Exploring the Influence of Growth-Associated Host Genetics on the Initial Gut Microbiota in Horses
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. 16S rRNA Amplicon Sequencing
2.3. Animal Genotyping
2.4. Taxonomic Assignment and Diversity
2.5. Growth-Associated Host Genetic Effects on Gut Microbiota
3. Results
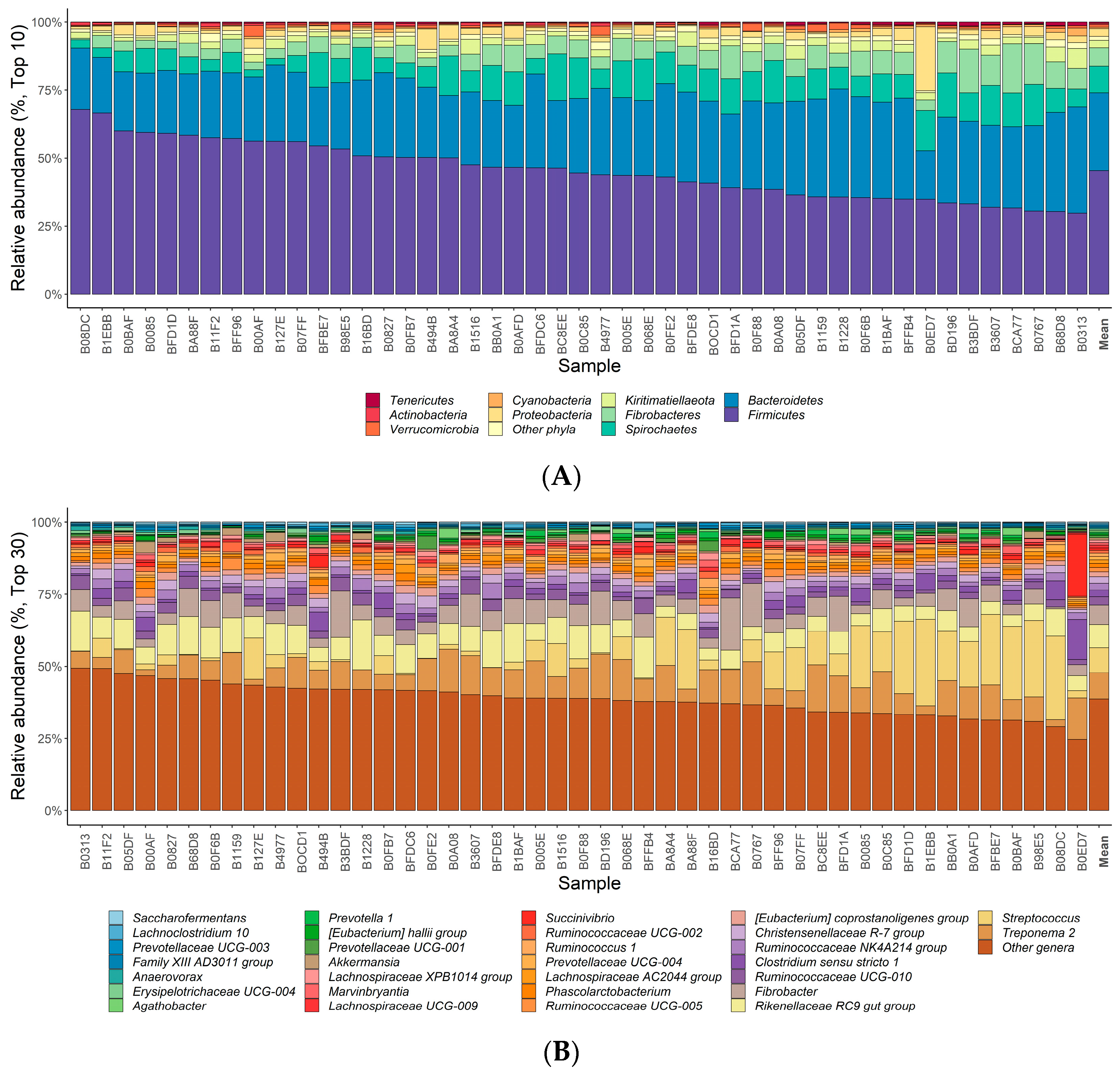
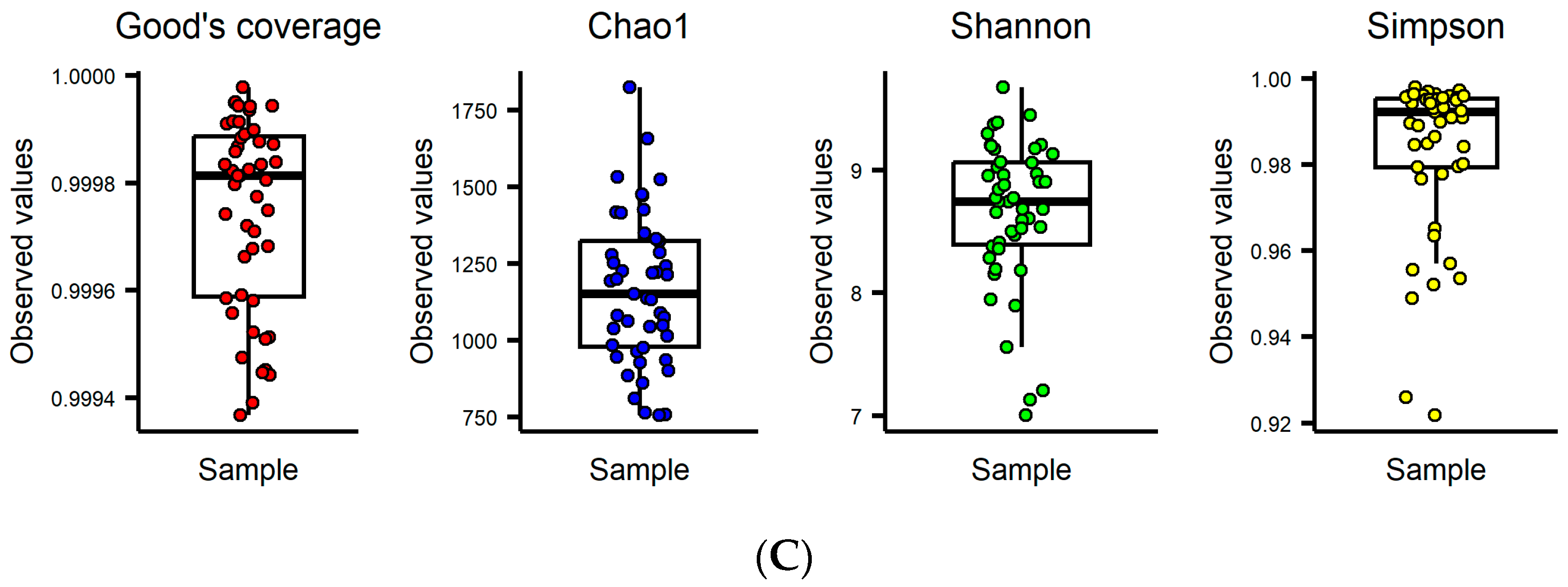
3.1. Gut Microbial Composition and α Diversity
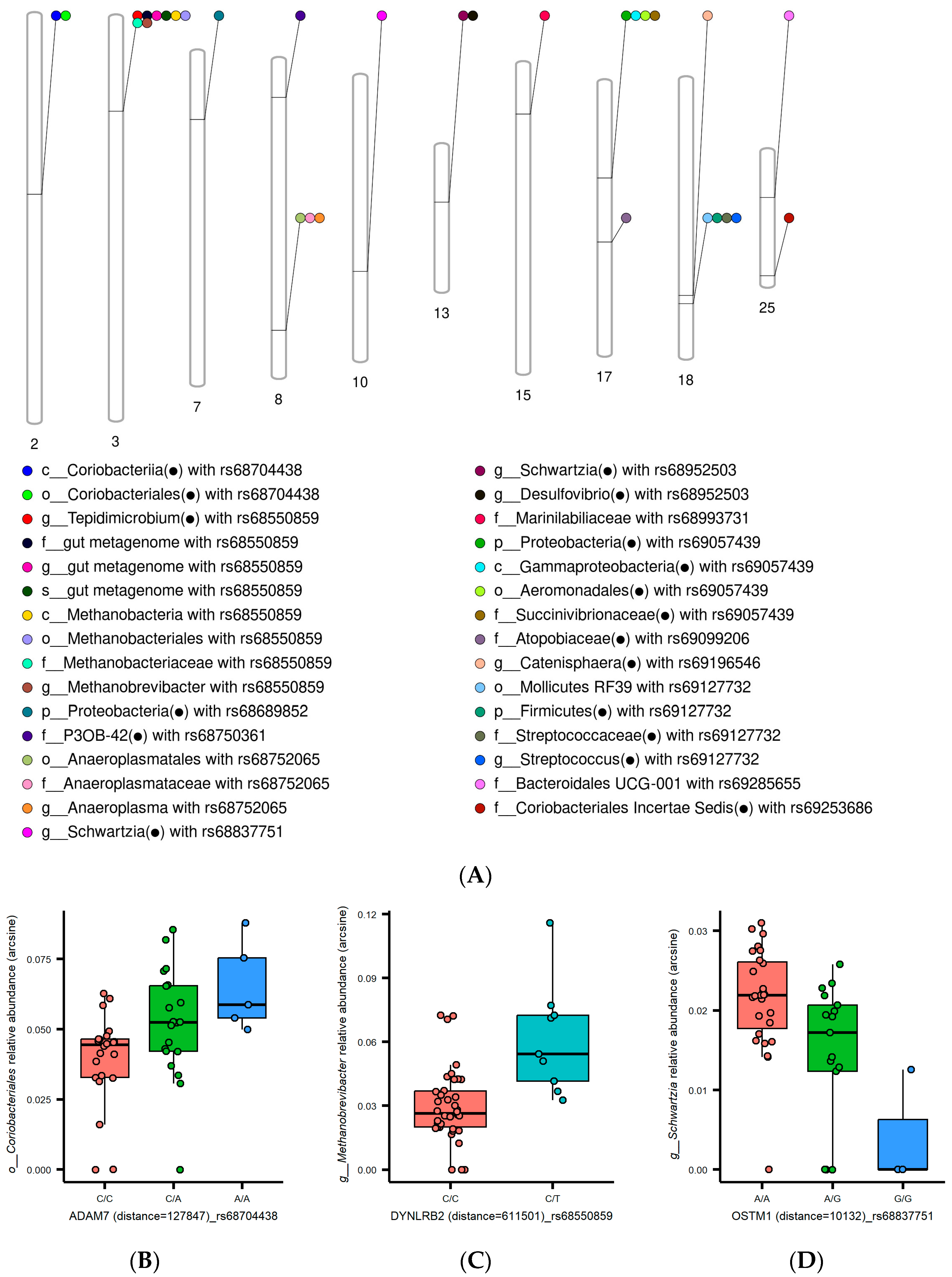
3.2. Interplay between Growth-Associated Variants and Gut Microbiota
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-J.; Hase, K. Gut microbiota–generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef]
- Su, S.; Zhao, Y.; Liu, Z.; Liu, G.; Du, M.; Wu, J.; Bai, D.; Li, B.; Bou, G.; Zhang, X. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments of Mongolian horses. Microbiologyopen 2020, 9, 1085–1101. [Google Scholar]
- Costa, M.C.; Weese, J.S. Understanding the intestinal microbiome in health and disease. Vet. Clin. Equine Pract. 2018, 34, 1–12. [Google Scholar] [CrossRef]
- Kauter, A.; Epping, L.; Semmler, T.; Antao, E.-M.; Kannapin, D.; Stoeckle, S.D.; Gehlen, H.; Lübke-Becker, A.; Günther, S.; Wieler, L.H. The gut microbiome of horses: Current research on equine enteral microbiota and future perspectives. Anim. Microbiome 2019, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [PubMed]
- Montgomery, T.L.; Künstner, A.; Kennedy, J.J.; Fang, Q.; Asarian, L.; Culp-Hill, R.; D’Alessandro, A.; Teuscher, C.; Busch, H.; Krementsov, D.N. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc. Natl. Acad. Sci. USA 2020, 117, 27516–27527. [Google Scholar]
- Zhang, Q.; Linke, V.; Overmyer, K.A.; Traeger, L.L.; Kasahara, K.; Miller, I.J.; Manson, D.E.; Polaske, T.J.; Kerby, R.L.; Kemis, J.H. Genetic mapping of microbial and host traits reveals production of immunomodulatory lipids by Akkermansia muciniphila in the murine gut. Nat. Microbiol. 2023, 8, 424–440. [Google Scholar] [CrossRef]
- Bay, V.; Gillespie, A.; Ganda, E.; Evans, N.; Carter, S.D.; Lenzi, L.; Lucaci, A.; Haldenby, S.; Barden, M.; Griffiths, B.E. The bovine foot skin microbiota is associated with host genotype and the development of infectious digital dermatitis lesions. Microbiome 2023, 11, 4. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Wen, C.; Yan, W.; Sun, C.; Ji, C.; Zhou, Q.; Zhang, D.; Zheng, J.; Yang, N. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. ISME J. 2019, 13, 1422–1436. [Google Scholar] [CrossRef] [PubMed]
- Chaucheyras-Durand, F.; Sacy, A.; Karges, K.; Apper, E. Gastro-Intestinal Microbiota in Equines and Its Role in Health and Disease: The Black Box Opens. Microorganisms 2022, 10, 2517. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.W.; Gee, E.K.; Dittmer, K.E. Growth and bone development in the horse: When is a horse skeletally mature? Animals 2021, 11, 3402. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Berri, M.; Estellé, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.J.; Chevaleyre, C.; Billon, Y.; Doré, J. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar] [CrossRef] [PubMed]
- Ryu, E.P.; Davenport, E.R. Host Genetic Determinants of the Microbiome Across Animals: From Caenorhabditis elegans to Cattle. Annu. Rev. Anim. Biosci. 2022, 10, 203–226. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Luo, C.; Zou, X.; Lv, X.; Xu, Y.; Shu, D.; Qu, H. Association of host genetics with intestinal microbial relevant to body weight in a chicken F2 resource population. Poult. Sci. 2019, 98, 4084–4093. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Xu, Y.; Lei, B.; Zhang, Q.; Lei, Y.; Li, C.; Li, X.; Yao, R.; Hu, R.; Liu, K.; Wang, Y. ADDAGMA: A database for domestic animal gut microbiome atlas. Comput. Struct. Biotechnol. J. 2022, 20, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef]
- Gupta, R.S.; Nanda, A.; Khadka, B. Novel molecular, structural and evolutionary characteristics of the phosphoketolases from bifidobacteria and Coriobacteriales. PLoS ONE 2017, 12, e0172176. [Google Scholar] [CrossRef]
- Misiukiewicz, A.; Gao, M.; Filipiak, W.; Cieslak, A.; Patra, A.; Szumacher-Strabel, M. Methanogens and methane production in the digestive systems of nonruminant farm animals. Animal 2021, 15, 100060. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, J.; Zheng, N.; Zhang, Y.; Jin, D. Different milk replacers alter growth performance and rumen bacterial diversity of dairy bull calves. Livest. Sci. 2020, 231, 103862. [Google Scholar] [CrossRef]
- Morais-Silva, F.O.; Rezende, A.M.; Pimentel, C.; Santos, C.I.; Clemente, C.; Varela–Raposo, A.; Resende, D.M.; da Silva, S.M.; de Oliveira, L.M.; Matos, M. Genome sequence of the model sulfate reducer Desulfovibrio gigas: A comparative analysis within the Desulfovibrio genus. MicrobiologyOpen 2014, 3, 513–530. [Google Scholar] [CrossRef]
- Hailemariam, S.; Zhao, S.; Wang, J. Complete genome sequencing and transcriptome analysis of nitrogen metabolism of Succinivibrio dextrinosolvens strain Z6 isolated from dairy cow rumen. Front. Microbiol. 2020, 11, 1826. [Google Scholar] [CrossRef]
- Grewe, B.S.; Richmond, J.E.; Featherstone, D.E. The spatial and developmental expression of mouse Vwa8 (von Willebrand domain-containing protein 8). Gene Expr. Patterns 2018, 29, 39–46. [Google Scholar] [CrossRef]
- Van Gylswyk, N.; Hippe, H.; Rainey, F. Schwartzia succinivorans gen. nov., sp. nov., another ruminai bacterium utilizing succinate as the sole energy source. Int. J. Syst. Bacteriol. 1997, 47, 155–159. [Google Scholar] [CrossRef]
- Antunes, K.H.; Stein, R.T.; Franceschina, C.; da Silva, E.F.; de Freitas, D.N.; Silveira, J.; Mocellin, M.; Leitão, L.; Fachi, J.L.; Pral, L.P. Short-chain fatty acid acetate triggers antiviral response mediated by RIG-I in cells from infants with respiratory syncytial virus bronchiolitis. EBioMedicine 2022, 77, 103891. [Google Scholar] [CrossRef]
- Bagchi, S.; Perland, E.; Hosseini, K.; Lundgren, J.; Al-Walai, N.; Kheder, S.; Fredriksson, R. Probable role for major facilitator superfamily domain containing 6 (MFSD6) in the brain during variable energy consumption. Int. J. Neurosci. 2020, 130, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Al Jassim, R.A.; Scott, P.T.; Trebbin, A.L.; Trott, D.; Pollitt, C.C. The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiol. Lett. 2005, 248, 75–81. [Google Scholar] [CrossRef]
- Morinaga, K.; Kusada, H.; Tamaki, H. Bile Salt Hydrolases with Extended Substrate Specificity Confer a High Level of Resistance to Bile Toxicity on Atopobiaceae Bacteria. Int. J. Mol. Sci. 2022, 23, 10980. [Google Scholar] [CrossRef] [PubMed]
- Kanno, M.; Katayama, T.; Morita, N.; Tamaki, H.; Hanada, S.; Kamagata, Y. Catenisphaera adipataccumulans gen. nov., sp. nov., a member of the family Erysipelotrichaceae isolated from an anaerobic digester. Int. J. Syst. Evol. Microbiol. 2015, 65, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Cazaudehore, G.; Monlau, F.; Gassie, C.; Lallement, A.; Guyoneaud, R. Active microbial communities during biodegradation of biodegradable plastics by mesophilic and thermophilic anaerobic digestion. J. Hazard. Mater. 2023, 443, 130208. [Google Scholar] [CrossRef]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Geor, R.J.; Nielsen, B.D.; Schott, H.C.; Elzinga, S.; Newbold, C.J. Characterisation of the faecal bacterial community in adult and elderly horses fed a high fibre, high oil or high starch diet using 454 pyrosequencing. PLoS ONE 2014, 9, e87424. [Google Scholar] [CrossRef]
- Metcalf, J.L.; Song, S.J.; Morton, J.T.; Weiss, S.; Seguin-Orlando, A.; Joly, F.; Feh, C.; Taberlet, P.; Coissac, E.; Amir, A. Evaluating the impact of domestication and captivity on the horse gut microbiome. Sci. Rep. 2017, 7, 15497. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Seong, H.-S.; Kim, D.C.; Park, N.G.; Yang, B.C.; Son, J.K.; Shin, S.M.; Woo, J.H.; Shin, M.C.; Yoo, J.H. Genome-wide analyses of the Jeju, Thoroughbred, and Jeju crossbred horse populations using the high density SNP array. Genes Genom. 2018, 40, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Stämpfli, H.; Allen-Vercoe, E.; Weese, J.S. Development of the faecal microbiota in foals. Equine Vet. J. 2016, 48, 681–688. [Google Scholar] [CrossRef]
- Lindenberg, F.; Krych, L.; Kot, W.; Fielden, J.; Frøkiær, H.; Van Galen, G.; Nielsen, D.; Hansen, A. Development of the equine gut microbiota. Sci. Rep. 2019, 9, 14427. [Google Scholar] [CrossRef] [PubMed]
- Huntington, P.; Brown-Douglas, C.; Pagan, J. Growth and development of Thoroughbred horses. Anim. Prod. Sci. 2020, 60, 2093–2102. [Google Scholar] [CrossRef]
- Hoffman, R.; Lawrence, L.; Kronfeld, D.; Cooper, W.; Sklan, D.; Dascanio, J.; Harris, P. Dietary carbohydrates and fat influence radiographic bone mineral content of growing foals. J. Anim. Sci. 1999, 77, 3330–3338. [Google Scholar]
- Gupta, R.S.; Chen, W.J.; Adeolu, M.; Chai, Y. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3379–3397. [Google Scholar] [PubMed]
- Arai, T. The development of animal nutrition and metabolism and the challenges of our time. Front. Media SA 2014, 1, 23. [Google Scholar] [CrossRef]
- Brøkner, C.; Austbø, D.; Næsset, J.; Blache, D.; Knudsen, K.B.; Tauson, A. Metabolic response to dietary fibre composition in horses. Animal 2016, 10, 1155–1163. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.-X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef]
- Xue, X.; Liu, B.; Hu, J.; Bian, X.; Lou, S. The potential mechanisms of lactate in mediating exercise-enhanced cognitive function: A dual role as an energy supply substrate and a signaling molecule. Nutr. Metab. 2022, 19, 52. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Kang, Y.-J.; Kim, Y.-K.; Choi, J.-Y.; Shin, S.-M.; Shin, M.-C. Exploring the Influence of Growth-Associated Host Genetics on the Initial Gut Microbiota in Horses. Genes 2023, 14, 1354. https://doi.org/10.3390/genes14071354
Lee J, Kang Y-J, Kim Y-K, Choi J-Y, Shin S-M, Shin M-C. Exploring the Influence of Growth-Associated Host Genetics on the Initial Gut Microbiota in Horses. Genes. 2023; 14(7):1354. https://doi.org/10.3390/genes14071354
Chicago/Turabian StyleLee, Jongan, Yong-Jun Kang, Yoo-Kyung Kim, Jae-Young Choi, Sang-Min Shin, and Moon-Cheol Shin. 2023. "Exploring the Influence of Growth-Associated Host Genetics on the Initial Gut Microbiota in Horses" Genes 14, no. 7: 1354. https://doi.org/10.3390/genes14071354
APA StyleLee, J., Kang, Y.-J., Kim, Y.-K., Choi, J.-Y., Shin, S.-M., & Shin, M.-C. (2023). Exploring the Influence of Growth-Associated Host Genetics on the Initial Gut Microbiota in Horses. Genes, 14(7), 1354. https://doi.org/10.3390/genes14071354




