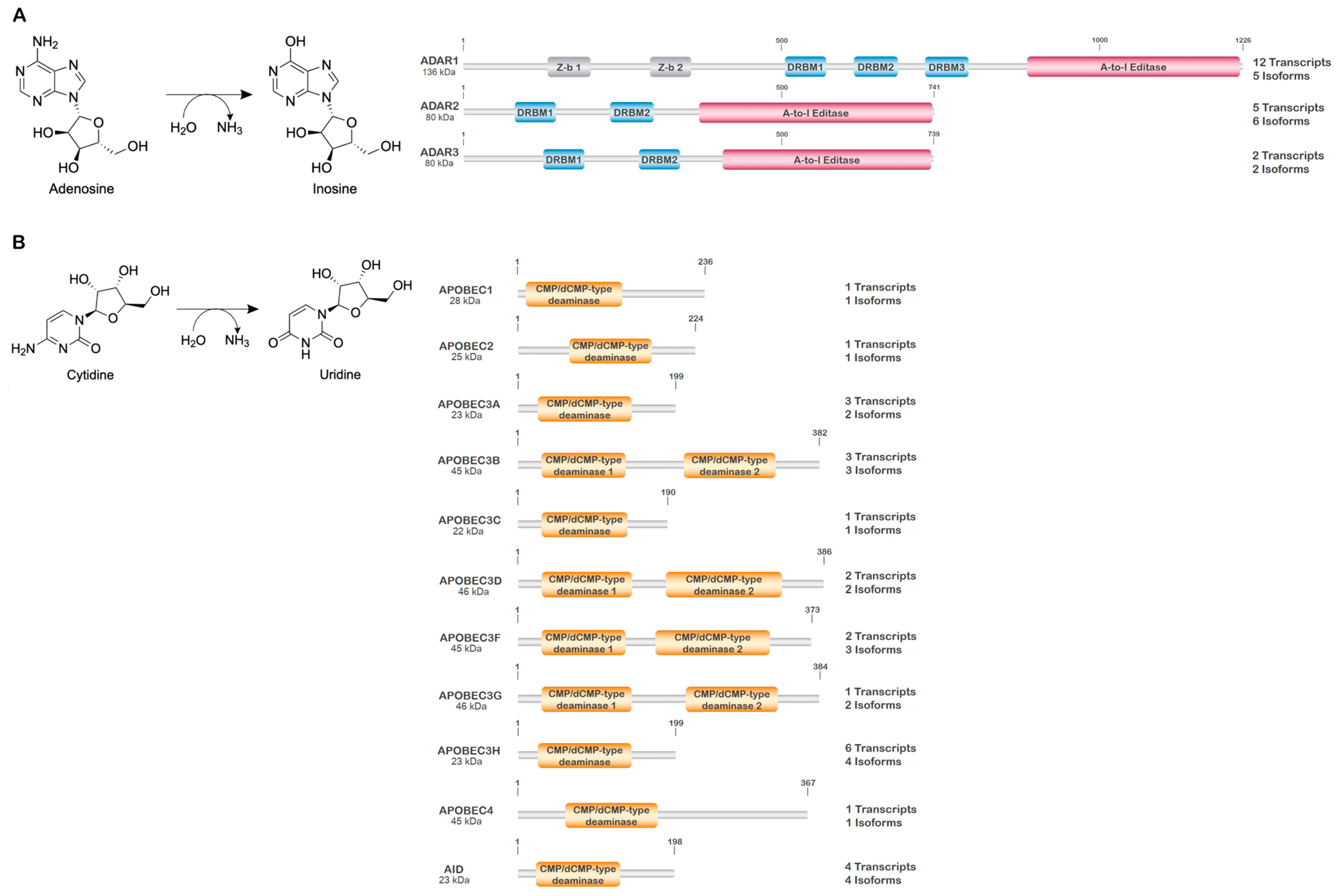
| Protein (UniProt ID) | Gene | Location | Function/Role | Localization | 1 Alternant Transcripts/Isoforms | 2 PTMs |
|---|---|---|---|---|---|---|
| * SREK1 (Q8WXA9) | SREK1 | Ch.5q12.3 | Regulates alternative splicing by influencing the activity of other splicing factors. Inhibits the splicing activity of SFRS1, SFRS2, and SFRS6, but augments the splicing activity of SFRS3. | Nuc | 3 trscpts/2 isoforms | 18 (uc) |
| * SRRM1 (Q8IYB3) | SRRM1 | Ch.1p36.11 | Part of the pre- and post-splicing mRNP complex. Promotes constitutive ESE-dependent splicing by bridging sequence-specific SRSFs, TRA2B, and basal snRNP factors. Ubiquitously expressed. | Nuc matrix, Speckles | 1 trscpt/2 isoforms # | 166 (uc) |
| * SRRM2 (Q9UQ35) | SRRM2 | Ch.16p13.3 | A required component of the pre-mRNA spliceosome. Mainly expressed in liver placenta and leukocytes. Associates with SRRM1, SRSF3, SRSF4, and SRSF5. | Nuc, Speckles | 3 trscpts/3 isoforms | 736 (uc) |
| SRSF1 (Q07955) | SRSF1 | Ch.17q22 | Prevents exon skipping to ensure the accuracy of splicing and alternative splicing by forming a bridge between the 5′- and 3′-splice site binding components, U1 snRNP, and U2AF. Isoform 1 acts as a splicing enhancer. Isoforms 2 and 3 act as splicing repressors. | Speckles, Cyto (shuttles) | 2 trscpts/3 isoforms | 65 (muc) |
| SRSF2 (Q01130) | SRSF2 | Ch.17q25.1 | Central to pre-mRNA splicing. Required for the formation of the ATP-dependent splicing complex. Forms a bridge between the 5′- and 3′-splice site binding components, U1 snRNP, and U2AF. | Nuc, Nucleoplasm, Speckles | 3 trscpts/2 isoforms | 45 (muc) |
| SRSF3 (P84103) | SRSF3 | Ch.6p21.31-21.2 | Specifically promotes exon inclusion during alternative splicing. Recruitment of SFRS3 to mRNA binding elements near N6-meythyladenosine (m6A) sites, leads to exon inclusion. | Nuc, Cyto, Speckles | 2 trscpts/2 isoforms* | 33 (muc) |
| SRSF4 (Q08170) | SRSF4 | Ch.1p35.3 | Plays a role in alternative splice site selection during pre-mRNA splicing. | Speckles | 1 trscpt/1 isoform | 67 (uc) |
| SRSF5 (Q13243) | SRSF5 | Ch.14q24.1 | Plays a role in constitutive splicing and can influence alternative splice site selection. | Nuc | 4 trscpts/2 isoforms # | 39 (uc) |
| SRSF6 (Q13247) | SRSF6 | Ch.20q13.11 | Plays a role in constitutive splicing and can influence alternative splice site selection. | Nuc, Speckles | 1 trscpt/3 isoforms # | 56 (uc) |
| SRSF7 (Q16629) | SRSF7 | Ch.2p22.1 | Required for pre-mRNA splicing. Can modulate alternative splicing in vitro. Prevalent in brain, kidneys, and lungs. | Nuc, Cyto | 5 trscpts/4 isoforms # | 53 (uc) |
| SRSF8 (Q9BRL6) | SRSF8 | Ch.11q21 | Involved in pre-mRNA alternative splicing. Highly expressed in pancreas, spleen, and prostate; poorly expressed in lungs, liver, and thymus. | Nuc | 1 trscpt/2 isoforms # | 44 (uc) |
| SRSF9 (Q13242) | SRSF9 | Ch.12q24.31 | Plays a role in constitutive splicing and can influence alternative splice site selection. Highly expressed in heart, kidneys, pancreas, and placenta; poorly expressed in brain, liver, lungs, and skeletal muscle. | Nuc | 1 trscpt/1 isoform | 49 (uc) |
| SRS10 (O75494) | SRSF10 | Ch.1p36.11 | Acts as a general repressor of pre-mRNA splicing when in its dephosphorylated form by interfering with the U1 snRNP 5′ splice recognition of SNRNP70. Required for splicing repression in M-phase cells and after heat shock. Promotes exon skipping during alternative splicing. May be involved in the regulation of alternative splicing in neurons, with Isoform 1 acting to promote alternative splicing and Isoform 3 acting to suppress alternative splicing. | Speckles, Cyto | 7 trscpts/6 isoforms | 46 (muc) |
| SRS11 (Q05519) | SRSF11 | Ch.1p31.1 | May function in pre-mRNA splicing. | Nuc | 4 trscpts/3 isoforms | 58 (uc) |
| SRS12 (Q8WXF0) | SRSF12 | Ch.6q15 | Acts as an antagonist to SR proteins in pre-mRNA splicing regulation. Mainly expressed in testis. | Nuc | 1 trscpt/1 isoform | 21 (uc) |
| * TIA1 (P31483) | TIA1 | Ch.2p13.3 | Regulates alternative pre-RNA splicing by binding U-rich sequences immediately downstream of 5′ splice sites in a U snRNP-dependent manner. Can promote atypical 5′ splice site selection by promoting splicing of exons with weak 5′ splice sites. Isoform 2 demonstrates enhanced splicing regulatory activity as compared to Isoform 1. Most prominently expressed in heart, small intestine, kidneys, liver, lungs, skeletal muscle, pancreas, ovary, and testis. Disease associated. | Nuc, Cyto, Stress granule | 5 trscpts/5 isoforms | 11 (uc) |
| * TRA2A (Q13595) | TRA2A | Ch.7p15.3 | Associates with pre-mRNA in a sequence-specific manner to regulate pre-mRNA splicing. Expression is ubiquitous. Associates with SR30, SRSF3, SRSF4, SRSF5, and SRSF6. | Nuc | 3 trscpts/3 isoforms | 63 (uc) |
| * TRA2B (P62995) | TRA2B | Ch.3q27.2 | Associates with pre-mRNA in a sequence-specific manner to promote or inhibit exon inclusion. Works by antagonizing splicing regulators belonging to the hnRNP class of proteins. Ubiquitously expressed with highest expression in heart, skeletal muscle, and pancreas; lowest expression in kidneys and liver. | Nuc | 2 trscpts/2 isoforms | 67 (uc) |
| Protein (UniProt ID) | Gene | Location | Function/Role | Localization | 1 Alternant Transcripts/Isoforms | 2 PTMs |
|---|---|---|---|---|---|---|
| FUS (P35637) | FUS | Ch.16p11.2 | Also referred to as hnRNP P2. DNA/RNA-binding protein that plays a role in transcription regulation, RNA splicing, RNA transport, and DNA damage response. Acts as a molecular mediator between RNA polymerase II and U1 small nuclear ribonucleoprotein, thus coupling transcription and splicing. Autoregulates its own expression through nonsense mediated decay. Disease associated. | Nuc | 2 trscpts/2 isoforms | 98 (46 wc) |
| ROA0 (Q13151) | HNRNPA0 | Ch.5q31.2 | Specifically binds AU-rich element (ARE)-containing mRNAs and is involved in post-transcriptional stability of diverse cytokine mRNAs. | Nuc | 1 trscpt/1 isoform | 62 (muc) |
| ROA1 (P09651) | HNRNPA1 | Ch.12q13.13 | Involved in the packaging of pre-mRNA into hnRNP particles, nucleo-cytoplasmic transport of poly(A) mRNA, and modulation of splice site selection. Disease associated. | Nuc, Cyto (shuttles) | 5 trscpts/3 isoforms | 91 (ssc) |
| RA1L2 (Q32P51) | HNRNPA1L2 | Ch.13q14.3 | Involved in the packaging of pre-mRNA into hnRNP particles, nucleo-cytoplasmic transport of poly(A) mRNA, and modulation of splice site selection. | Nuc, Cyto | 1 trscpt/1 isoform | 37 (uc) |
| ROAA (Q99729) | HNRNPAB | Ch.5q35.3 | Has high affinity for G-rich and U-rich regions of ss hnRNA. Binds to APOB transcripts around the APOBEC C-to-U RNA editing site. | Nuc, Cyto | 4 trscpts/4 isoforms | 43 (uc) |
| ROA2 (P22626) | HNRNPA2B1 | Ch.7p15.2 | Associates with pre-mRNAs in a sequence-dependent fashion to package the transcripts. Packaging plays a role in transcription, pre-mRNA processing, RNA nuclear export, subcellular location, mRNA translation, and stability of mature mRNAs. Promotes pri-miRNA processing and sorting. Involved in innate immune response activation. Disease associated. | Nuc, Cyto, Nucleoplasm, Cyto granules, secreted | 7 trscpts/2 isoforms | 107 (muc) |
| ROA3 (P51991) | HNRNPA3 | Ch.2q31.2 | Has a role in cytoplasmic trafficking of RNA. May be involved in pre-mRNA splicing. | Nuc | 5 trscpts/2 isoforms | 94 (muc) |
| HNRPC (P07910) | HNRNPC | Ch.14q11.2 | Nucleates assembly of 40S hnRNP particles by binding pre-mRNA. May play a role in the early steps of spliceosome assembly and pre-mRNA splicing. N6-methyladenosine (m6A) has been shown to influence mRNA splicing by enhancing HNRNPC binding. | Nuc | 9 trscpts/4 isoforms | 82 (ssc) |
| HNRC1 (O60812) | HNRNPCL1 | Ch.1p36.21 | May play a role in nucleosome assembly. Specifically found in skeletal muscle tissue. | Nuc | 1 trscpt/1 isoform | 23 (uc) |
| HNRC2 (B2RXH8) | HNRNPCL2 | Ch.1p36.21 | May play a role in nucleosome assembly. Tissue expression is unclear. | Nuc | 1 trscpt/1 isoform | 4 (uc) |
| HNRC3 (B7ZW38) | HNRNPCL3 | Ch.1p36.21 | Role unknown. Expressed mainly in the cortical plate, blood, and hindlimb stylopod muscle. | Nuc | 1 trscpt/1 isoform | 11 (uc) |
| HNRPD (Q14103) | HNRNPD | Ch.4q21.22 | Binds with high affinity to AU-rich elements (AREs) found within the 3′-UTR of many proto-oncogenes and cytokine mRNAs. Also functions as a transcription factor. May be involved in translationally coupled mRNA turnover. | Nuc, Cyto (shuttles with circadian clock) | 5 trscpts/4 isoforms | 76 (muc) |
| HNRDL (O14979) | HNRNPDL | Ch.4q21.22 | Promotes transcriptional activation or repression in a context-dependent manner. Binds AU-rich elements (AREs) found within the 3′-UTR of many proto-oncogenes and cytokine mRNAs with high affinity. Preferentially expressed in heart, brain, placenta, lungs, liver, skeletal muscle, kidneys, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon, and leukocytes. Disease associated. Elevated expression observed in diverse cancers. | Nuc, Cyto (shuttles) | 6 trscpts/3 isoforms | 61 (uc) |
| HNRPF (P52597) | HNRNPF | Ch.10q11.21 | Component of the heterogeneous nuclear ribonucleoprotein (hnRNP) complexes. Plays a role in regulating alternative splicing events. Maintains target RNA in an unfolded state. | Nuc, Nucleoplasm | 6 trscpts/1 isoform | 65 (muc) |
| HNRH1 (P31943) | HNRNPH1 | Ch.5q35.3 | Component of the heterogeneous nuclear ribonucleoprotein (hnRNP) complexes. Regulates pre-mRNA alternative splicing. Disease associated. | Nuc, Nucleoplasm | 4 trscpts/2 isoforms | 64 (muc) |
| HNRH2 (P55795) | HNRNPH2 | Ch.Xq22.1 | Component of the heterogeneous nuclear ribonucleoprotein (hnRNP) complexes. Disease associated. | Nuc, Nucleoplasm | 1 trscpt/1 isoform | 48 (muc) |
| HNRH3 (P31942) | HNRNPH3 | Ch.10q21.3 | Participates in early heat shock-induced splicing arrest. Different isoforms suspected of possessing distinct functions in splicing. | Nuc | 2 trscpts/6 isoforms | 60 (uc) |
| HNRPK (P61978) | HNRNPK | Ch.9q21.32 | A major pre-mRNA-binding protein that binds to poly(C) sequences. Plays a vital role in the p53/TP53 response to DNA damage. Disease associated. | Nuc, Cyto, Nucleoplasm, Cellular Projections | 15 trscpts/3 isoforms | 132 (23 wc) |
| HNRPL (P14866) | HNRNPL | Ch.19p13.2 | Component of heterogeneous nuclear ribonucleoprotein (hnRNP) complexes. Binds to exonic or intronic sites to function as an activator or repressor of exon inclusion. Regulates its own expression through the inclusion of a "poison" exon. | Nuc, Cyto, Nucleoplasm | 2 trscpts/2 isoforms | 90 (ssc) |
| HNRPM (P52272) | HNRNPM | Ch.19p13.2 | Pre-mRNA binding protein involved in splicing. Acts as a receptor for carcinoembryonic antigen in Kupffer cells to initiate signaling events, leading to the induction of IL-1 alpha, IL-6, IL-10, and TNFα. | Nuc, Nucleolus | 3 trscpts/2 isoforms | 125 (muc) |
| HNRPR (O43390) | HNRNPR | Ch.1q36.12 | Component of ribonucleosomes. Role unknown. Disease associated. | Nuc, Cyto, Nucleoplasm, Microsome | 6 trscpts/5 isoforms | 76 (uc) |
| HNRPU (Q00839) | HNRNPU | Ch.1q44 | DNA-/RNA-binding protein involved in nuclear chromatin organization, telomere-length regulation, transcription, mRNA alternative splicing and stability, transcriptional silencing, and mitotic cell progression. Required for embryonic development. Disease associated. | Nuc, Cyto, Nuc Matrix, Speckles, cytoskeleton | 3 trscpts/3 isoforms | 185 (ssc) |
| PCBP1 (Q15365) | PCBP1 | Ch.2p13.3 | Also referred to as hnRNP E1. Binds ssRNA and ssDNA. Together with PCBP2, may regulate erythropoiesis through mRNA splicing. Predominantly expressed in skeletal muscle, thymus, and peripheral blood leukocytes; lower expression observed in prostate, spleen, testis, ovary, small intestine, heart, liver, and adrenal and thyroid glands. | Nuc, Cyto (shuttles) | 1 trscpt/1 isoform | 49 (muc) |
| PCBP2 (Q15366) | PCBP2 | Ch.12q13.13 | Also referred to as hnRNP E2. Binds ssRNA and ssDNA. Negatively regulates innate immune antiviral responses mediated by MAVS, and the cGAS-STING pathway. Together with PCBP1, may regulate erythropoiesis through mRNA splicing. | Nuc, Cyto (shuttles) | 9 trscpts/8 isoforms | 42 (ssc) |
| PTBP1 (P26599) | PTBP1 | Ch.19p13.3 | Polypyrimidine tract binding protein (PTBP)-1 (also known as hnRNP I) interacts with polypyrimidine (PP) stretches at the branch point region. Plays a role in pre-mRNA splicing, alternative splicing, and alternate 5′-3′ splice site usage. Promotes exon skipping of its own pre-mRNA during muscle cell differentiation. Can sequester miRNAs. | Nuc | 4 trscpts/4 isoforms | 65 (uc) |
| RBMX (P38159) | RBMX | Ch.Xq26.3 | Also referred to as hnRNP G. Involved in the regulation of pre- and post-transcriptional processes. A component of the supraspliceosome complex that regulates pre-mRNA alternative splice site selection. Can activate or suppress exon inclusion. Ubiquitously expressed. Disease associated. | Nuc | 3 trscpts/3 isoforms | 118 (uc) |
| HNRPQ (O60506) | SYNCRIP | Ch.6q14.3 | Component of the CRD-mediated complex. Isoform 1 binds to APOB mRNA AU-rich sequences and is a regulatory part of the APOB mRNA editosome complex. May be involved in translationally coupled mRNA turnover. Isoform 3 is a component of the GAIT (gamma interferon-activated inhibitor of translation) complex, which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. The GAIT complex binds GAIT elements in the 3′-UTR of diverse inflammatory mRNAs to suppress their translation. | Nuc, Cyto, Microsome, ER (Isoforms 1-3 preferentially localize in the nucleoplasm) | 9 trscpts/4 isoforms | 87 (uc) |
| Protein (UniProt ID) | Gene | Location | 1 Substrate | Function/Role | Localization | 2 Alternant Transcripts/Isoforms | 3 PTMs |
|---|---|---|---|---|---|---|---|
| ADAR Family of Adenosine Deaminases | |||||||
| DSRAD (P55265) | ADAR | Ch.1q21.3 | dsRNA, Z-DNA | Catalyzes the deamination of A-to-I in RNA of complex secondary structure. May affect gene expression by altering protein coding, pre-mRNA splicing, RNA stability, RNA transport, miRNA targeting, and RNA–protein interactions. Edits both host and foreign RNAs. May have a role in DNA strand break repair. Isoform 1 is interferon-inducible, primarily localizes to the cytoplasm, and demonstrates elevated activity toward foreign (viral) RNAs. Suppresses PKR activation and the induction of the integrated stress response. Can associate with Z-DNA or Z-RNA. Isoform 5 is constitutively expressed and localizes primarily to the nucleus. Ubiquitously expressed with highest expression in brain and lungs. Disease associated. Overexpressed in many cancers; expression associated with cancer aggressiveness. | Nuc, Nucleolus, Cyto (shuttles) | 12
trscpts/5 isoforms # These numbers may double due to alternate exon 1 usage. | 125 (ssc) |
| RED1 (P78563) | ADARB1 | Ch.22q22.3 | dsRNA | Catalyzes the deamination of A-to-I in RNA of complex secondary structure. May affect gene expression by altering protein coding, pre-mRNA splicing, RNA stability, RNA transport, miRNA targeting, and RNA–protein interactions. Edits both host and foreign RNAs. Suppresses PKR activation and the induction of the integrated stress response. Involved in the RNA editing of RNA/DNA hybrids during DNA double-strand break repair. Disease associated. Ubiquitously expressed. Highly expressed in brain, heart, lower placenta; moderately expressed in lungs, liver, and kidneys. Isoform 5 is seen in hippocampus and colon. Decreased activity in astrocytomas correlated with disease severity. | Nuc | 5 trscpts/6 isoforms # | 23 (ssc) |
| RED2 (Q9NS39) | ADARB2 | Ch.10q15.3 | dsRNA | ADAR family member that lacks editing activity. Likely alters/regulates ADAR1 and ADAR2. Binds both ss and dsRNA. Expression is brain-specific. | Nuc | 2 trscpts/2 isoforms | 13 (uc) |
| Cytidine Deaminases: APOBEC Family and AID | |||||||
| ABEC1 (P41238) | APOBEC1 | Ch.12p13.31 | ssRNA, ssDNA (h) | Catalyzes the C-to-U post-transcriptional editing of several mRNAs, including APOB and NF1. Acts as a homodimer. May play a role in the epigenetic regulation of gene expression. Expressed exclusively in small intestine. | Nuc, Cyto | 1 trscpt/1 isoform # | 2 (wc) |
| ABEC2 (Q9Y235) | APOBEC2 | Ch.6p21.1 | ss/dsDNA (h) (binding only) | A potential C-to-U editing enzyme with no known substrate and poor deaminase activity. May have a role in epigenetic regulation of gene expression. Exclusively expressed in heart and skeletal muscle. | Nuc, Cyto | 1 trscpt/1 isoform | 2 (uc) |
| ABC3A (P31941) | APOBEC3A | Ch.22q13.1 | ssRNA, ssDNA | Demonstrates C-to-U editing activity toward ssRNA and ssDNA. Best effectiveness against ssDNAs of foreign origin (viral). Deaminates both C and meC and can hyper-edit nuclear and mitochondrial DNA. May have a role in epigenetic regulation of gene expression. Expressed in peripheral leukocytes and CD14+ phagocytic cells. Highly expressed in keratinocytes and peripheral blood monocytes. Present at detectable levels in other lymphoid tissue, lungs, bladder, urinary tract, and adipose tissue. Enhanced expression observed in diverse tumor tissues. Induced by interferon and CpG ssDNA. | Nuc, Cyto | 3 trscpts/2 isoforms # | 4 (uc) |
| ABC3B (Q9UH17) | APOBEC3B | Ch.22q13.1 | ssDNA | Demonstrates C-to-U editing activity. Selectively targets ssDNAs of foreign origin (viral). Has the ability to hyper-edit nuclear and mitochondrial DNA. Acts as a homodimer. Interacts with APOBEC3G. Expressed in peripheral blood leukocytes, bone marrow, spleen, kidneys, bladder, urinary tract, testes, prostate, heart, thymus, ovary, and gastrointestinal tract. Expression is induced by interferon. | Nuc | 3 trscpts/3 isoforms | 10 (muc) |
| ABC3C (Q9NRW3) | APOBEC3C | Ch.22q13.1 | ssDNA (v) | Demonstrates C-to-U editing activity. Selectively targets ssDNAs of foreign origin (viral). May play a role in epigenetic regulation of gene expression. Expressed in peripheral blood mononuclear cells, bone marrow, spleen, thymus, respiratory tract, kidneys, bladder, urinary tract, testes, prostate, skin, muscle, heart, ovary, endocrine tissues, liver, and gastrointestinal tract. Expression is induced by interferon. | Nuc, Cyto | 1 trscpt/1 isoform | 5 (uc) |
| ABC3D (Q96AK3) | APOBEC3D | Ch.22q13.1 | ssDNA (v) | Demonstrates C-to-U editing activity. Selectively targets ssDNAs of foreign origin (viral). Antiviral activity occurs through both deaminase-dependent and -independent mechanisms. Can homodimerize or form heterodimers with APOBEC3F and APOBEC3G. Expressed in peripheral blood mononuclear cells, bone marrow, lymphoid tissues, gastrointestinal tract, and female reproductive system. | Cyto, P-bodies | 2 trscpts/2 isoforms | 4 (uc) |
| ABC3F (Q8UX4) | APOBEC3F | Ch.22q13.1 | ssDNA (v) | Demonstrates C-to-U editing activity. Selectively targets ssDNAs of foreign origin (viral). Exhibits antiviral activity toward wide spectrum of viruses through both deaminase-dependent and -independent mechanisms. Has not been shown to target nuclear or mitochondrial DNA. May play a role in epigenetic regulation of gene expression. Widely expressed with highest expression observed in ovary. Interacts with APOBEC3G. Expression is induced by interferon. | Cyto, P-bodies | 2 trscpts/3 isoforms # | 3 (muc) |
| ABC3G (Q9HC16) | APOBEC3G | Ch.22q13.1 | ssRNA, ssDNA (v) | Demonstrates C-to-U editing activity. Selectively targets ssDNAs of foreign origin (viral). Antiviral activity occurs through both deaminase-dependent and -independent mechanisms. Acts as a homodimer or homo-oligomer. Can bind RNA and form inactive high molecular weight (HMM) or active low molecular weight (LMM) ribonucleoprotein complexes. Expressed in peripheral blood mononuclear cells, bone marrow, lymphoid tissue, ovary, testis, bladder, urinary tract, gastrointestinal tract, and liver respiratory tract. Interacts with APOBEC3B, APOBEC3F, MOV10, AGO2, EIF4E, EIF4ENIF1, DCP2, and DDX6 in an RNA-dependent manner. Expression is induced by interferon. Upregulated in certain tumors. | Nuc, Cyto, P-bodies | 1 trscpt/2 isoforms # | 11 (ssc) |
| ABC3H (Q6NTF7) | APOBEC3H | Ch.22q13.1 | ssDNA (v) | Demonstrates C-to-U editing activity. Selectively targets ssDNAs of foreign origin (viral). Antiviral activity occurs through both deaminase-dependent and -independent mechanisms. Expressed in peripheral blood mononuclear cells, bone marrow, lymphoid tissue, lungs, testis, ovary, and skin. | Nuc, Cyto, P-bodies | 6 trscpts/4 isoforms | 1 (uc) |
| ABEC4 (Q8WW27) | APOBEC4 | Ch.1q25.3 | ND | Possible C-to-U editing enzyme with no known substrate. Expression is restricted to testis. | Nuc (predicted) | 1 trscpt/1 isoform | 1 (uc) |
| AICDA (Q9GZX7) | AICDA | 12p13.31 | ssRNA (binding only), ssDNA (h), RNA/DNA hybrid | Demonstrates C-to-U editing activity. Involved in somatic hyper-editing, gene conversion, and class-switch recombination in B-lymphocytes. Necessary for B-cell differentiation and antibody maturation. Hyper-edits other sites in the genome in several pathologies. May have a role in epigenetic regulation of gene expression. Disease associated. Highly expressed in lymphoid tissues and germinal center B-cells. | Nuc, Cyto (predominant) | 4 trscpts/4 isoforms | 7 (2 wc) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piazzi, M.; Bavelloni, A.; Salucci, S.; Faenza, I.; Blalock, W.L. Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing. Genes 2023, 14, 1386. https://doi.org/10.3390/genes14071386
Piazzi M, Bavelloni A, Salucci S, Faenza I, Blalock WL. Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing. Genes. 2023; 14(7):1386. https://doi.org/10.3390/genes14071386
Chicago/Turabian StylePiazzi, Manuela, Alberto Bavelloni, Sara Salucci, Irene Faenza, and William L. Blalock. 2023. "Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing" Genes 14, no. 7: 1386. https://doi.org/10.3390/genes14071386
APA StylePiazzi, M., Bavelloni, A., Salucci, S., Faenza, I., & Blalock, W. L. (2023). Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing. Genes, 14(7), 1386. https://doi.org/10.3390/genes14071386







